
Human-Machine Collaboration for the Visual Exploration and Analysis
of High-Dimensional Spatial Simulation Ensembles
Mai Dahshan
1 a
, Nicholas F. Polys
2 b
, Leanna House
3 c
, Karim Youssef
2 d
and Ryan Pollyea
4 e
1
School of Computing, University of North Florida. U.S.A.
2
Department of Computer Science, Virginia Tech, U.S.A.
3
Department of Statistics, Virginia Tech, U.S.A
4
Department of Geosciences, Virginia Tech, U.S.A
Keywords:
Simulation Ensembles, Spatial Data, Visual Analytics, Large Scale Visualization, Gaussian Process.
Abstract:
Continuous improvements in supercomputing have given scientists from various fields the ability to conduct
large-scale multi-dimensional numerical simulation ensembles. A simulation ensemble involves running mul-
tiple simulations, each with slight variations in model settings, such as input parameters, initial conditions,
or boundary values. Exploring and analyzing these ensembles facilitates understanding parameter sensitivity
and the correlations between different ensemble members. To capture these relationships, visual analytical
tools are used to extract important features from the ensemble. In many cases, however, these visualizations
highlight the differences in the ensemble using aggregated or descriptive statistics, ignoring the correlations
and local differences between different spatial regions, which could hinder the exploration process. This paper
proposes a visual analytical approach, SpatialGLEE, to interactively explore the spatial variability in the sim-
ulation ensemble. The proposed approach uses Gaussian Process Regression (GPR) and Semantic Interaction
(SI) to help scientists explore the impact of input parameters on the ensemble and find the commonalities
and differences across ensemble members and regions of interest (ROI). GPR models the spatial correlation
structure in the ensemble. The modeled data is then inputted into the visualization pipeline for analysis and
exploration with SI. The effectiveness of SpatialGLEE is demonstrated using a real-life case study.
1 INTRODUCTION
Numerical simulations are used in many scientific do-
mains such as geosciences, meteorology, or compu-
tational fluid dynamics (Winsberg, 2013; Cappello
et al., 2015). These simulations help in understand-
ing complex real-world phenomena. However, deter-
mining the optimal initial conditions and model inputs
is difficult due to the complexity of the studied phe-
nomena. To address this uncertainty, multiple runs
are carried out by slightly modifying initial condi-
tions or model parameters, resulting in an ensemble
(Wang et al., 2016; Potter et al., 2009). An ensem-
ble allows scientists to investigate commonalities and
differences across runs, determine parameter sensitiv-
ity, and find optimal settings. Continuous improve-
ments in computing power allow performing large-
a
https://orcid.org/0000-0002-5758-4890
b
https://orcid.org/0000-0002-8503-970X
c
https://orcid.org/0009-0003-2848-4131
d
https://orcid.org/0000-0003-4544-9613
e
https://orcid.org/0000-0001-5560-8601
scale simulation ensembles on high-resolution grids
in a few hours. However, the analysis and exploration
of large ensembles still pose challenges. Therefore,
an appropriate representation of ensembles is needed
for a more intuitive understanding of the simulated
model.
Visualization has a crucial role in understanding
large volumes of data at a glance. Visual exploration
and analysis of multidimensional ensembles enable
scientists to gain insight, identify hidden patterns, and
make discoveries, contrasting with traditional manual
analysis methods that are exhausting and error-prone.
The majority of ensemble visualization literature fo-
cuses on analyzing aggregated or sampled ensemble
members(Wang et al., 2018; Athawale et al., 2020;
Chen et al., 2019). While these visualization ap-
proaches have shown promising results, their high ab-
straction involves losing much ensemble data, which
could potentially hide important patterns or trends.
Moreover, ensemble members and their parameters
cannot be directly examined. Additionally, many of
these approaches do not account for spatial character-
istics in the data and assume prior knowledge of data
678
Dahshan, M., Polys, N., House, L., Youssef, K. and Pollyea, R.
Human-Machine Collaboration for the Visual Exploration and Analysis of High-Dimensional Spatial Simulation Ensembles.
DOI: 10.5220/0012405100003660
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 19th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2024) - Volume 1: GRAPP, HUCAPP
and IVAPP, pages 678-689
ISBN: 978-989-758-679-8; ISSN: 2184-4321
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

patterns, restricting the analysis process.
By collaborating closely with domain experts, we
observed their interest in understanding spatial varia-
tion in the ensemble, identifying and comparing ma-
jor patterns between ensemble members, and track-
ing parameter sensitivity and optimization. Exploring
and analyzing spatial variability involves identifying
spatial regions that exhibit variability and pinpointing
common features and patterns within these regions.
Therefore, we propose an interactive approach, Spa-
tialGLEE, for simultaneously exploring multidimen-
sional spatial ensemble members and their parame-
ters. Our approach helps scientists understand the
ensemble by exploring ”what-if” scenarios to vali-
date hypotheses about the ensemble and its parame-
ters. It involves examining ensemble variability, se-
lecting subsets of ensemble members, and selecting
spatial sub-regions within ensemble members for fur-
ther analysis. To achieve this, we use Gaussian Pro-
cess Regression (GPR) to encode the spatial struc-
ture of each ensemble member, preserving the spatial
trends, outliers, variability, and autocorrelation in the
data. This paper focuses on the exploration and anal-
ysis of 2D spatial ensembles.
This paper presents SpatialGLEE, an expanded
interactive approach built on the GLEE visualiza-
tion tool (Dahshan et al., 2020) to explore and an-
alyze multidimensional spatial ensembles. Spatial-
GLEE specifically addresses spatial variability, un-
like GLEE, which focuses on derived or summary
statistics. SpatialGLEE has two main steps: 1) mod-
eling input parameters and simulation outputs while
preserving the spatial structure; 2) interactive analy-
sis and exploration of ensemble members, ROI, and
simulation inputs and outputs. SpatialGLEE lever-
ages statistical modeling and visual analytics tech-
niques into interactive coordinated visualizations to
help scientists simultaneously explore and make sense
of spatial ensemble and parameter spaces, considering
scientists’ visual reasoning, complexity, and struc-
ture of data. This enables scientists to visually deter-
mine complex insights, including spatial correlations
among ensemble members and ROI, as well as param-
eter sensitivity and optimization. To summarize, the
following are the contributions of the paper:
• Refining GLEE’s SI pipeline and developing a
visualization pipeline to support SpatialGLEE in
exploring spatial ensemble and parameter spaces.
• Demonstrating that GPR can preserve spatial
characteristics in spatial ensembles, leading to po-
tentially meaningful scientific insights.
• Implementing a parallelized version of maximum
likelihood estimation (MLE) for GPR to enhance
scalability with spatial grid sizes, achieving a 21×
speedup.
• Demonstrating the effectiveness of our proposed
approach in exploring and analyzing multidimen-
sional spatial ensembles using real-world data and
domain expert feedback.
2 RELATED WORK
Ensemble visualization approaches have been pro-
posed to examine ensemble member correlations, pa-
rameter optimization, and sensitivity (Wang et al.,
2018). Common methodologies for ensemble visual-
ization either aggregate ensemble members by calcu-
lating the statistical properties of the ensemble (Pot-
ter et al., 2009) or transform ensemble members into
more abstract representations (i.e., isocontours, iso-
surfaces, pathlines, streamlines, etc.) using major
trends in the ensemble (Kumpf et al., 2021; Zhang
et al., 2020). The former methodology is most rele-
vant to our approach. Aggregated ensembles are usu-
ally represented using different statistical displays, in-
cluding but not limited to box plots (Mirzargar et al.,
2014), parallel coordinates (Wang et al., 2016), and
line charts (Demir et al., 2014). However, these tech-
niques often hinder many details about the ensemble,
resulting in the loss of significant information about
the data. Moreover, they are prone to visual clutter-
ing.
To overcome these limitations, improved tech-
niques were developed to reveal variations and de-
tailed information about the data distribution using
histograms (Ahmed et al., 2019), statistical dependen-
cies (Li et al., 2017), and circular treemaps (Huang
et al., 2023). Moreover, clustering techniques have
been employed to detect major patterns by grouping
location points that follow certain distributions (Shu
et al., 2016). However, these techniques are limited in
their application to multidimensional parameter set-
tings. Therefore, they would not provide scientists
with a complete picture of both ensemble and param-
eter spaces.
Recently, ensemble visualization approaches have
tried to address the exploration and analysis of mul-
tidimensional spatial ensembles. Several efforts at-
tempted to capture ensemble spatiality using di-
verse techniques, including confidence intervals (Vi-
etinghoff et al., 2022), hyper-slicer (Evers and Lin-
sen, 2022), neural network-latent-based surrogate
model (Shi et al., 2022), deep neural networks (Hues-
mann and Linsen, 2022), critical points (Favelier
et al., 2018), function plots, (Fofonov and Lin-
sen, 2018), similarity measure (Fofonov and Linsen,
Human-Machine Collaboration for the Visual Exploration and Analysis of High-Dimensional Spatial Simulation Ensembles
679
2019) and uncertainty calculation (Liu et al., 2018).
However, the majority of these approaches are pri-
marily focused on parameter space exploration or an-
alyzing a few ensemble members at once, with less
emphasis on simultaneously exploring both parameter
and ensemble spaces (Orban et al., 2018). Therefore,
our approach focused on the exploration of spatial en-
sembles by integrating visualization with a human-
machine collaboration technique to empower the vi-
sual analysis of parameter and ensemble spaces.
3 APPROACH
3.1 System Design
SpatialGLEE is designed to help scientists gain
insights and find discoveries about simulated data for
more effective exploration and analysis. Therefore,
our proposed approach and its manifestation in a
visual analytics tool result from a long-term collab-
oration with geoscientists. We studied scientists’
conventional analysis workflows through interviews
and focus groups while developing the method and
tool. This identified the analysis tasks scientists need
to understand spatial ensembles. The main analysis
goals are as follows:
Goal 1: Parameter Optimization and Sensitivity
Analysis. Parameter sensitivity analysis examines
the relationship between ensemble members and
the model parameters. Scientists aim to explore the
parameter space to identify 1) key input parameters
that contribute more to explaining simulation outputs
and those with little or no impact; 2) the association
between multiple input parameters; and 3) input pa-
rameter correlation(s) with spatial trends or features.
Concurrently, parameter optimization determines the
optimal parameter settings for given objectives.
Goal 2: Interactive Exploration and Comparison
of Ensemble Members. Exploring individual
ensemble members can uncover the commonalities
and differences between different groups. Scientists
explore the ensemble space to identify and interpret:
1) the locations and reasons behind similarities or
differences among ensemble members; 2) prevailing
patterns, trends, and anomalies; and 3) the spatial
correlation and variability among subsets of ensemble
members.
Goal 3: Interactive Exploration of Subsets of
Ensemble Members and ROI. Understanding
the ensemble’s inherent spatial structure enables
scientists to investigate the dynamics of simulated
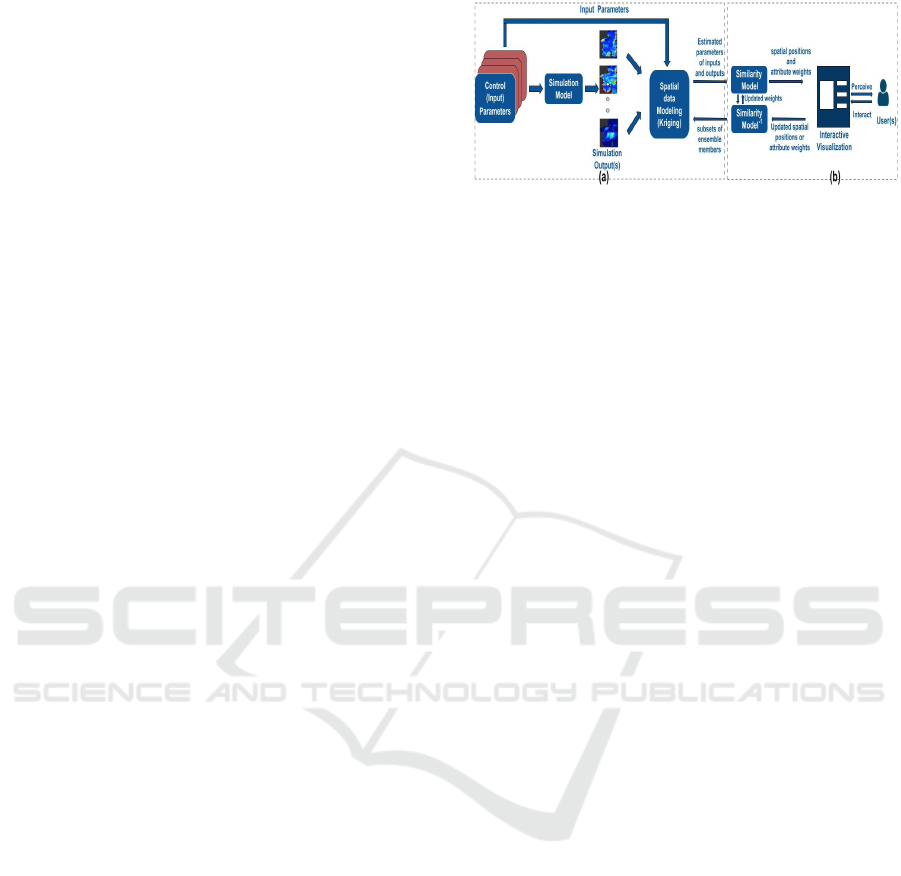
Figure 1: The workflow of our proposed visual analytical
approach. Our approach has two main steps: a) Statistical
modeling of the ensemble while considering spatial char-
acteristics in the data. b) Interactive visual exploration and
analysis of ensemble members, simulation parameters, and
spatial patterns.
models. Scientists aim to understand and analyze: 1)
ROI’s spatial characteristics within the ensemble to
find features and patterns across members; 2) how
specific parameter or parameter settings affect ROI;
and 3) subsets of ensemble members to determine if
the relationship between input parameters holds for
the entire ensemble or only within specific subsets,
and vice versa.
Goal 4: Interactive Exploration and Comparison
of Spatial Distributions. Exploring the statistical at-
tributes of raw spatial data to uncover correlations and
static properties. This exploration involves analyzing
the data distribution to test hypotheses and verify find-
ings.
3.2 System Overview
This section introduces our visual exploration frame-
work for the multidimensional spatial ensemble. The
proposed framework and its visualization components
align with the aforementioned tasks. Figure 1 pro-
vides a high-level overview of our approach. Our ap-
proach begins with an ensemble E of M members. In-
dividual ensemble members consist of input parame-
ters and simulation outputs (i.e., grid). Initially, we
model the simulation ensemble using GPR to esti-
mate spatial parameters that characterize each mem-
ber’s input parameters and simulation outputs. GPR
employs local MLE to determine various spatial pro-
cess features. These features are spatially smoothed
to capture the correlation structure between nearby
grid points. GPR does not require prior knowledge
of the ensemble member distribution. Thus, it can
uncover each ensemble member’s variability, main
trends, and autocorrelations. The estimated spatial
parameters are subsequently inputted into the visual-
ization pipeline of SpatialGLEE.
SpatialGLEE’s coordinated multi-views (Fig-
ure 2), coupled with their supported interaction tech-
IVAPP 2024 - 15th International Conference on Information Visualization Theory and Applications
680

niques, empower scientists to explore and analyze
multidimensional spatial ensembles. Ensemble view
(Figure 2a) visualizes ensemble members by project-
ing them from higher-dimensional space to lower-
dimensional space (i.e., 2D) via a projection tech-
nique (e.g., MDS, t-SNE, etc.), using estimated spa-
tial parameters for simulation outputs and input pa-
rameters and weights associated with them. The po-
sitioning of ensemble members in the ensemble view
reflects relative distances, where members with sim-
ilar estimated attributes are placed close together,
while those with dissimilar attributes are positioned
farther apart.
Scientists explore and compare ensemble mem-
bers and data spatially within the ensemble view us-
ing two main interactions: observation-level inter-
action (OLI), subsetting of ensemble members, and
ROI selection within ensemble members. OLI is an
interaction technique built on the SI principles (En-
dert et al., 2012). It allows scientists to directly ma-
nipulate ensemble members to investigate and un-
derstand their commonalities and differences. The
subsetting feature allows scientists to navigate sub-
sets of ensemble members for a more in-depth anal-
ysis. This capability enables scientists to seamlessly
switch between overview-first and detailed-first anal-
ysis modes. Moreover, scientists can utilize the ROI
selection feature to identify specific spatial regions of
interest within the data. This feature is valuable when
dealing with a large grid where not all regions are
equally significant to the analysis. This allows a fo-
cused exploration of regions with similar patterns or
trends, enhancing the effectiveness of the analysis.
Parameter view (Figure 2b) offers scientists the
opportunity to investigate parameter sensitivity using
ensemble attributes (i.e., input parameters and out-
puts). Each attribute is represented on a horizontal
slider. The slider’s value represents the weight of
the attribute in the model, thereby marking its im-
portance. Using a Parametric Level Interaction (PLI),
scientists are able to manipulate the slider in order to
interact with model attributes. PLI allows scientists
to investigate associations and relationships between
input parameters and explore their impact on the sim-
ulation outputs.
Conversely, the statistical view (Figure 2c) helps
find the optimal parameter settings. This view allows
scientists to explore raw data through various statisti-
cal representations, such as parallel coordinates, scat-
terplots, and boxplots. By leveraging these differ-
ent displays, scientists can determine variability in
the data, validate conclusions, identify hidden corre-
lations not found by other views, and gain insights
into the distribution of different ensemble members.
4 SPATIAL ENSEMBLE
MODELING
Simulation inputs and outputs serve as entry points
to SpatialGLEE’s visualization pipeline. Passing spa-
tial ensemble raw data directly to the visualization
pipeline would capture the spatiality in the data dur-
ing the exploration and analysis. However, it poses
computational challenges due to the complexity and
size of the ensemble. Therefore, there is a need for
modeling the spatial ensemble data while preserving
the underlying spatial structure.
Given a multidimensional spatial ensemble E=
{K
1
,K
2
,...K
N
} with M members, where each K
i
∈ E
is of a 2D grid G. Every grid point within K
i
is linked
to input and output values obtained from spatial simu-
lation assessments or measurements conducted across
the entire grid. K
i
represents a spatial stochastic pro-
cess {Q(s) : s ∈ G} with the spatial domain G ⊂ R
d
,
d≥1 (d vector of coordinates)(Dahshan, 2021). We
model Q(s) by
Q(s) = X (s)β+ w(s) + ε, (1)
so that Q(s) has mean X(s) β and error w(s) + ε. The
mean is the result of the product between the coef-
ficients β and X(s), which represents a vector of p
co-variates at locations S. The additive error adds a
spatial-dependent error term, w(s), and an indepen-
dent measurement error term, ε, characterized by a
zero mean and variance τ
2
. The spatial dependence is
imposed by modeling w(s) as a stationary, mean-zero
spatial process with a covariance function
C
S
(s − s
′
) = σ
2
g(||Σ
−1/2
(s − s
′
)||), (2)
g() represents a spatial kernel covariance function that
operates on the distance between two grid points, and
σ
2
serves as a covariance inflation term. Spatial co-
variance kernel functions establish the characteristics
and degree of spatial dependence within the spatial
process. For instance, they can establish greater de-
pendence between spatial outcomes when they are
close compared to those at a greater distance.
Based on the discussions with our collaborators,
we learned that their primary focus lies in understand-
ing spatial autocorrelation and variability in the en-
semble. According to Tobler’s 1st Law of Geography
(Miller, 2004), spatial autocorrelation expects nearby
spatial grid points to be more similar than far apart
ones. Therefore, our proposed approach fits GPR
(L
´
azaro-Gredilla et al., 2010), also known as Krig-
ing, to learn the characteristics of spatial processes.
Kriging provides a method for interpolating between
grid points. These interpolations maintain the spatial
correlation and variability present in the data.
Human-Machine Collaboration for the Visual Exploration and Analysis of High-Dimensional Spatial Simulation Ensembles
681

Figure 2: SpatialGLEE’s main interface:(a) Ensemble view shows the WMDS projection of the simulation ensemble in 2D
space. Ensemble members are spatially arranged so that similar ensemble members are near each other and dissimilar en-
semble members are far apart. Scientists can interactively manipulate image thumbnails representing ensemble members to
explore and analyze them. (b) Parameter view presents the weights assigned to both input parameters and simulation outputs.
Scientists can alter the slider values to examine the impact of the ensemble attributes. (c) Statistical view offers multiple
statistical displays to explore and understand the distributions, patterns, trends, and outliers in ensemble raw data.
With simulation measurements Y (s
i
) of N grid lo-
cations s
i
(i ∈ {1,...,N}). Estimating the process Y at
a new location s
0
, denoted Y
∗
(s
0
) is a two-step pro-
cess. The process begins by fitting a variogram to as-
certain the spatial covariance structure and parameters
based on observed data. Subsequently, it calculates
weights, denoted as λ
i
, from the covariances between
each observed location s
i
and the new location s
0
. The
value Y
∗
(s
0
) is then derived from a weighted average.
Y
∗
(s
0
) =
N
∑
i=1
λ
i
Y (s
i
). (3)
In our approach, we model a second-order station-
arity Gaussian Process (GP) with an isotropic Matern
covariance kernel. (Nychka et al., 2002). The Matern
covariance function is
C(d|κ,ν,σ
2
) = σ
2
2
ν−1
Γ(ν)
(d/κ)
ν
K
ν
(d/κ) (4)
where d is the Euclidean distance between s and s
′
(||s − s
′
||), Γ(.) is the gamma function, ν is a smooth-
ing parameter that controls the mean square differen-
tiability of the process (ν > 0), κ is a range parameter,
and K ν(.) is the modified Bessel function of second
kind order. We selected the Matern covariance be-
cause it offers significant flexibility in modeling spa-
tially correlated random processes. The smoothness
parameter associated with Matern covariance allows
control over the level of smoothness in the spatial pro-
cess.
To estimate model parameters, Kriging maximizes
the likelihood of the simulation measurements gen-
erating five estimates: nugget variance, scalar MLE
for kappa, anisotropy parameters (lam1, lam2), and
process variance. The computation of these estimates
relies heavily on matrix operations. The distance ma-
trices and covariance utilized in these operations cap-
ture the pairwise correlations among the locations of
the grid. As the spatial grid size increases, the com-
putational cost of kriging escalates, restricting its fea-
sibility with large datasets. To tackle this challenge,
various approaches have emerged, falling into two
categories: sparse approximations (Kaufman et al.,
2008) and local approximations (Wackernagel, 2013).
Sparse approximations involve approximating the co-
variance matrix with sparse matrices. Conversely, lo-
cal approximations partition input data into local and
independent regions, with each spatial region encom-
passing a relatively small number of local points clos-
est to the prediction point. Despite their ability to han-
dle large datasets, these approximation approaches
rely on some form of approximation, introducing the
risk of information loss and errors, thereby limiting
the advantages of using kriging.
Our aim is to implement a scalable high-
performance parallel and distributed version of the
MLE of the Kriging model. This implementation
leveraged multi-core and multi-node architectures to
boost computational performance. We leveraged
the Aniso fit() API from the ConvoSPAT package of
R (Risser and Calder, 2015) for our implementation.
IVAPP 2024 - 15th International Conference on Information Visualization Theory and Applications
682

We modified the Aniso fit() MLE implementation to
evaluate the maximization function and the function
returning the gradient for the same parameter value in
parallel. We used the Parallel package from R, which
supports multi-core and multi-node parallelization.
5 SPATIAL ENSEMBLE VISUAL
EXPLORATION
SpatialGLEE’s modeling and visualization aim to in-
vestigate similar behaviors and key parameters in the
ensemble, aligning with design goals. Scientists can
use SpatialGLEE’s multi-linked views and interac-
tion techniques to explore and analyze spatial ensem-
bles. SpatialGLEE integrates scientists’ intuition and
expertise with machine learning and statistics, facil-
itating the analysis and exploration of multidimen-
sional spatial ensembles. The visualization pipeline
of SpatialGLEE is comprised of three main compo-
nents: the input source, similarity models, and co-
ordinated visual interfaces. The spatially estimated
simulation outputs and input parameters serve as the
input source, processed through similarity models for
2D projection and manipulation. Coordinated multi-
views facilitate analysis, providing a comprehensive
understanding of spatial relationships and patterns in
the data.
5.1 Spatial Ensemble Attributes
The SpatialGLEE visualization pipeline employs the
spatial estimates (i.e., anisotropy parameters (lam1,
lam2), nugget variance, process variance, and scalar
MLE for kappa) as a foundation for spatial ensemble
analysis and exploration. To ensure an accurate unbi-
ased representation of the data during the exploration
and analysis of the spatial ensemble, these estimates
are z-score normalized. This normalization process
helps standardize the data and ensures that each es-
timate is considered in the context of its distribution,
preventing any skewed or misleading interpretations.
In addition, an initial weight of (1/ d) is assigned to
each attribute (i.e., inputs and outputs), where d is the
total number of simulation outputs and input parame-
ters. The weight of each attribute is evenly distributed
among its estimates, resulting in a weight of (1/5d)
for each individual estimate.
5.2 Similarity Models
The similarity models in SpatialGLEE manage the
visualization and interaction of simulation ensem-
ble data through two models: forward and back-
ward. The forward model utilizes weighted mul-
tidimensional scaling (WMDS) to project multidi-
mensional ensemble data into two-dimensional space.
This weighted projection integrates a weighted dis-
tance function to capture commonalities and distinc-
tions between ensemble members based on spatially
estimated attributes. The determination of the dis-
tance function in SpatialGLEE depends on the data
characteristics, the task, and the projection technique.
The primary interface offers scientists a range of dis-
tance functions to choose from, including weighted
Cosine, weighted Euclidean, and weighted Manhat-
tan, with the default being the weighted Mahalanobis
distance. The Mahalanobis distance measures the dis-
tance between any point in space and the distribu-
tion’s center, taking into account correlations between
attributes.
To explore ensemble members described by spa-
tial estimates of d parameters, WMDS is applied us-
ing weighted distance D
w
(i, j), for ensemble mem-
bers i and j ( i, j ∈ {1,...,M}), with weight w
a
rep-
resenting the weight applied to each spatial estimate
to denote its significance in the projection. The pair-
wise distance function result between ensemble mem-
bers is subsequently inputted into WMDS. WMDS
determines the position of each individual member in
the low dimensions by minimizing the mean squared
error between the pairwise distances in two dimen-
sions and the corresponding distances in the high-
dimensional space.
The backward model is activated when scientists
interact with SpatialGLEE’s different views using dif-
ferent interaction techniques (i.e., OLI, PLI, brush-
ing, or subsetting). OLI enables scientists to create
a customized spatialization of multidimensional en-
semble members based on their intuition and domain
expertise. For instance, the expertise of the scien-
tists may contradict the spatialization of the ensem-
ble members, or they may observe interesting patterns
in the data. In response, they perform an OLI by
dragging subsets of ensemble members into groups.
These groupings signify scientists’ hypothesized sim-
ilarity between these ensemble members. The back-
ward model is then triggered, calling upon a semi-
supervised metric learning model. This model at-
tempts to learn new weights that correspond to the
identified similarity and subsequently adjusts the pro-
jection. Thus, OLI facilitates the exploration of re-
lationships and associations between ensemble mem-
bers by allowing scientists to shape the spatialization
based on their insights and hypotheses.
PLI empowers scientists to explore parameter sen-
sitivity by directly manipulating the attribute’s weight
Human-Machine Collaboration for the Visual Exploration and Analysis of High-Dimensional Spatial Simulation Ensembles
683

on the slider. This interaction results in an updated
weight vector and a new projection of ensemble mem-
bers based on the manipulated weight on the slider.
Given that all weights are constrained to add up to
one, increasing the weight of one attribute necessi-
tates decreasing the weights of all other attributes,
and vice versa. Thus, the updated projection ampli-
fies the similarities and differences among data points,
intensifying with an increase in weight and diminish-
ing with a decrease in weight. This enables scientists
to offer parametric feedback to the backward model
regarding which attribute they consider to be signifi-
cant and to observe how this attribute affects ensem-
ble members’ low-dimensional grouping. This inter-
action facilitates the exploration of the influence of
individual attributes on the spatial representation, al-
lowing scientists to gain insights into parameter sen-
sitivity within the ensemble.
The average size of a spatial simulation grid may
surpass millions of grid cells. In this case, scientists
might be interested in examining specific regions, like
those near wells or aquifers. The SpatialGLEE ROI
selection feature enables the interactive selection of
an ROI within the ensemble member within thumb-
nails in the ensemble view. Scientists can explore ROI
in two ways: either exclusively focusing on an ROI or
assigning a higher weight to it compared to other re-
gions on the grid. Exploring a specific ROI across all
ensemble members triggers the backward model, up-
dating the visualization pipeline with a newly updated
weight vector for spatial estimates of the chosen sub-
region of interest and thus adjusting the projection ac-
cordingly. On the other hand, increasing the weight of
a particular ROI triggers the backward model, which
updates the weight vector by dividing the weights be-
tween ensemble estimated attributes and subregion of
interest estimated attributes based on a percent deter-
mined by the scientist.
5.3 Coordinated Multi-View
Visualizations
SpatialGLEE’s multi-coordinated views, Figure 2,
contain an ensemble view, a parameter view, and a
statistical view.
Ensemble View: visualizes multidimensional ensem-
ble members in two-dimensional space using WMDS
to provide an overview of spatial ensemble data.
Each ensemble member is presented through a two-
dimensional image of the simulation output. The en-
semble view offers two main interactions: OLI and
ensemble members and ROI selection. These two in-
teractions provide scientists with an interactive explo-
ration environment for analyzing simulation ensem-
ble members, thereby contributing to achieving the
second and third design goals.
Parameter View: represents simulation input param-
eters and output parameters on horizontal sliders, with
each attribute represented by a single slider whose
value is the attribute’s weight. The weight of each
attribute is the sum of the weights of all its estimates.
Scientists utilize PLI within parameter view to inves-
tigate parameter sensitivity, achieving the first design
goal.
Statistical View: supports other views by allowing
scientists to validate assumptions and refine hypothe-
ses derived from other views. It offers three statisti-
cal displays—parallel coordinates, boxplot, and scat-
ter plot—supporting univariate, bivariate, and multi-
variate analyses. It can also be used to gain insights
into data distributions, identify the variability across
different regions, discover new patterns undiscovered,
or/and produce novel hypotheses that other views
could confirm, thereby contributing to achieving the
fourth design goal. Statistical View offers statistical
displays for individual ensemble members, multiple
ensemble members, and specific regions within en-
semble members. For instance, scientists can explore
correlations between ensemble members or analyze
the distribution of single or multiple ensemble mem-
bers across the entire grid or specific subsets.
6 EVALUATION
We evaluated our proposed approach using a 2D spa-
tial ensemble from geologic CO2 sequestration. Dur-
ing the evaluation, we examined the effectiveness of
the proposed approach in aiding scientists to explore
and analyze multidimensional spatial ensembles. Our
emphasis was on gauging how well the approach fa-
cilitates the exploration of parameter sensitivity and
optimization, as well as the examination of similar-
ities and differences among ensemble members and
spatial regions of interest. Specifically, we investi-
gated the extent to which the statistically estimated
parameters could preserve the spatial structure during
exploration and analysis.
Three geoscience domain scientists (two gradu-
ate students and a faculty member) evaluated the pro-
posed approach. The faculty member supplied the en-
semble data used in the experiment. To initiate the
evaluation, the scientists received instructions on how
to use SpatialGLEE and its interaction techniques.
Subsequently, they were tasked with utilizing Spatial-
GLEE’s interaction techniques and visual interfaces
to explore and analyze the ensemble. Throughout the
evaluation, we recorded both the duration to complete
IVAPP 2024 - 15th International Conference on Information Visualization Theory and Applications
684

each task and the level of task completion. Addition-
ally, we timed every interaction that the domain ex-
perts carried out.
Figure 3: The initial projection of the multidimensional
CO2 flow ensemble using spatially estimated simulation at-
tributes.
6.1 Case Study
The simulation ensemble study examines the multi-
phase fluid dynamics of CO2 flow in a basalt frac-
ture network (Gierzynski and Pollyea, 2017). The
model domain has a 2-D fracture network based on
high-resolution LiDAR scans of a basalt outcrop in
the Columbia River Plateau. The 5 m x 5 m model
domain is divided into 40,000 2.5 cm x 2.5 cm Carte-
sian grid cells. The study constructed a simulation
ensemble that randomly assigns fracture permeabil-
ity to each grid cell from a basalt core sample per-
meability distribution because centimeter-scale frac-
ture permeability is unknown. Thus, the ensemble has
25 equally probable model domains with permeabil-
ity spatial distribution as the random variable. Using
e-type estimations, this simulation ensemble exam-
ined how permeability impacts buoyant CO2’s flow
characteristics during geologic CO2 sequestration in
a basalt reservoir as it phases from supercritical fluid
to subcritical gas.
Figure 3 shows the initial projection of a multidi-
mensional CO2 flow ensemble in 2D space. The sci-
entist didn’t observe any interesting patterns or groups
in the initial projection. The scientist was interested
in understanding the parametric controls of the phase
change to gas because the gas phase CO2 is sig-
nificantly more buoyant and therefore has a greater
chance of leaking out of the CO2 storage reservoir.
To investigate this, the scientist started grouping en-
semble members into two groups based on how much
CO2 has moved to the upper portion of the model,
which is where the CO2 undergoes a phase change to
a gas phase, performing an OLI (Figure 4 a).
The reprojection of the ensemble revealed that
there was a notable increase in the weight of gas-
phase CO2, permeability, and fluid pressure, as il-
lustrated in Figure 4b. This led the scientist to con-
clude that fluid pressure is the dominant control for
this grouping. The rationale behind this conclusion
is that the phase change from supercritical CO2 to
gas-phase CO2 occurs as the CO2 floats to shallower
depths, where fluid pressure is lower. Additionally,
the scientist gained insights from this reprojection that
permeability affects CO2 flow; this necessitates addi-
tional experiments to determine how permeability dis-
tributions vary between ensemble members that ex-
hibit phase change and those that do not.
Figure 4: Investigating migration CO2 flow using OLI: a)
semantically grouping ensemble members from the CO2
flow ensemble into two clusters based on how much CO2
has moved to the upper portion of the model; b) The re-
sulted projection shows permeability and fluid pressure (P)
have a dominant role in the phase change from supercritical
CO2 to gas-phase CO2.
The scientist wanted to investigate the effect of
fluid pressure alone on the ensemble, so s/he con-
ducted PLI by increasing the fluid pressure’s weight.
However, the reprojection result was inconclusive.
So, the scientist decided to explore if the tempera-
ture has any influence on the ensemble by perform-
ing PLI (Figure 5). The resulted projection was a
linear projection in which ensemble members with
high CO2 gas at shallow depths close to the top of
the workspace. This discovery led the scientist to ob-
serve that there is a thermal control on the CO2 phase
change, but it is much more subtle in the ensemble.
This was a new discovery that required a more de-
tailed analysis.
The scientist was interested in determining
whether log permeability affects the ensemble. So,
Human-Machine Collaboration for the Visual Exploration and Analysis of High-Dimensional Spatial Simulation Ensembles
685

Figure 5: Increasing the weight of the temperature attribute
while performing PLI to explore its impact on the ensemble.
The resulted projection led to the conclusion that there is a
thermal control on the CO2 phase change.
s/he increased the weight of log permeability per-
forming an PLI (Figure 6). The projection resulted
in the separation of ensemble members into distinct
piles. The pile on the right has low CO2 gas at shal-
low depths, whereas the pile on the left has high CO2
gas concentrations at shallow depths. This led the sci-
entist to conclude that the pile on the right has low
permeability in the conductive fracture shallow depth,
which keeps fluid pressure high and prevents the CO2
from undergoing phase change to CO2 gas.
After interacting with SpatialGLEE through var-
ious interactions, the scientist aimed to identify po-
tential patterns among parameters and investigate
whether the distribution of raw data for different pa-
rameters varied across runs. To explore this, the sci-
entist employed statistical view displays (Figure 7).
The use of a boxplot showed that the distribution of
log permeability remained consistent across all runs.
Furthermore, a scatter plot uncovered a moderately
positive correlation between pressure and CO2 se-
questration, as well as a positive correlation between
CO2 sequestration and CO2 total. By employing par-
allel coordinates, the scientist detected a correlation
among log permeability, temperature, and CO2 gas
across specific runs.
The scientist aimed to investigate regions of inter-
est (ROI) within the reservoir. To facilitate this ex-
ploration, s/he resets the ensemble view to obtain a
new projection. Subsequently, s/he selected a partic-
ular run and identified a ROI. SpatialGLEE supports
two exploration options for ROI: either allocate a per-
centage of the weight vector to this ROI and explore
it concurrently with the entire reservoir or assign the
entire weight vector exclusively to this specific ROI.
The scientist explored both options while using Spa-
tialGLEE interaction techniques and determined that
specific characteristics in these ROIs required further
investigation through higher-fidelity simulations (Fig-
ure 8.
Figure 6: Performing a PLI to investigate the effect of in-
creasing the log permeability weight on the ensemble. The
resulted projection led to the conclusion that low permeabil-
ity in the conductive fracture shallow depth keeps fluid pres-
sure high and prevents CO2 from undergoing phase change
to CO2 gas.
6.2 Domain Expert Evaluation
We discussed the SpatialGLEE tool and its interac-
tion techniques with the domain experts who pro-
vided the ensemble data for the case study. We so-
licited their feedback on the usability and utility of
SpatialGLEE. Scientists confirmed that by using Spa-
tialGLEE, they were able to reach conclusions in sig-
nificantly less time compared to the traditional anal-
ysis process. SpatialGLEE coordinated views allow
them to simultaneously visualize, explore, and under-
stand parameter and ensemble spaces in the absence
of prior knowledge. The ensemble view allows an-
alyzing both ensemble members and spatial regions
within members. This helps them identify signifi-
cant regions in the grid. The parameter view permits
them to directly investigate input parameters’ effects
on simulation outputs. The statistical view supports
scientists in locating interesting patterns in raw spatial
data and exploring them using SpatialGLEE’s views
and interaction techniques. It also helps in finding the
optimal parameter settings. SpatialGLEE has the po-
tential to aid in the discovery of new insights that ne-
cessitate additional experiments for in-depth analysis.
6.3 Discussion
SpatialGLEE Showed the Potential to Improve
Spatial Ensemble Exploration over Traditional
Approaches. SpatialGLEE presents an approach for
exploring multidimensional spatial ensembles when
scientists do not have an in-depth understanding of the
simulated model. Traditionally, scientists utilize visu-
alizations of simulation outputs and summary statis-
tics to investigate the variability of an individual pa-
rameter (whether input or output) throughout the en-
tire ensemble. This traditional approach often ne-
IVAPP 2024 - 15th International Conference on Information Visualization Theory and Applications
686

Figure 7: Exploring the distributions and correlations be-
tween parameters of raw spatial data utilizing the statistical
view. This view shows statistical distributions and proper-
ties of data using univariate, bivariate, and multivariate sta-
tistical displays.
Figure 8: Spatial Region of Interest (ROI) Selection: a) The
scientist was interested in exploring a certain region within
the reservoir, so s/he selected the ROI and increased its im-
portance over other regions in the grid by 100%. b) The
ensemble members are re-projected based on the newly up-
dated weight vector.
cessitates the implementation of several programs or
scripts for data visualization, which takes a substan-
tial amount of time and increases the risk of errors.
In our comparison between the insights and conclu-
sions generated by SpatialGLEE and those obtained
through the scientist’s regular analysis process, we
found that SpatialGLEE led to the same conclusions
as the manual analysis process but in significantly less
time. Furthermore, SpatialGLEE facilitated the dis-
covery of new phenomena and insights that would be
challenging to uncover using traditional methods.
The qualitative analysis of the study reveals that
using OLI, the scientist was able to figure out com-
monalities and differences across ensemble members
and within ROI in ensemble members based on
specific patterns or hypotheses. The resulted projec-
tion from OLI provides scientists with the ability to
identify which parameter(s) guide the grouping of
ensemble members. However, OLI is a technique
for exploratory interaction; therefore, it would not
always generate significant outcomes. Obtaining
significant insights or even discoveries would be
possible if the grouped ensemble members shared
high-dimensional features that could be captured
by the metric learning model. Utilizing the PLI,
scientists managed to identify the sensitivity of input
parameters to the simulation output. This facilitated
the identification of crucial parameters and those
that could be set as constants in the simulation. On
the other hand, through the use of statistical view
displays, scientists were able to explore and analyze
raw data from spatial ensembles. This helped them
understand the distribution and variability of the data,
leading to the identification of optimal parameter
settings for the input parameters.
SpatialGLEE Performance. Our quantitative mea-
surements demonstrate that the interaction techniques
of SpatialGLEE allow scientists to accomplish all
preliminary exploration tasks. However, when
interacting with SpatialGLEE, additional exploratory
inquiries emerged. The scientists addressed some
of these inquiries, but others demanded simulations
of greater fidelity. We computed the total number
of interactions needed to answer the preliminary
exploration tasks. This count varied among scien-
tists, depending on the nature of the interactions
they performed within the ensemble. For example,
completing the preliminary exploration tasks needed
on average took 1–5 interactions. Advanced tasks
that emerged during the analysis exhibit variability in
the number of interactions, ranging from an average
of 3 to 8. Additionally, SpatialGLEE responded
to the interactions of scientists within a reasonable
time frame. Responding to OLI, PLI, and ensemble
member and ROI selection required less than 5s, 3s,
and 1s respectively.
Spatial Ensemble Modeling. Modeling spatial en-
sembles using GPR preserves the spatiality of the
data during exploration and analysis of both ensme-
ble members and ROI within the ensemble. Us-
ing the modeled data with SpatialGLEE interaction
techniques revealed that grouping ensemble members
captured the intrinsic structures and spatial charac-
teristics of the data. This provided scientists with
additional insights that might be challenging to ob-
tain using traditional visualization methods reliant
on summary statistics (i.e., standard deviation and
mean). Moreover, the parallel implementation of
MLE demonstrates superior computational perfor-
mance compared to conventional MLE. Our approach
is scalable, functioning effectively across both the
same node and multiple nodes (Figure 9). We con-
ducted our scaling evaluation on an Intel SkyLake
Xeon Gold cluster with 24 cores and 384 GB of mem-
Human-Machine Collaboration for the Visual Exploration and Analysis of High-Dimensional Spatial Simulation Ensembles
687

ory per node. We observed that while our implemen-
tation scales on multiple cores of the same node, using
multiple nodes leverages the aggregate cluster mem-
ory, resulting in further performance improvement.
On 8 nodes and 128 cores, our parallel implementa-
tion achieved approximately 8× speedup compared to
2 nodes and 16 cores. The total speedup compared to
the sequential Aniso fit() implementation is 21×.
Figure 9: Execution time of parallel MLE on various cores
(i.e., 16, 32, 64, 128) across distributed nodes (i.e.,2, 4, 8)
Distance Function Learning Model. SpatialGLEE
is designed to assist scientists in exploring spatial
simulation ensembles. The projection of ensemble
members from high-dimensional space to 2D space
and their grouping in the ensemble view significantly
influence the exploration process. In the forward
similarity SI model, the distance function can be
described as an interactive distance function learning
model. The selection of the distance function plays a
pivotal role in the forward similarity SI model and,
consequently, impacts the projection of ensemble
members. The interactive nature of the distance
function suggests that it may learn from the specific
characteristics of the data, contributing to a more
dynamic approach to determining similarities be-
tween the ensemble members. While the choice of
distance function is based on the characteristics of
the data and the task at hand, we observed different
performances of different distance functions (i.e.,
Euclidean, Manhattan, and Mahalanobis) when
using spatial ensembles. The Euclidean distance
only captures the general spatial arrangement in the
ensemble, providing limited insights to scientists.
Conversely, manhattan distance outperforms eu-
clidean distance due to its sensitivity to multivariate
outliers. In contrast, when compared to other distance
functions, Mahalanobis distance demonstrates greater
accuracy in grouping ensemble members. This can
be attributed to its consideration of the multivariate
covariance structure during distance calculation.
However, Mahalanobis distance comes with a notable
drawback: its computational complexity is high for
large datasets.
Limitations and Future Work. Increasing the num-
ber of ensemble members to the hundreds may result
in visual cluttering within the ensemble view. One po-
tential solution to the problem is using larger displays
that are capable of accommodating a greater number
of ensemble members. Based on scientists’ feedback,
it was observed that scientists typically opt for an en-
semble size that is smaller than one hundred. Our cur-
rent approach and its parallel implementation are tai-
lored for 2D spatial grids and do not currently provide
support for 3D grids. In our future work, we plan to
expand SpatialGLEE to add support for 3D grids and
incorporate the capability for handling time-varying
simulation ensembles.
7 CONCLUSION
In this paper, we proposed SpatialGLEE, a visual ex-
ploration approach for multi-dimensional spatial en-
sembles. The proposed approach modeled the spa-
tiality of data in ensemble members using Gaussian
Process Regression (GPR) and explored its feasibil-
ity for visual exploration with Semantic Interaction.
SpatialGLEE interactive visual interfaces and interac-
tions enabled scientists to explore commonalities and
distinctions across ensemble members, subsets of the
ensemble, and ROI within the ensemble. Addition-
ally, they were able to determine parameter sensitivity
and optimization, as well as analyze the statical prop-
erties of the raw spatial data of ensemble members
and their parameters. The effectiveness of our pro-
posed approach was evaluated through experiments
involving domain experts. We found that by employ-
ing the SpatialGLEE approach, scientists could effec-
tively explore spatial simulation parameter and en-
semble spaces simultaneously, potentially leading to
the generation of new findings and discoveries.
REFERENCES
Ahmed, K., Sachindra, D. A., Shahid, S., Demirel, M. C.,
and Chung, E.-S. (2019). Selection of multi-model
ensemble of general circulation models for the simula-
tion of precipitation and maximum and minimum tem-
perature based on spatial assessment metrics. Hydrol-
ogy and Earth System Sciences, 23(11):4803–4824.
Athawale, T., Maljovec, D., Yan, L., Johnson, C., Pascucci,
V., and Wang, B. (2020). Uncertainty visualization
of 2d morse complex ensembles using statistical sum-
mary maps. IEEE Transactions on Visualization and
Computer Graphics.
Cappello, F., Constantinescu, E., Hovland, P., Peterka, T.,
Phillips, C., Snir, M., and Wild, S. (2015). Improv-
ing the trust in results of numerical simulations and
scientific data analytics. Technical report, Argonne
National Lab.(ANL), Argonne, IL (United States).
IVAPP 2024 - 15th International Conference on Information Visualization Theory and Applications
688

Chen, X., Shen, L., Sha, Z., Liu, R., Chen, S., Ji, G., and
Tan, C. (2019). A survey of multi-space techniques in
spatio-temporal simulation data visualization. Visual
Informatics, 3(3):129–139.
Dahshan, M., Polys, N. F., Jayne, R., and Pollyea, R. M.
(2020). Making sense of scientific simulation ensem-
bles with semantic interaction. In Computer Graphics
Forum, volume 39, pages 325–343. Wiley Online Li-
brary.
Dahshan, M. M. S. I. (2021). Visual analytics for high di-
mensional simulation ensembles.
Demir, I., Dick, C., and Westermann, R. (2014). Multi-
charts for comparative 3d ensemble visualization.
IEEE Transactions on Visualization and Computer
Graphics, 20(12):2694–2703.
Endert, A., Fiaux, P., and North, C. (2012). Semantic inter-
action for sensemaking: inferring analytical reasoning
for model steering. IEEE Transactions on Visualiza-
tion and Computer Graphics, 18(12):2879–2888.
Evers, M. and Linsen, L. (2022). Multi-dimensional
parameter-space partitioning of spatio-temporal sim-
ulation ensembles. Computers & Graphics, 104:140–
151.
Favelier, G., Faraj, N., Summa, B., and Tierny, J. (2018).
Persistence atlas for critical point variability in en-
sembles. IEEE transactions on visualization and com-
puter graphics, 25(1):1152–1162.
Fofonov, A. and Linsen, L. (2018). Multivisa: Visual anal-
ysis of multi-run physical simulation data using inter-
active aggregated plots. In VISIGRAPP (3: IVAPP),
pages 62–73.
Fofonov, A. and Linsen, L. (2019). Projected field similar-
ity for comparative visualization of multi-run multi-
field time-varying spatial data. In Computer Graphics
Forum, volume 38, pages 286–299. Wiley Online Li-
brary.
Gierzynski, A. O. and Pollyea, R. M. (2017). Three-phase
co2 flow in a basalt fracture network. Water Resources
Research, 53(11):8980–8998.
Huang, Q., Chen, Q., Liu, G., and Cui, Z. (2023). Visual-
ization facilitates uncertainty evaluation of multiple-
point geostatistical stochastic simulation. Visual In-
telligence, 1(1):12.
Huesmann, K. and Linsen, L. (2022). Similaritynet: A deep
neural network for similarity analysis within spatio-
temporal ensembles. In Computer Graphics Forum,
volume 41, pages 379–389. Wiley Online Library.
Kaufman, C. G., Schervish, M. J., and Nychka, D. W.
(2008). Covariance tapering for likelihood-based esti-
mation in large spatial data sets. Journal of the Amer-
ican Statistical Association, 103(484):1545–1555.
Kumpf, A., Stumpfegger, J., Hartl, P. F., and Westermann,
R. (2021). Visual analysis of multi-parameter distri-
butions across ensembles of 3d fields. IEEE Transac-
tions on Visualization and Computer Graphics.
L
´
azaro-Gredilla, M., Quinonero-Candela, J., Rasmussen,
C. E., and Figueiras-Vidal, A. R. (2010). Sparse spec-
trum gaussian process regression. The Journal of Ma-
chine Learning Research, 11:1865–1881.
Li, W., Duan, Q., Miao, C., Ye, A., Gong, W., and Di, Z.
(2017). A review on statistical postprocessing meth-
ods for hydrometeorological ensemble forecasting.
Wiley Interdisciplinary Reviews: Water, 4(6):e1246.
Liu, L., Padilla, L., Creem-Regehr, S. H., and House, D. H.
(2018). Visualizing uncertain tropical cyclone predic-
tions using representative samples from ensembles of
forecast tracks. IEEE transactions on visualization
and computer graphics, 25(1):882–891.
Miller, H. J. (2004). Tobler’s first law and spatial analysis.
Annals of the Association of American Geographers,
94(2):284–289.
Mirzargar, M., Whitaker, R. T., and Kirby, R. M. (2014).
Curve boxplot: Generalization of boxplot for ensem-
bles of curves. IEEE transactions on visualization and
computer graphics, 20(12):2654–2663.
Nychka, D., Wikle, C., and Royle, J. A. (2002). Multires-
olution models for nonstationary spatial covariance
functions. Statistical Modelling, 2(4):315–331.
Orban, D., Keefe, D. F., Biswas, A., Ahrens, J., and Rogers,
D. (2018). Drag and track: A direct manipulation in-
terface for contextualizing data instances within a con-
tinuous parameter space. IEEE transactions on visu-
alization and computer graphics, 25(1):256–266.
Potter, K., Wilson, A., Bremer, P.-T., Williams, D., Dou-
triaux, C., Pascucci, V., and Johnson, C. R. (2009).
Ensemble-vis: A framework for the statistical visual-
ization of ensemble data. In 2009 IEEE International
Conference on Data Mining Workshops, pages 233–
240. IEEE.
Risser, M. D. and Calder, C. A. (2015). Local likelihood
estimation for covariance functions with spatially-
varying parameters: the convospat package for r.
arXiv preprint arXiv:1507.08613.
Shi, N., Xu, J., Li, H., Guo, H., Woodring, J., and Shen, H.-
W. (2022). Vdl-surrogate: A view-dependent latent-
based model for parameter space exploration of en-
semble simulations. IEEE Transactions on Visualiza-
tion and Computer Graphics, 29(1):820–830.
Shu, Q., Guo, H., Liang, J., Che, L., Liu, J., and Yuan, X.
(2016). Ensemblegraph: Interactive visual analysis of
spatiotemporal behaviors in ensemble simulation data.
In 2016 IEEE Pacific Visualization Symposium (Paci-
ficVis), pages 56–63. IEEE.
Vietinghoff, D., Bottinqer, M., Scheuermann, G., and
Heine, C. (2022). Visualizing confidence intervals
for critical point probabilities in 2d scalar field ensem-
bles. In 2022 IEEE Visualization and Visual Analytics
(VIS), pages 145–149. IEEE.
Wackernagel, H. (2013). Multivariate geostatistics: an
introduction with applications. Springer Science &
Business Media.
Wang, J., Hazarika, S., Li, C., and Shen, H.-W. (2018). Vi-
sualization and visual analysis of ensemble data: A
survey. IEEE transactions on visualization and com-
puter graphics, 25(9):2853–2872.
Wang, J., Liu, X., Shen, H.-W., and Lin, G. (2016). Multi-
resolution climate ensemble parameter analysis with
nested parallel coordinates plots. IEEE transactions
on visualization and computer graphics, 23(1):81–90.
Winsberg, E. (2013). Computer simulations in science.
Zhang, M., Chen, L., Li, Q., Yuan, X., and Yong, J. (2020).
Uncertainty-oriented ensemble data visualization and
exploration using variable spatial spreading. IEEE
Transactions on Visualization and Computer Graph-
ics, 27(2):1808–1818.
Human-Machine Collaboration for the Visual Exploration and Analysis of High-Dimensional Spatial Simulation Ensembles
689
