
Medi-CAT: Contrastive Adversarial Training for Medical Image
Classification
Pervaiz Iqbal Khan
1,2 a
, Andreas Dengel
1,2 b
and Sheraz Ahmed
1 c
1
German Research Center for Artificial Intelligence (DFKI), Kaiserslautern, Germany
2
RPTU Kaiserslautern-Landau, Germany
Keywords:
Adversarial Training, Contrastive Learning, Medical Image Classification, Vision Transformers, FGSM.
Abstract:
There are not many large medical image datasets available. Too small deep learning models can’t learn useful
features, so they don’t work well due to underfitting, and too big models tend to overfit the limited data. As a
result, there is a compromise between the two issues. This paper proposes a training strategy to overcome the
aforementioned issues in medical imaging domain. Specifically, it employs a large pre-trained vision trans-
formers to overcome underfitting and adversarial and contrastive learning techniques to prevent overfitting.
The presented method has been trained and evaluated on four medical image classification datasets from the
MedMNIST collection. Experimental results indicate the effectiveness of the method by improving the ac-
curacy up-to 2% on three benchmark datasets compared to well-known approaches and up-to 4.1% over the
baseline methods. Code can be accessed at: https://github.com/pervaizniazi/medicat.
1 INTRODUCTION
The classification of medical images aids healthcare
professionals in evaluating the images in a quick and
error-free manner. It uses the discriminative features
present in the images to distinguish between differ-
ent images. Traditionally, convolutional neural net-
works (CNNs) have been employed to learn the image
features and hence improve computer-aided diagnosis
systems (Lo and Hung, 2022; Hu et al., 2022b; Hu
et al., 2022a; Yang and Stamp, 2021). CNNs learn
the discriminative features from the images to per-
form tasks such as classification, object detection, etc.
However, CNNs learn these features by exploiting
local image structure, and they cannot capture long-
range dependencies present within the image. Re-
cently, transformer methods (Vaswani et al., 2017;
Devlin et al., 2018; Yang et al., 2019; Radford et al.,
2018) have revolutionized natural language process-
ing (NLP) field by employing a self-attention mech-
anism to capture global dependencies present in the
text. The success in NLP tasks has led to the sug-
gestion of a transformer architecture for vision tasks.
Vision Transformer (ViT) (Dosovitskiy et al., 2020)
a
https://orcid.org/0000-0002-1805-335X
b
https://orcid.org/0000-0002-6100-8255
c
https://orcid.org/0000-0002-4239-6520
converts an image into 16 × 16 patches ( like tokens
in NLP tasks), and takes them as input to generate its
feature representation. It has shown superior perfor-
mance over the CNNs in various studies (Wang et al.,
2021).
Large models like ViT may be prone to overfitting
the smaller datasets by retaining the training exam-
ples and may fail to perform well when faced with
unknown information. This can be particularly prob-
lematic in the medical imaging, where data is scarce.
Despite the large number of training samples in some
datasets (Yang et al., 2023), the per-class samples are
still small due to the large number of classes.
In this paper, we propose a training methodology
to overcome the overfitting issue by utilizing adver-
sarial training and contrastive learning. We primarily
use the Fast Gradient Sign Method (FGSM) (Good-
fellow et al., 2014) to generate adversarial examples.
Then we jointly train the clean and adversarial exam-
ples to learn their representations. In addition, we use
a contrastive learning method (Zbontar et al., 2021)
that improves image representation by bringing the
clean and adversarial example pairs closer and push-
ing the other examples away from them. The main
contributions of this paper are:
• It proposes a novel method for avoiding overfit-
ting by jointly minimizing the training objective
for the clean and adversarial examples.
832
Khan, P., Dengel, A. and Ahmed, S.
Medi-CAT: Contrastive Adversarial Training for Medical Image Classification.
DOI: 10.5220/0012396500003636
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 16th International Conference on Agents and Artificial Intelligence (ICAART 2024) - Volume 3, pages 832-839
ISBN: 978-989-758-680-4; ISSN: 2184-433X
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

• It performs experimentation on four public
datasets in the domain of medical image classi-
fication to evaluate the effectiveness of our pro-
posed training method.
• The proposed approach exceeds the well-known
approaches in the literature on three out of four
datasets.
2 RELATED WORK
2.1 Convolutional Neural Networks
Convolutional neural networks (CNNs) have made
great progress in the domain of computer vision due
to their ability to learn useful image feature represen-
tation. GoogLenet (Szegedy et al., 2015) used the in-
ception network to improve feature learning. ResNet
(He et al., 2016) employed residual connections to
overcome the vanishing gradient problem. MobileNet
(Howard et al., 2017) enhanced the efficiency of
CNNs by employing both depth-wise separable con-
volutions and point-wise convolutions. DenseNet
(Huang et al., 2017) used skip-connections between
every two successive layers and concatenated their
features instead of their summation. ConvNext (Liu
et al., 2022b) applied 7x7 depth-wise convolutions
and achieved comparable performance to ViT.
2.2 Vision Transfomers
After achieving significant success in NLP, transform-
ers in the image domain, i.e., vision transformers
(ViT) have been successfully implemented in vari-
ous tasks, including image classification (Dosovitskiy
et al., 2020), image segmentation (Zheng et al., 2021),
and object detection (Carion et al., 2020). ViT di-
vides an image into patches, which resemble tokens in
NLP, and then applies transformer layers to uncover
the correlation between these patches. This way, it
learns useful features for the downstream tasks. Many
improvements have been proposed over the standard
ViT. To strengthen the local structural relationship be-
tween the patches, T2T-ViT (Yuan et al., 2021) gen-
erates tokens and then combines neighboring tokens
into a single token. Swin Transformer (Liu et al.,
2021) learns the in-window and cross-window rela-
tionships by applying self-attention in the local win-
dow with the shifted window. The pooling-based vi-
sion transformer (PiT) (Heo et al., 2021) uses a newly
designed pooling layer in the transformer architecture
to reduce spatial size similar to CNNs and empirically
shows the improvement.
2.3 Medical Image Classification
MedMNIST (Yang et al., 2023) comprises of 12
datasets related to 2D images and 6 datasets related
to 3D images. The authors presented baseline re-
sults on these datasets using various models such as
ResNet-18 (He et al., 2016), ResNet-50 (He et al.,
2016), auto-sklearn (Feurer et al., 2015), AutoKeras
(Jin et al., 2019), and Google AutoML Vision (Bisong
et al., 2019). MedViT (Manzari et al., 2023) pro-
posed a hybrid model that combines the capabilities
of CNNs to model local representations with the ca-
pabilities of transformers to model the global rela-
tionship. Their attention mechanisms use efficient
convolution to solve the problem of quadratic com-
plexity. A novel mixer, known as a C-Mixer (Zheng
and Jia, 2023) incorporates a pre-training mechanism
to address the uncertainty and inefficient information
problem in label space. This mixer employs an incen-
tive imaginary matrix and a self-supervised method
with random masking to overcome the uncertainty
and inefficient information problem in label space.
BioMedGPT (Zhang et al., 2023), is a generalized
framework for multi-modal tasks in the medical do-
main, such as images and clinical notes. It first em-
ploys pre-training using masked language molding
(MLM), masked image infilling, question answering,
image captioning, and object detection to learn di-
verse types of knowledge. Then it is fine-tuned to the
downstream tasks to show the efficacy of the model
for transferring knowledge to other tasks.
3 METHODOLOGY
In this section, we present our proposed training
method for medical image classification. As shown
in Figure 1, our method consists of three main com-
ponents. (1) Transformer-based image encoder that
extracts features from the input image; (2) image en-
coder that takes images with perturbations generated
by FGSM (Goodfellow et al., 2014) and extracts fea-
tures; (3) Contrastive loss that takes the average patch
embeddings of the clean and perturbed images as in-
put and further improves their features in the repre-
sentation space.
3.1 Image Encoder
The pre-trained ViT (Dosovitskiy et al., 2020) is cho-
sen as the image encoder to encode the image in
the representation space. An image is first split into
16 × 16 patches as tokens, and then these tokens are
passed as inputs to the ViT. At the end of its forward
Medi-CAT: Contrastive Adversarial Training for Medical Image Classification
833
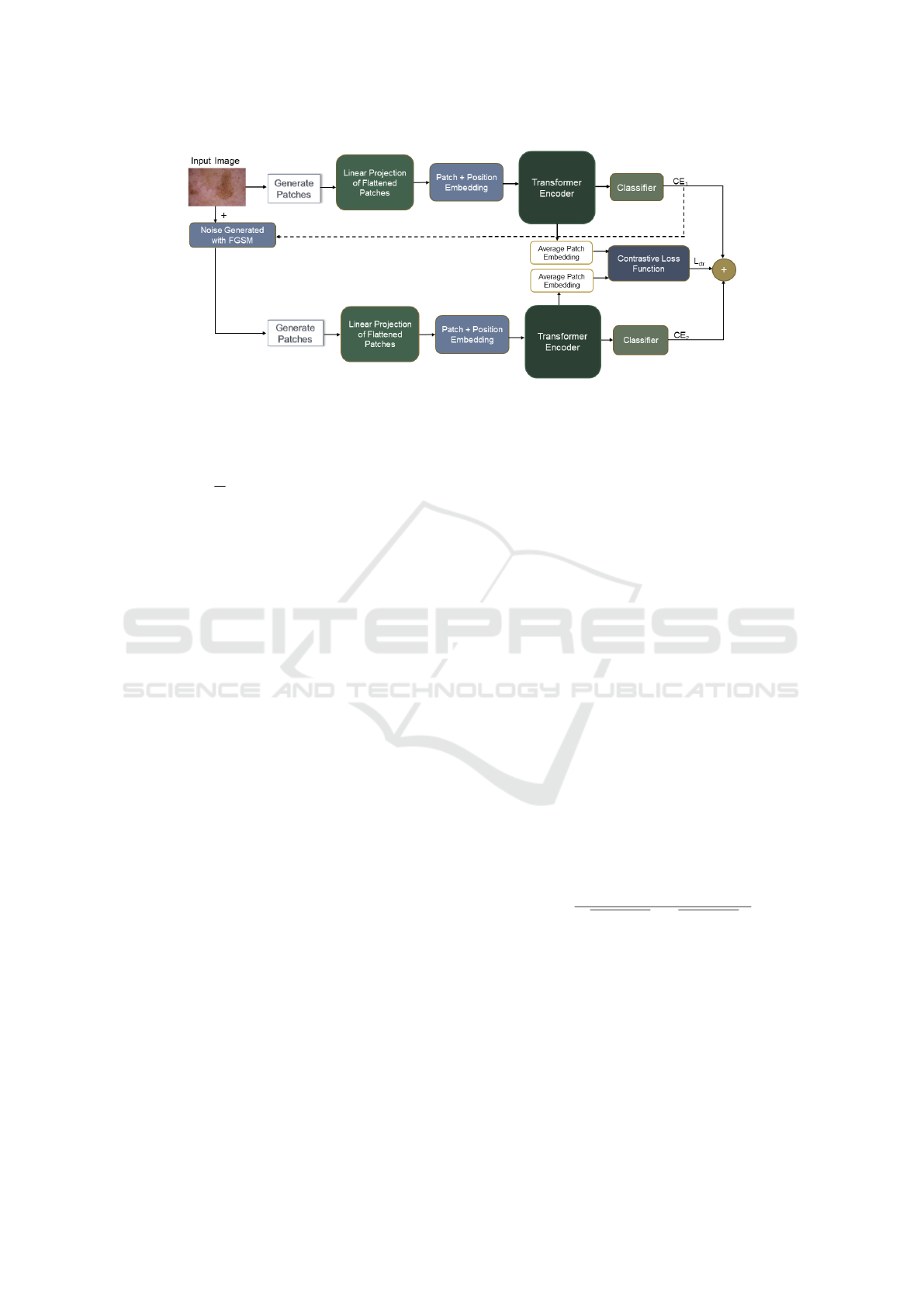
Figure 1: Proposed training methodology for medical image classification to overcome underfitting and overfitting.
pass, ViT returns the classification loss computed us-
ing cross-entropy as given by the following equation:
L
CE
= −
1
N
N
∑
i=1
C
∑
c=1
y
i,c
log(p(y
i
, c|s
i
[CLS]
)) (1)
where s
i
[CLS]
is the final hidden representation for the i-
th training example in the batch, ‘N’ is the number of
training examples in the batch, and ‘C’ is the number
of classes.
3.2 Adversarial Examples
Adversarial examples are generated by adding a small
amount of perturbations in the images from the train-
ing set. We utilize the FGSM (Goodfellow et al.,
2014) to generate the amount of noise η. Let f
θ
(x
i
, y
i
)
be a neural network parameterized by θ where x
i
, and
y
i
represent the input example and its corresponding
label, respectively. Let, L represent the loss at the
end of the forward pass as calculated using equation
1. Then perturbation η generated by FGSM is given
as follows:
η = −εsign(∇
x
i
L( f
θ
(x
i
), y
i
)) (2)
In equation 2, ∇ is the gradient of the loss L w.r.t in-
put x
i
. ε is the hyperparameter controlling the amount
of noise. The generated noise η is added to the input
image to generate an adversarial example. The gen-
erated adversarial example is passed to the image en-
coder as discussed in section 3.1, where another for-
ward pass is completed and another classification loss
is computed as given by equation 1. We use the shared
image encoder to extract the representations for the
clean and perturbed images.
3.3 Contrastive Learning
We employed Barlow Twins (Zbontar et al., 2021) as
a contrastive learning method that takes two inputs,
i.e., encoding of the clean image, and encoding of
its perturbed version that are generated by image en-
coder. The encoding of the last hidden state of the im-
age encoder can be represented as H ∈ R
p×d
. Here, p
is the number of patches, i.e. 16, and d is the number
hidden units of ViT, i.e., 1024. We average the en-
coding of all the patches for both clean and perturbed
examples and then pass it to Barlow Twins (Zbontar
et al., 2021) loss function that improves their repre-
sentations by pulling the pair of clean and perturbed
encoding closer while pushing them away from other
image encoding in the training batch.
Let E
o
and E
p
represent the averaged encoding
of the original and its perturbed version, respectively.
Then, the Barlow Twins (Zbontar et al., 2021) im-
proves their representations by using following objec-
tive function:
L
C T R
=
∑
i=1
(1 − X
ii
)
2
+ λ
∑
i=1
∑
j̸=i
X
2
i j
(3)
where
∑
i=1
(1 − X
ii
)
2
, and
∑
i=1
∑
j̸=i
X
2
i j
are the in-
variance, and redundancy reduction terms respec-
tively, and λ controls weights between the two terms.
The matrix X computes the cross-correlation between
E
o
, and E
p
. It is computed as follows:
X
i j
=
∑
N
b=1
E
o
b,i
E
p
b,i
q
∑
N
b=1
(E
o
b,i
)
2
q
∑
N
b=1
(E
p
b,i
)
2
(4)
where b is the batch size, and X
i j
represents the entry
of the i-th row and j-th column of X. Both E
o
and E
p
∈ R
1×1024
3.4 Training Objective
The training objective of our proposed method con-
sists of three parts:(1) Minimizing the classification
loss of the clean images; (2) minimizing the classifi-
cation loss of the perturbed images; (3) minimizing
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
834

the contrastive loss for the clean and perturbed image
encoding.
Total loss L is given as follows:
L =
(1 − α)
2
(L
CE
1
+ L
CE
2
) + αL
CT R
(5)
where L
CT R
is the contrastive loss, L
CE
1
, and L
CE
2
are
two classification losses for the clean and perturbed
images, and α is the trade-off parameter between the
three losses. A higher value of α means more weight
to the contrastive loss.
4 EXPERIMENTS
4.1 Datasets
MedMNIST (Yang et al., 2023) is a collection of 2D
and 3D medical images related to ordinal regression,
multi-label, and multi-class classification. We per-
formed experimentation on four multi-class classifi-
cation datasets from this collection to validate the per-
formance of our proposed training strategy. The de-
tails of each dataset are given in Table 1.
4.2 Evaluation Metrics
Following (Zhang et al., 2023), we use accuracy as an
evaluation metric. Accuracy is based on the thresh-
old used to evaluate the discrete label prediction and
is sensitive to class imbalance. As there is no class
imbalance in the datasets we used in experimentation,
accuracy is a good metric. On each dataset, we report
the average accuracy score for two random runs with
seeds of 42, and 44 respectively.
4.3 Training Details
We conducted training on each of the datasets men-
tioned in the section 4.1 for 50 epochs, with a batch
size of 48. Before the training, all images were re-
sized to 224x224 pixels. We used the same param-
eters as in (Yang et al., 2023) to normalize all the
images. We used a fixed learning rate of 1e
−4
and
AdamW (Loshchilov and Hutter, 2018) as an opti-
mizer in all our experiments. The cross-entropy and
the Barlow Twins (Zbontar et al., 2021) were em-
ployed as classification loss and contrastive loss, re-
spectively. The default hyperparameters were used
for contrastive loss, and unlike the original imple-
mentation, we did not use a projection network for
its two inputs. We performed a grid search for
α ∈ {0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9} and ε ∈
{0.0001, 0.001, 0.0005, 0.001} and used the valida-
tion set model with the highest accuracy for test set
evaluation.
5 RESULTS AND ANALYSIS
In this section, we present the results and analysis
of our proposed approach. Furthermore, we com-
pare our results with well known approaches in the
literature and also discuss the effect of various hyper-
parameters on the model performance.
5.1 Comparison with Existing Methods
Table 2 shows that our proposed method outperforms
the existing methods on three datasets, whereas it
remains second-best on the fourth one. These en-
hancements can be attributed to adversarial training
and contrastive learning, which enhance the general-
ization of the model by avoiding overfitting. How-
ever, these improvements come with additional train-
ing costs, which are incurred by gradient calculations
in FGSM (Goodfellow et al., 2014) method and ad-
ditional training passes with perturbed images. How-
ever, accuracy can be more important in health-related
tasks than training costs.
5.2 Analysis of Noise Amount and
Trade-off Parameter
Figure 2 shows the effect of trade-off parameter α and
noise controlling parameter ε on the validation sets of
four datasets. For simplicity, these results are taken
from one of the training runs. All the plots show that
the accuracy for the smaller values of α is generally
higher, whereas it decreases sharply for α ≥ 0.6. This
implies giving more weight to contrastive loss after
a certain degree negatively affects performance. For
values of α < 0.6 there is only a slight change in the
performance of the model. As shown in Figure 2g
and 2h, DermaMNIST (Yang et al., 2023) is more
sensitive to both α and ε values as compared to other
datasets.
5.3 Effectiveness of Proposed Method
Table 3 illustrates the effectiveness of our proposed
method. The results show that incorporating adver-
sarial training enhances the model’s precision on the
DermaMNIST (Yang et al., 2023). Furthermore, in-
corporating contrastive learning further improves the
performance of the model. This performance en-
Medi-CAT: Contrastive Adversarial Training for Medical Image Classification
835

Table 1: Statistics of datasets from MedMNIST (Yang et al., 2023) collection used in our experiments.
Name Modality # Classes # Samples Train/validation/Test
DermaMNIST (Yang et al., 2023) Dermatoscope 7 10,015 7,007/1,003/2,005
OrganAMNIST (Yang et al., 2023) Abdominal CT 11 58,850 34,581/6,491/17,778
OrganCMNIST (Yang et al., 2023) Abdominal CT 11 23,660 13,000/2,392/8,268
OrganSMNIST (Yang et al., 2023) Abdominal CT 11 25,221 13,940/2,452/8,829
Table 2: Compares the results of our proposed method with existing methods in literature on DermaMNIST, OrganAMNIST,
OrganCMNIST, and OrganSMNIST (Yang et al., 2023) datasets in terms of accuracy score. Similar to (Zhang et al., 2023),
we only present SotA approaches if they provided open-source code for reproducibility. The proposed method outperforms
existing methods on three out of four datasets.
Methods DermaMNIST OrganAMNIST OrganCMNIST OrganSMNIST
ResNet-18 (28) (Yang et al., 2023) 0.735 0.935 0.900 0.782
ResNet-18 (224) (Yang et al., 2023) 0.754 0.951 0.920 0.778
ResNet-50 (28) (Yang et al., 2023) 0.735 0.935 0.905 0.770
ResNet-50 (224) (Yang et al., 2023) 0.731 0.947 0.911 0.785
auto-sklearn (Yang et al., 2023) 0.719 0.762 0.829 0.672
AutoKeras (Yang et al., 2023) 0.749 0.905 0.879 0.813
Google AutoML Vision (Yang et al., 2023) 0.768 0.886 0.877 0.749
FPVT (Liu et al., 2022a) 0.766 0.935 0.903 0.785
MedVIT-T (224) (Manzari et al., 2023) 0.768 0.931 0.901 0.789
MedVIT-S (224) (Manzari et al., 2023) 0.780 0.928 0.916 0.805
MedVIT-L (224) (Manzari et al., 2023) 0.773 0.943 0.922 0.806
Complex Mixer (Zheng and Jia, 2023) 0.833 0.951 0.922 0.810
BioMed-GPT (Zhang et al., 2023) 0.786 0.952 0.931 0.823
Ours 0.824 0.961 0.940 0.843
Table 3: Shows accuracy scores on four datasets for the proposed method. Here, AT stands for adversarial training.
Method DermaMNIST OrganAMNIST OrganCMNIST OrganSMNIST
ViT
Large
(Dosovitskiy et al., 2020) (Baseline) 0.783 0.954 0.937 0.841
AT Only 0.817 0.949 0.942 0.841
AT + Contrastive (Proposed) 0.824 0.961 0.940 0.843
hancement of over 4% can be attributed to adversar-
ial and contrastive training. Since the original dataset
size is smaller as compared to other datasets, the
FGSM (Goodfellow et al., 2014) generates new train-
ing samples with small perturbations, and then adver-
sarial training and contrastive learning improve fea-
ture representations. For the OrganAMNIST (Yang
et al., 2023), adversarial training results in a decrease
in model performance, whereas the addition of con-
trastive training enhances the performance compared
to the baseline model. The inclusion of contrastive
learning results in a slight decrease in performance
compared to adversarial training for the OrganCM-
NIST (Yang et al., 2023). Our method for OrganSM-
NIST (Yang et al., 2023) only makes a small improve-
ment over the standard model. The difficulty of the
dataset itself might be the reason for this, as it doesn’t
allow noise to improve performance.
6 CONCLUSIONS
In this paper, a training method is proposed to over-
come the problems of underfitting and overfitting in
medical image classification. The proposed method
used the power of a vision transformer to learn the
features for different classes by fine-tuning it on the
downstream classification task. To fix the overfitting,
perturbations were added to the training images, and
then both clean and perturbed images were jointly
trained. To further improve the feature representa-
tion, contrastive loss was added, which pushes the
clean and perturbed versions of the sample closer and
farther than the other samples in the representation
space. Extensive experiments on the four benchmark
medical image classification datasets demonstrate the
effectiveness of the proposed method. In the future,
we intend to apply the proposed method to object de-
tection and segmentation tasks.
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
836
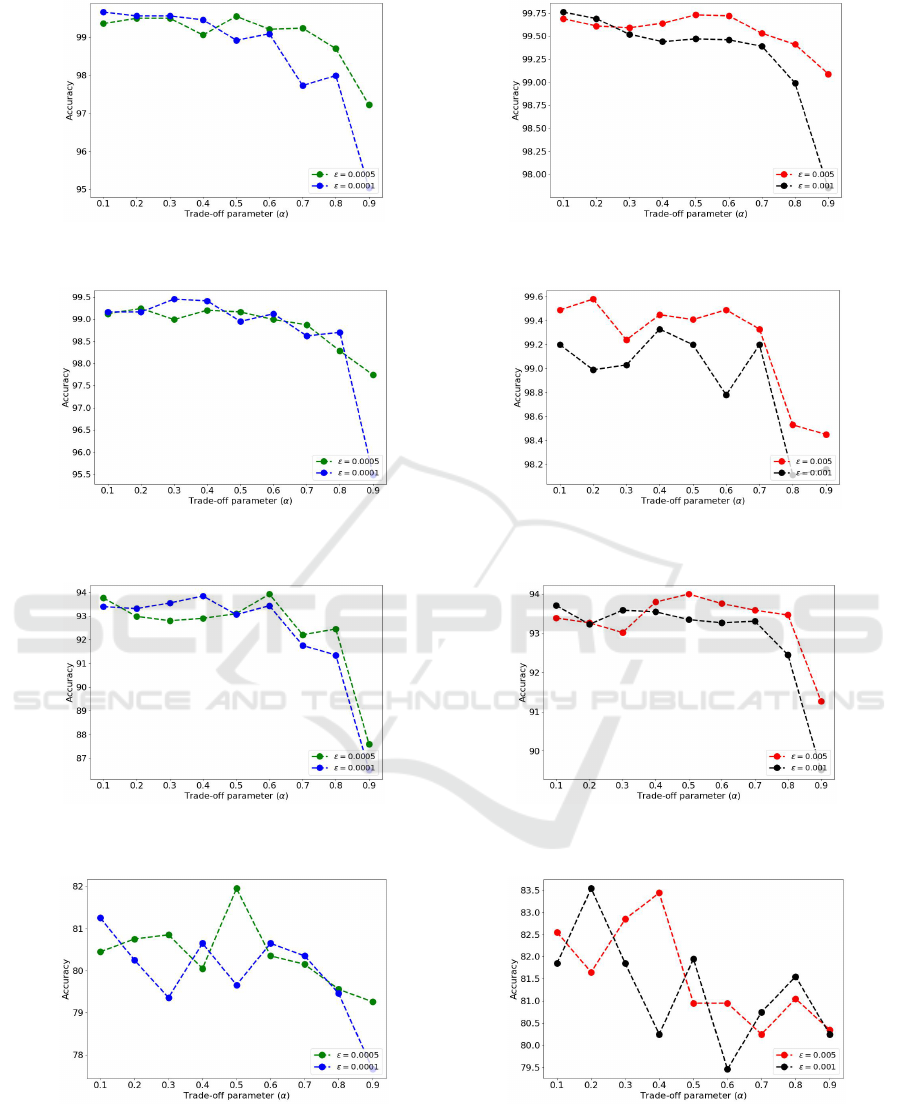
(a) Plots showing the effect of trade-off parameter α
and noise controlling parameter ε ∈ (0.0005, 0.0001) on
OrganAMNIST.
(b) Plots showing the effect of trade-off parameter α
and noise controlling parameter ε ∈ (0.005, 0.001) on
OrganAMNIST.
(c) Plots showing the effect of trade-off parameter α
and noise controlling parameter ε ∈ (0.0005, 0.0001) on
OrganCMNIST.
(d) Plots showing the effect of trade-off parameter α
and noise controlling parameter ε ∈ (0.005, 0.001) on
OrganCMNIST.
(e) Plots showing the effect of trade-off parameter α
and noise controlling parameter ε ∈ (0.0005, 0.0001) on
OrganSMNIST.
(f) Plots showing the effect of trade-off parameter α
and noise controlling parameter ε ∈ (0.005, 0.001) on
OrganSMNIST.
(g) Plots showing the effect of trade-off parameter α
and noise controlling parameter ε ∈ (0.0005, 0.0001) on
DermaMNIST.
(h) Plots showing the effect of trade-off parameter α
and noise controlling parameter ε ∈ (0.005, 0.001) on
DermaMNIST.
Figure 2: Accuracy plots on the validation set for MedMNIST datasets showing the effect of trade-off parameter and noise
amount.
Medi-CAT: Contrastive Adversarial Training for Medical Image Classification
837

REFERENCES
Bisong, E. et al. (2019). Building machine learning
and deep learning models on Google cloud platform.
Springer.
Carion, N., Massa, F., Synnaeve, G., Usunier, N., Kirillov,
A., and Zagoruyko, S. (2020). End-to-end object de-
tection with transformers. In European conference on
computer vision, pages 213–229. Springer.
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2018). Bert: Pre-training of deep bidirectional trans-
formers for language understanding. arXiv preprint
arXiv:1810.04805.
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn,
D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer,
M., Heigold, G., Gelly, S., et al. (2020). An image is
worth 16x16 words: Transformers for image recogni-
tion at scale. arXiv preprint arXiv:2010.11929.
Feurer, M., Klein, A., Eggensperger, K., Springenberg, J.,
Blum, M., and Hutter, F. (2015). Efficient and robust
automated machine learning. Advances in neural in-
formation processing systems, 28.
Goodfellow, I. J., Shlens, J., and Szegedy, C. (2014). Ex-
plaining and harnessing adversarial examples. arXiv
preprint arXiv:1412.6572.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resid-
ual learning for image recognition. In Proceedings of
the IEEE conference on computer vision and pattern
recognition, pages 770–778.
Heo, B., Yun, S., Han, D., Chun, S., Choe, J., and Oh, S. J.
(2021). Rethinking spatial dimensions of vision trans-
formers. In Proceedings of the IEEE/CVF Interna-
tional Conference on Computer Vision, pages 11936–
11945.
Howard, A. G., Zhu, M., Chen, B., Kalenichenko, D.,
Wang, W., Weyand, T., Andreetto, M., and Adam,
H. (2017). Mobilenets: Efficient convolutional neu-
ral networks for mobile vision applications. arXiv
preprint arXiv:1704.04861.
Hu, Q., Chen, C., Kang, S., Sun, Z., Wang, Y., Xiang, M.,
Guan, H., Xia, L., and Wang, S. (2022a). Applica-
tion of computer-aided detection (cad) software to au-
tomatically detect nodules under sdct and ldct scans
with different parameters. Computers in Biology and
Medicine, 146:105538.
Hu, W., Li, C., Li, X., Rahaman, M. M., Ma, J., Zhang,
Y., Chen, H., Liu, W., Sun, C., Yao, Y., et al.
(2022b). Gashissdb: A new gastric histopathology
image dataset for computer aided diagnosis of gas-
tric cancer. Computers in biology and medicine,
142:105207.
Huang, G., Liu, Z., Van Der Maaten, L., and Weinberger,
K. Q. (2017). Densely connected convolutional net-
works. In Proceedings of the IEEE conference on
computer vision and pattern recognition, pages 4700–
4708.
Jin, H., Song, Q., and Hu, X. (2019). Auto-keras: An ef-
ficient neural architecture search system. In Proceed-
ings of the 25th ACM SIGKDD international confer-
ence on knowledge discovery & data mining, pages
1946–1956.
Liu, J., Li, Y., Cao, G., Liu, Y., and Cao, W. (2022a). Fea-
ture pyramid vision transformer for medmnist classifi-
cation decathlon. In 2022 International Joint Confer-
ence on Neural Networks (IJCNN), pages 1–8. IEEE.
Liu, Z., Lin, Y., Cao, Y., Hu, H., Wei, Y., Zhang, Z., Lin,
S., and Guo, B. (2021). Swin transformer: Hierar-
chical vision transformer using shifted windows. In
Proceedings of the IEEE/CVF international confer-
ence on computer vision, pages 10012–10022.
Liu, Z., Mao, H., Wu, C.-Y., Feichtenhofer, C., Darrell, T.,
and Xie, S. (2022b). A convnet for the 2020s. In Pro-
ceedings of the IEEE/CVF conference on computer vi-
sion and pattern recognition, pages 11976–11986.
Lo, C.-M. and Hung, P.-H. (2022). Computer-aided diagno-
sis of ischemic stroke using multi-dimensional image
features in carotid color doppler. Computers in Biol-
ogy and Medicine, 147:105779.
Loshchilov, I. and Hutter, F. (2018). Fixing weight
decay regularization in adam. arXiv preprint
arXiv:2011.08042v1.
Manzari, O. N., Ahmadabadi, H., Kashiani, H., Shokouhi,
S. B., and Ayatollahi, A. (2023). Medvit: a ro-
bust vision transformer for generalized medical image
classification. Computers in Biology and Medicine,
157:106791.
Radford, A., Narasimhan, K., Salimans, T., and Sutskever,
I. (2018). Improving language understanding by gen-
erative pre-training.
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S.,
Anguelov, D., Erhan, D., Vanhoucke, V., and Rabi-
novich, A. (2015). Going deeper with convolutions.
In Proceedings of the IEEE conference on computer
vision and pattern recognition, pages 1–9.
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones,
L., Gomez, A. N., Kaiser, Ł., and Polosukhin, I.
(2017). Attention is all you need. Advances in neural
information processing systems, 30.
Wang, W., Xie, E., Li, X., Fan, D.-P., Song, K., Liang, D.,
Lu, T., Luo, P., and Shao, L. (2021). Pyramid vi-
sion transformer: A versatile backbone for dense pre-
diction without convolutions. In Proceedings of the
IEEE/CVF international conference on computer vi-
sion, pages 568–578.
Yang, J., Shi, R., Wei, D., Liu, Z., Zhao, L., Ke, B., Pfis-
ter, H., and Ni, B. (2023). Medmnist v2-a large-scale
lightweight benchmark for 2d and 3d biomedical im-
age classification. Scientific Data, 10(1):41.
Yang, X. and Stamp, M. (2021). Computer-aided diagno-
sis of low grade endometrial stromal sarcoma (lgess).
Computers in Biology and Medicine, 138:104874.
Yang, Z., Dai, Z., Yang, Y., Carbonell, J., Salakhutdinov,
R. R., and Le, Q. V. (2019). Xlnet: Generalized au-
toregressive pretraining for language understanding.
Advances in neural information processing systems,
32.
Yuan, L., Chen, Y., Wang, T., Yu, W., Shi, Y., Jiang, Z.-H.,
Tay, F. E., Feng, J., and Yan, S. (2021). Tokens-to-
token vit: Training vision transformers from scratch
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
838

on imagenet. In Proceedings of the IEEE/CVF inter-
national conference on computer vision, pages 558–
567.
Zbontar, J., Jing, L., Misra, I., LeCun, Y., and Deny, S.
(2021). Barlow twins: Self-supervised learning via
redundancy reduction. In International Conference on
Machine Learning, pages 12310–12320. PMLR.
Zhang, K., Yu, J., Yan, Z., Liu, Y., Adhikarla, E., Fu,
S., Chen, X., Chen, C., Zhou, Y., Li, X., et al.
(2023). Biomedgpt: A unified and generalist biomed-
ical generative pre-trained transformer for vision,
language, and multimodal tasks. arXiv preprint
arXiv:2305.17100.
Zheng, S., Lu, J., Zhao, H., Zhu, X., Luo, Z., Wang, Y., Fu,
Y., Feng, J., Xiang, T., Torr, P. H., et al. (2021). Re-
thinking semantic segmentation from a sequence-to-
sequence perspective with transformers. In Proceed-
ings of the IEEE/CVF conference on computer vision
and pattern recognition, pages 6881–6890.
Zheng, Z. and Jia, X. (2023). Complex mixer for
medmnist classification decathlon. arXiv preprint
arXiv:2304.10054.
Medi-CAT: Contrastive Adversarial Training for Medical Image Classification
839
