
Diversifying Knowledge Enhancement of Biomedical Language Models
Using Adapter Modules and Knowledge Graphs
Juraj Vladika, Alexander Fichtl and Florian Matthes
Department of Computer Science, Technical University of Munich,
Boltzmannstraße 3, 85748 Garching bei M
¨
unchen, Germany
Keywords:
Natural Language Processing (NLP), Pre-Trained Language Models, Knowledge Graphs, Domain
Knowledge, Knowledge Enhancement, Adapters, Biomedicine, Biomedical NLP.
Abstract:
Recent advances in natural language processing (NLP) owe their success to pre-training language models on
large amounts of unstructured data. Still, there is an increasing effort to combine the unstructured nature
of LMs with structured knowledge and reasoning. Particularly in the rapidly evolving field of biomedical
NLP, knowledge-enhanced language models (KELMs) have emerged as promising tools to bridge the gap be-
tween large language models and domain-specific knowledge, considering the available biomedical knowledge
graphs (KGs) curated by experts over the decades. In this paper, we develop an approach that uses lightweight
adapter modules to inject structured biomedical knowledge into pre-trained language models (PLMs). We
use two large KGs, the biomedical knowledge system UMLS and the novel biochemical ontology OntoChem,
with two prominent biomedical PLMs, PubMedBERT and BioLinkBERT. The approach includes partitioning
knowledge graphs into smaller subgraphs, fine-tuning adapter modules for each subgraph, and combining the
knowledge in a fusion layer. We test the performance on three downstream tasks: document classification,
question answering, and natural language inference. We show that our methodology leads to performance
improvements in several instances while keeping requirements in computing power low. Finally, we provide a
detailed interpretation of the results and report valuable insights for future work.
1 INTRODUCTION
The field of natural language processing (NLP) has
been marked by impressive advancements in recent
years. The appearance of new model architectures,
including the emergence of generative transformers
and pre-trained language models (PLMs), has brought
along with it widespread usage and attention. Still,
most of these models were trained on large amounts
of web content, and while they excel at tasks in a
general-purpose setting, there is still a performance
gap when it comes to domain-specific challenges.
One of these challenging domains is bio-
medicine, which centers around the study of the hu-
man body, diseases, drugs, and treatments. Biomed-
ical text is often characterized as highly complex
because of its advanced terminology, which fre-
quently includes names of chemical compounds,
long-spanning relations, and other jargon not com-
monly used in everyday language. For NLP mod-
els trained on general corpora to work well in the
biomedical domain, researchers have turned to trans-
fer learning methods and domain adaption. The
most common approach to domain adaptation is to
continue the initial general pre-training of language
models with data from domain-specific medical cor-
pora. Examples of models adapted in this way are
BioBERT (Lee et al., 2019) and SciBERT (Beltagy
et al., 2019), which drew the additional training data
from biomedical and computer science research ab-
stracts. Dropping the mixed-domain approach from
previous frameworks, models like PubMedBERT (Gu
et al., 2020) and BioLinkBERT (Yasunaga et al.,
2022) were instead trained solely on PubMed research
articles, with BioLinkBERT even leveraging links (ci-
tations) to other research articles.
While domain fine-tuning of whole PLMs has
proven to increase the performance on downstream
biomedical NLP tasks, additional pre-training can of-
ten be resource-intensive and infeasible for smaller re-
search groups and situations where computing power
is limited. A promising research direction has
emerged in the form of knowledge-enhanced lan-
guage models (KELMs) (Hu et al., 2023). It refers
376
Vladika, J., Fichtl, A. and Matthes, F.
Diversifying Knowledge Enhancement of Biomedical Language Models Using Adapter Modules and Knowledge Graphs.
DOI: 10.5220/0012395200003636
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 16th International Conference on Agents and Artificial Intelligence (ICAART 2024) - Volume 2, pages 376-387
ISBN: 978-989-758-680-4; ISSN: 2184-433X
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

to any set of methods that try to incorporate external
knowledge into language models, usually by inject-
ing it into the model’s input, architecture, or output.
In a sea of knowledge-enhancement methods, an es-
pecially interesting one is the utilization of adapters.
Broadly speaking, adapters are small bottleneck
feed-forward layers inserted within each layer of a
transformer-based language model (Houlsby et al.,
2019; Pfeiffer et al., 2020b). The small amount of
additional parameters allows for the injection of new
data or knowledge without requiring the whole model
to be fine-tuned. Adapters plugged on top of large lan-
guage models will often only have around 1% of the
number of training parameters compared to the trans-
former. The transformer model’s learned parameters
(weights) are frozen and left unchanged, and only the
adapter is fine-tuned. Other than being lightweight on
resources, this approach also helps avoid the problem
of catastrophic forgetting, where language models
forget their existing knowledge from the pre-training
corpora when they are fine-tuned on a new, smaller
corpus (Colon-Hernandez et al., 2021).
This paper specifically focuses on using adapters
to inject structured biomedical knowledge from large
knowledge graphs into PLMs. We provide an
overview of existing adapter approaches for the
biomedical domain, as well as existing biomedi-
cal language models. We perform extensive ex-
periments to test the performance of knowledge-
enhanced, adapter-based biomedical language models
on a number of representative biomedical classifica-
tion tasks (document classification, question answer-
ing, natural language inference). We show that the
model performance is improved in several instances
on downstream tasks and provide a deeper look into
the resulting change in model predictions. Finally,
our experiments demonstrate that the OntoChem on-
tology (Irmer et al., 2013), which has not been used
for knowledge enhancement yet, is a viable alterna-
tive to other prominent knowledge sources.
2 RELATED WORK
2.1 Knowledge-Enhanced PLMs
PLMs are trained on enormous corpora of training
data, ranging from 3.3 billion tokens in the case of the
original BERT (Devlin et al., 2019), all the way to 3.5
trillion tokens in the case of the recent Falcon-180B
model (Almazrouei et al., 2023). The power of the
model architecture, combined with transfer learning,
has led to these models showing impressive capabili-
ties on most NLP tasks. While the textual data used
for the model training is usually completely unstruc-
tured in nature, research has shown that models like
BERT do encode, to some extent, syntactic structures,
hierarchical concepts, and certain semantic concep-
tual relations (Rogers et al., 2021). Still, other studies
have shown weakness in modeling tasks dealing with
structured knowledge, such as hyponymy relations
(Ravichander et al., 2020) or preserving the associ-
ation between text and meaning (Di Sciullo, 2018).
In most cases, the knowledge we find and gather,
especially scientific knowledge, can be represented in
a structured manner. This is the underlying idea of
knowledge graphs (KGs), a data structure that mod-
els concepts (entities) and relations between them
in a graph-like format (Ji et al., 2021). KGs have
been used in the field of NLP to enhance the per-
formance of NLP models in many downstream NLP
tasks (Schneider et al., 2022). There are multiple
ways to combine KGs with PLMs. The knowledge
triples from KGs can be embedded as vector represen-
tations such as TransE (Wang et al., 2014) or TuckER
(Balazevic et al., 2019) and then combined with the
vectors encoding text. Alternatively, the triples from
KGs can be converted to sentences, and, in turn, these
textual representations can then be used to fine-tune
PLMs in the same way as with any other text. This
approach was followed by COMET (Bosselut et al.,
2019), which utilized the knowledge graph Concept-
Net (Speer et al., 2017) to enhance the performance
on commonsense reasoning tasks. Besides knowledge
graphs, lexicons are sometimes used for knowledge
enhancement (Hoang et al., 2022).
While there are numerous ways to inject struc-
tured knowledge into PLMs such as adding it to
the input and output of models (Wei et al., 2021),
an especially promising approach is adding adapters
to the architecture of the model (Colon-Hernandez
et al., 2021). Adapters are small layers that are in-
serted within a language model and are subsequently
fine-tuned to a specific task. The major benefit of
adapters is that they add a minimal amount of ad-
ditional parameters, thus significantly reducing the
needed training time. Combined with freezing orig-
inal model weights, adapters can avoid catastrophic
forgetting, where the PLM’s performance deteriorates
when all of its weights are fine-tuned with a new
knowledge source. Adapters have been used for nu-
merous purposes such as learning hierarchical rep-
resentation (Chronopoulou et al., 2022), transferring
models trained on English to low-resource languages
(Wang et al., 2021), and in the domain of efficient
transformers as low-rank adapters (LoRA) (Hu et al.,
2022). General knowledge-enhanced PLMs utiliz-
ing adapters include, for example, KnowBERT (Pe-
Diversifying Knowledge Enhancement of Biomedical Language Models Using Adapter Modules and Knowledge Graphs
377

ters et al., 2019) and K-Adapter (Wang et al., 2020).
A practical tool emerged that combines well-known
adapter architectures in one place, called AdapterHub
(Pfeiffer et al., 2020b).
2.2 Biomedical Knowledge-Enhanced
PLMs
A major focus of knowledge enhancement in PLMs
is in domain adaption to expert domains such as the
biomedical domain. So far, most of the advance-
ments have focused on utilizing the knowledge graph
UMLS (Bodenreider, 2004) for this purpose. Exam-
ples include BERT-MK (He et al., 2020) and KeBi-
oLM (Yuan et al., 2021), which both fine-tune the
whole weights of the base language model by using
masked language modeling of triples from UMLS.
Biomedical PLMs can then be used for various NLP
tasks, such as biomedical text summarization (Abacha
et al., 2021), named entity recognition (Sung et al.,
2022), medical fact-checking (Vladika and Matthes,
2023), information retrieval (Luo et al., 2022), or
health question answering (Vladika et al., 2023).
There are also existing approaches using adapters
for biomedical knowledge enhancement. Represen-
tative works are DAKI (Lu et al., 2021), which fine-
tunes the adapters with entity prediction task, and KE-
BLM (Lai et al., 2023), which fine-tunes the adapters
on three different knowledge types from UMLS and
PubChem (Kim et al., 2019), namely entity descrip-
tions, entity-entity relations, and entity synonyms.
The most similar approach to ours and a direct inspi-
ration was the Mixture-of-Partitions (MoP) approach
(Meng et al., 2021), where the adapters were fine-
tuned on smaller subgraphs of UMLS.
Figure 1: Triplet from the OntoChem Fact Finder
1
.
In essence, our work builds on the present foun-
dations of adapter-based biomedical models and uses
the yet unexplored knowledge graph OntoChem,
which is rich with chemical knowledge. For our ex-
periments, we use the well-known biomedical PLM
1
https://sciwalker.com/analytics/factfinder
PubMedBERT as well as the yet unexplored but pow-
erful BioLinkBERT base model. Following the sug-
gestions of (Meng et al., 2021), we use only the
triplets corresponding to the 20 most frequent rela-
tions of OntoChem for the knowledge injection. An
example of an OntoChem triplet can be seen in Figure
1. Finally, we provide a deeper qualitative analysis
of learned structured knowledge on a specific dataset.
Notably, our work achieves the SOTA (averaged)
performance on the question-answering BioASQ-7b
dataset.
3 METHODOLOGY
In this section, we will explain the training method-
ology we used for the experiments in this paper. It is
depicted in Figure 2.
3.1 Knowledge Graph Representation
A central element of our method is the knowledge
graph (KG). This KG is a structured representation of
information denoted as a collection of ordered triples
(Ji et al., 2021). We denote these triples as (s, r, o),
where s is a subject, r is a relation, and o is an object.
Both s and o are entities that come from an entity set
E, while relations come from a relation set R. Each
entity and relation in the KG is associated with its cor-
responding textual surface form. This form can take
the shape of a single word or a compound term (e.g.,
for names of chemicals) or even a concise phrase, es-
pecially in case of relations. This textual association
is critical as it bridges the gap between the structured
KG and natural language, allowing for easier injec-
tion of KG knowledge into the language models and
associated fine-tuning.
The primary objective is to enhance the capabili-
ties of a pre-trained language model, denoted as LM,
by integrating the knowledge contained within the
KG. To achieve this, we need a training objective that
effectively incorporates the KG knowledge into the
model. Most encoder-only PLMs based on the origi-
nal BERT use masked language modeling (MLM) as
one of its pre-training objectives. This task consists of
masking a certain word in a given sentence and hav-
ing the model predict which word would fit the best
in the place of the masked token. We follow the es-
tablished approach of using an entity prediction ob-
jective, where we mask one of the entities and have
the model predict which token would best fit. In this
way, the model incorporates the structured knowledge
of (s, r, o) triples into its internal weights.
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
378
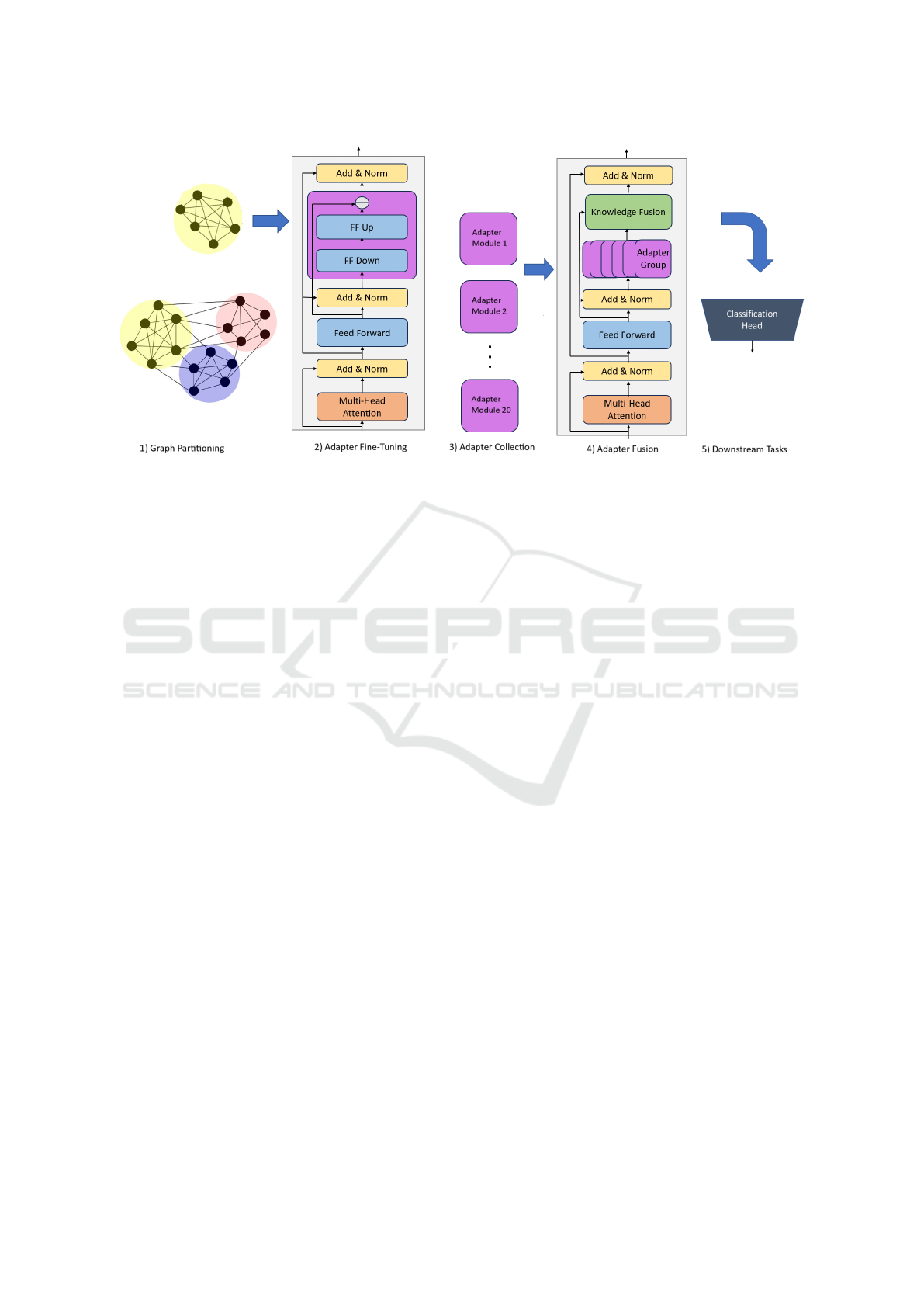
Figure 2: Methodology used to construct the final model and run the experiments.
3.2 Graph Partitioning
During the prediction of the masked token, the model
produces a probability distribution (with a softmax
function) over all of the entities from KG’s entity
set E. Considering the massive size of the biomedi-
cal KGs we use in the paper, computing the softmax
function over all its entities carries a lot of compu-
tation complexity. This issue can significantly slow
down model training and inference. To bridge this
challenge, some approaches have been suggested in
the literature. We opt for the approach of (Meng
et al., 2021), which involves partitioning the KG into
smaller subgraphs, which are then trained on indepen-
dently, and later, their knowledge combined to unified
knowledge representations.
The process of dividing a KG yields smaller sub-
graphs that we denote as G
1
, G
2
, ..., G
k
. We set k in
final experiments to be 20, following empirical ob-
servations and previous literature, which balances ef-
ficiency and graph coverage well. Ideally, these 20
smaller subgraphs should be almost equal-sized com-
ponents, meaning nodes are balanced across parti-
tions. Additionally, the capacity of edges between dif-
ferent components should be minimized to maximize
the retention of factual knowledge. This is a problem
known as balanced graph partitioning and is known
to be an NP-complete problem (Andreev and R
¨
acke,
2004). Several good approximations have been devel-
oped to determine the exact solution despite its com-
putational complexity. We opt for the METIS algo-
rithm (Karypis and Kumar, 1997), which was used in
other works dealing with large-scale KG partitioning
(Zheng et al., 2020).
3.3 Adapter Module Learning
Once the KG is appropriately partitioned, the pro-
cess of fine-tuning the LM can be started. We deploy
adapter modules for this purpose. As mentioned pre-
viously, adapters are newly initialized feed-forward
networks inserted between the transformer model’s
layers. Notably, the training of adapter modules does
not require fine-tuning the existing parameters of the
pre-trained model. Instead, it focuses solely on updat-
ing the parameters within the adapters. This strategy
ensures that the pre-trained model’s core knowledge
remains intact while enabling the model to specialize
in the biomedical domain by adapting to the specific
knowledge contained in the KG.
There are multiple adapter module configurations,
such as (Houlsby et al., 2019) and (Bapna and Firat,
2019). The adapter module configuration used in the
paper is based on the one by (Pfeiffer et al., 2020a),
the so-called Pfeiffer architecture. In this configura-
tion, only one adapter module is added as a down-
projection and up-projection, unlike the Houlsby ar-
chitecture, where there are two projections. While
the Houlsby architecture has more learning capac-
ity, it comes with training and inference speed costs.
Previous studies showed no significant difference in
performance between the model architectures, mak-
ing Pfeiffer architecture a very lightweight choice that
brings powerful learning capabilities.
As already mentioned, masked language model-
ing is used to fine-tune the adapter modules. More
precisely, it is a task of entity prediction since a miss-
ing entity from the graph triple is being predicted.
Given a subgraph G
k
and its triples (s,r, o), each triple
has a textual representation. The object entity o is
Diversifying Knowledge Enhancement of Biomedical Language Models Using Adapter Modules and Knowledge Graphs
379

removed from each triple, and the remaining two el-
ements of the triple are transformed into a textual
representation like: ”[CLS] s [SEP] r [SEP]”. The
adapter module is then trained to predict the missing
object entity using the representation of the [CLS] to-
ken. The parameters of the adapter module are opti-
mized by minimizing the cross-entropy loss.
3.4 Knowledge Fusion
Finally, with a set of knowledge-encapsulated adapter
modules at hand, we need to fuse their knowledge to-
gether into a final representation. For this, we use
the so-called AdapterFusion mixture layers (Pfeiffer
et al., 2020a). These layers serve the purpose of com-
bining knowledge from various adapters to enhance
the model’s performance on downstream tasks. It is
a relatively recent approach designed to effectively
learn how to combine information from a set of task-
specific adapters. It does so by employing a softmax
attention mechanism that assigns contextual mixture
weights over the adapters. These weights are then
used to predict task labels in the final layer. The
composition of these layers and their interactions ulti-
mately contribute to the model’s ability to generalize
and perform well on a range of tasks.
4 EXPERIMENTS
In this section, we describe our approach to lever-
aging data from OntoChem’s SciWalker platform to-
gether with adapters to improve existing approaches
to biomedical KELMs. For reproducibility, we
made the code for the experiment runs available on
GitHub.
2
4.1 Datasets
All of our datasets, with the exception of MedNLI,
originated from a collection of common biomedical
NLP tasks known as BLURB – Biomedical Language
Understanding and Reasoning Benchmark.
3
Inspired
by a similar suite of tasks for general-purpose natu-
ral language understanding (NLU) known as GLUE
(Wang et al., 2018), BLURB covers a wide-range of
tasks related to biomedical NLU. This means no tasks
include text generation and are all essentially classifi-
cation tasks, which makes them convenient to evalu-
ate with common classification metrics such as preci-
2
https://github.com/alexander-fichtl/diversifying
KELMs.git
3
https://microsoft.github.io/BLURB/index.html
sion, recall, accuracy, and F1 score. The four datasets
are described in continuation.
MedNLI (Romanov and Shivade, 2018) is a dataset
for natural language inference (NLI). It consists of
14,049 unique sentence pairs, where one sentence is
a hypothesis, and the other one is a premise. The task
is to infer whether the premise entails the hypothesis,
contradicts it, or is in a neutral relation with respect
to it. The premises were collected from MIMIC-III
(Johnson et al., 2016), the largest repository of pub-
licly available clinical data (patient notes).
BioASQ-7b (Nentidis et al., 2020) is a biomedical
question answering (QA) benchmark dataset contain-
ing questions in English, along with golden stan-
dard (reference) answers and related material. It
has been designed to reflect real information needs
of biomedical experts. Other than only exact an-
swers, the BioASQ dataset also includes ideal an-
swers (summaries). Researchers working on para-
phrasing and textual entailment can also measure the
degree to which their methods improve the perfor-
mance of biomedical QA systems. The dataset is a
part of the ongoing shared challenge with the same
name (Tsatsaronis et al., 2015), while our dataset (7b)
is from the 2019 challenge.
PubMedQA (Jin et al., 2019) is a different QA
dataset collected from PubMed abstracts, the largest
collection of biomedical research papers (White,
2020). The task of PubMedQA is to answer research
questions with yes/no/maybe using the corresponding
abstracts. The dataset has 1,000 expert-annotated in-
stances of question-answer pairs. Each PubMedQA
instance is composed of a question, a context (abstract
without the conclusion), a long answer (conclusion of
the abstract), and a yes/no/maybe label that summa-
rizes the conclusion.
The Hallmarks of Cancer (HOC) Corpus (Baker
et al., 2015) consists of 1852 PubMed publication ab-
stracts manually annotated by experts according to a
taxonomy. The taxonomy consists of 37 classes in a
hierarchy. Zero or more class labels are assigned to
each sentence in the corpus. These hallmarks refer
to the alterations in cell behavior that characterize the
cancer cell. Proposed as a strategy to capture the com-
plexity of cancer in a few basic principles, it provides
an organized framework comprising of ten hallmarks
(Baker et al., 2017).
4.2 Knowledge Sources
The Unified Medical Language System (UMLS) is a
set of resources and tools developed by the US Na-
tional Library of Medicine (NLM) to facilitate the in-
tegration and retrieval of biomedical and clinical in-
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
380

Table 1: Twenty most common relations in each of the three KGs used in the experiments.
UMLS20 #Triples Onto20Fused #Triples Onto20Type #Triples
has finding site 367,237 relates to 708,076 [protein] relates to [disease] 295,841
has method 275,398 induces 502,512 [substance] induces [physiology] 282,721
has associated morphology 269729 modulates 326,534 [food] contains [compound] 269,211
has procedure site 129,686 treats 225,279 [substance] treats [disease] 247,348
possibly equivalent to 91,446 inhibits 219,720 [biomarker] of [disease] 205,604
has causative agent 86,780 is analyzed by 195,291 [substance] is analyzed by [method] 130,275
interprets 84,533 produces 173,979 [plant] produces [compound] 102,270
has direct procedure site 83,749 increases activity of 148,673 [protein] induces [physiology] 85,411
has active ingredient 63,792 contains 133,241 [compound] increases activity of [protein] 85,196
has pathological process 54,639 increases 110,803 [compound] decreases activity of [protein] 72,311
has occurrence 40,154 detects 93,373 [substance] inhibits [physiology] 68,728
has dose form 30,940 decreases activity of 85,425 [protein] is a [biomarker] 65,558
has direct morphology 29,667 prevents 82,574 [anatomy] produces [protein] 64,206
has intent 25,907 increases expression of 80,771 [substance] prevents [disease] 60,260
has interpretation 24,624 expresses 62,142 [protein] induces [disease] 59,577
has direct substance 23,042 attenuates 54,865 [substance] modulates [protein] 54,533
has direct device 17,726 decreases expression of 51,152 [protein] is analyzed by [method] 54,250
moved to 17,507 binds to 49,206 [method] treats [disease] 35,768
has temporal context 17,195 is a 47,435 [method] detects [physiology] 33,504
has subject relationship context 16,926 affects expression of 37,399 [protein] modulates [physiology] 24,332
Total 1,750,677 3,388,450 2,296,904
formation from various sources (Bodenreider, 2004).
Created in 1986 and continuously developed over the
decades, it can be viewed as a comprehensive the-
saurus and ontology of biomedical concepts, making
it easier to connect and use medical terminology in
research, clinical practice, and healthcare information
systems. We use the most recent SNOMED CT, US
Edition vocabulary from September 2023.
4
The second knowledge graph, more precisely on-
tology, that we use, is the OntoChem Ontology
(Irmer et al., 2013). The ontology contains more
than 900 complex relationships between two or more
named entities. Entities include chemical compounds,
diseases, drug combinations, chemical reactions, bi-
ological activities, adverse reactions, etc. Relation-
ships can be downloaded as RDF files. The data orig-
inates from MedLine,
5
a bibliographic database from
the US National Library of Medicine’s (NLM), that
contains more than 30 million journal articles focus-
ing on medicine and life sciences. The KG triples can
be interactively queried and also downloaded from the
SciWalker platform with the Fact Finder tool.
6
4.3 KG Subsets
The versions of the KGs from the two knowledge
sources we use in this work are subsets of their re-
spective full KGs – we use versions including only
4
https://www.nlm.nih.gov/healthit/snomedct/
us edition.html
5
https://www.nlm.nih.gov/medline/index.html
6
https://sciwalker.com/analytics/factfinder
the top 20 most common relations. This was done
to increase the efficiency of training but also because
initial experiments showed this smaller version does
not hurt the performance on downstream tasks. For
UMLS, the list of most common relations was taken
from MoP and SNOMED, a systematically organized
collection of medical terms providing codes, terms,
synonyms and definitions used in clinical documenta-
tion and reporting. We label this KG as UMLS20.
The relations provided by OntoChem are unique
to the type of entities that the relation connects, so
there can be several types of the same relation. For
example, the relation ”induces” can have a ”sub-
stance” as a subject and a ”disease” as an object, so
the full relation becomes ”[substance] induces [dis-
ease]”, while another one is with a ”physiology” as
a subject and a ”disease” as an object, producing
”[physiology] induces [disease]”. To test the perfor-
mance between these two types, we produce both a
KG with top 20 fused relations (independent of entity
types) and with top 20 typed relations (dependent on
entity types). We call these two KGs Onto20Fused
and Onto20Type.
The top 20 relations in each of the three KGs is
shown in Table 1. This also gives a good insight into
what kind of structured knowledge is actually con-
tained in these manually curated biomedical knowl-
edge bases. While there are certain overlaps between
top relations UMLS and OntoChem, a lot of them re-
fer to different types of interactions between entities.
Therefore, a promising research avenue that we did
not explore in this work would be to merge these two
knowledge bases into a unified KG and use both to
Diversifying Knowledge Enhancement of Biomedical Language Models Using Adapter Modules and Knowledge Graphs
381

fine-tune the adapters.
4.4 Setup
Task-specific fine-tuning is carried out for the four
chosen benchmark downstream tasks. We aligned
our hyperparameters with the settings recommended
by the BLURB creators (Gu et al., 2020): We de-
ploy the Adam optimizer (Zhang, 2018) alongside
the typical slanted triangular learning rate schedule,
with a warm-up for the initial 10 percent of steps
and a cool-down for the subsequent 90 percent, and
set the dropout probability at 0.1. Furthermore, we
followed (Pfeiffer et al., 2020a) and (Meng et al.,
2021) by introducing mixture layers and AdapterFu-
sion to route valuable knowledge from the adapters to
downstream tasks automatically. Given the random
initialization of the task-specific model and dropout,
outcomes can fluctuate based on different random
seeds, particularly for the small PubMedQA and
BioASQ7b datasets. For a more accurate represen-
tation, we present average results from ten iterations
for BioASQ7b and PubMedQA, five iterations HoC,
and three for MedNLI, as done in related biomedical
NLP papers benchmarking these tasks.
The training was carried out on Google Colab,
with V100 and T4 GPUs provided on the platform.
Specific hyperparameters and settings used in our ex-
periments are shown in Table 2. Run seeds are re-
ported on GitHub.
Table 2: Settings and hyperparameters used for training
each of the datasets of the downstream tasks.
Setting/Task HoC PubMedQA BioASQ7b MedNLI
repeat runs 5 10 10 3
epochs 20 30 25 20
patience 3 4 5 3
batch size 16 4 4 8
learning rate 1e-5 0.5e-5 0.5e-5 0.5e-5
max. seq. len. 128 512 512 256
5 RESULTS
This section describes the detailed experiment results.
We provide both a numerical analysis and a qualita-
tive analysis of the results.
5.1 Numerical Analysis
Table 3 shows the final results of the experiments.
Each section first shows the performance of the
base biomedical model on its own, namely SciB-
ERT (Beltagy et al., 2019), BioBERT (Lee et al.,
2019), PubMedBERT (Gu et al., 2020), and Bi-
oLinkBERT (Yasunaga et al., 2022). Afterwards, in-
dentended rows show the performance of knowledge-
enhanced versions of the models. For SciBERT
and BioBERT, we report on competing approaches
that use structured knowledge integration: MoP
(Meng et al., 2021), DAKI (Lu et al., 2021), and
KEBLM (Lai et al., 2023). For PubMedBERT
and BioLinkBERT, we report on the knowledge-
enhanced versions as described in our paper, aug-
mented with structured knowledge from knowledge
graphs UMLS20, Onto20Fused, and Onto20Type. It
should be noted that the BioLinkBERT results differ
from the ones in the original publication because we
report on averaged experiment results over multiple
runs, unlike the best single run in the original paper.
The results demonstrate that our knowledge en-
hancement approach improved PubMedBERT in six
instances and the BioLinkBERT model in eight in-
stances, either with the UMLS data or the OntoChem
data. Notably, there is a difference in the margin of
improvement between the datasets. For HoC, the im-
provement is either negligible or 1% in the best case.
This shows that the task of trying to classify document
abstracts according to cancer properties is mostly de-
pendent on the document context itself and does not
noticeably benefit from external knowledge. Simi-
lar is the case for MedNLI, which either deteriorates
or improves less than 1%, showing that entailment
recognition is mostly tied to the reasoning capabili-
ties of a language model and not the deeper medical
knowledge.
On the other hand, the two question-answering
datasets experience noticeable improvements. This
makes sense considering the knowledge-intensive na-
ture of QA, where factual knowledge is at its core.
Especially for PubMedQA, both base PLMs get a
7% jump in accuracy with different KGs. An im-
pressive result is the BioLinkBert-base + Onto20Type
model achieving state-of-the-art performance on the
BioASQ7b dataset (when looking at the averaged per-
formance over 10 runs). When looking at the dif-
ference between the two styles of OntoChem rela-
tions, the fused version was superior for PubMedQA
(by 3%), while the more detailed, typed version per-
formed better for BioASQ (by 1.5%). We attribute
this to the slight difference in the domain of these
two datasets – BioASQ contains more questions re-
lating to chemical knowledge, where specific types
could come into play, while PubMedQA covers di-
verse medical diagnoses and treatments.
An interesting result that we have to investigate
further is the relatively worse performance of our ap-
proach with OntoChem KGs on PubMedBERT com-
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
382

Table 3: Final results of the model experiments: The metric for HoC is Micro F1, while for the other three it is accuracy.
The best results for every task are in bold. ”↑” denotes that improvements are observed when compared to the base model.
“†” denotes a statistically significant better result over the base model (T-test, p < 0.05). The results in italic are taken from
previous works, while the rest of results comes from our experiments.
↓ model—dataset → HoC PubMedQA BioASQ7b MedNLI
SciBERT-base 80.52
±0.60
57.38
±4.22
75.93
±4.20
81.19
±0.54
+ MoP 81.79
†
±0.66
↑ 54.66
±3.10
78.50
†
±4.06
↑ 81.20
±0.37
+ KEBLM / 59.0 / 82.14
BioBERT-base 81.41
±0.59
60.24
±2.32
77.50
±2.92
82.42
±0.59
+ MoP 82.53
†
±1.08
↑ 61.04
±4.81
↑ 80.79
†
±4.40
↑ 82.93
±0.55
↑
+ KEBLM / 68.00 ↑ / 84.24 ↑
+ DAKI / / / 83.41 ↑
PubMedBERT-base 82.25
±0.46
55.84
±1.78
87.71
±4.25
84.18
±0.19
+ UMLS20 83.26
†
±0.32
↑ 62.84
†
±2.71
↑ 90.64
†
±2.43
↑ 84.70
±0.19
↑
+ Onto20Type 82.17
±0.62
55.40
±5.57
86.36
±3.07
83.94
±0.63
+ Onto20Fused 82.39
±0.65
↑ 56.12
±2.91
↑ 84.36
±4.73
83.97
±0.59
BioLinkBERT-base 82.21
±0.87
56.76
±3.00
91.29
±3.18
84.1
±0.03
+UMLS20 82.36
±0.57
↑ 63.62
†
±5.31
↑ 91.50
±2.25
↑ 83.78
±0.09
+Onto20Type 82.37
±0.42
↑ 60.46
±5.81
↑ 92.14
±2.30
↑ 82.84
±0.34
+Onto20Fused 82.24
±1.25
↑ 63.28
†
±4.46
↑ 90.57
±3.14
83.69
±0.55
pared to BioLinkBERT, even when factoring in the
stronger base performance of BioLinkBERT. When
the base models don’t match, it is hard to distin-
guish whether performance gains or losses come from
the difference in base models or the difference in the
adapter-based approaches. Here, the base models of
BioLinkBERT generally perform better than those of
PubMedBERT or SciBERT over a variety of tasks.
Therefore, whenever we use BioLinkBERT, we can-
not say how much of the performance gains come
from the superiority of our approach versus the su-
periority of the base model.
5.2 Qualitative Analysis
To investigate the performance of our knowledge-
enhanced models on a deeper level, we decided to
look at the classification performance on an instance
level and singled out some interesting examples. Ta-
ble 4 shows two instances from the BioASQ dataset
where our knowledge-enhanced model predicted the
answer correctly, unlike the base model. Instances in
BioASQ consist of a question and context, and the
goal is to answer the question with a yes/no verdict.
The first row contains a question on the rela-
tionship between Diazepam and traumatic brain in-
jury. While the vanilla BioLinkBERT answered the
question incorrectly, our knowledge-enhanced Bi-
oLinkBERT + Onto20Type model gave the correct
answer. Diazepam (first marketed as Valium) is listed
as an entity in the OntoChem KG, where it has a direct
relation to brain injuries – the full triple is ”diazepam
[substance] treats [disease] brain injury” (see also fig-
ure 1. It is likely that, thanks to the injection of this
knowledge, the enhanced model was able to deduce
the answer, while the base model was not.
The second row shows a question about axitinib
and its relation to pancreatic cancer. Here, the base
version of BioLinkBERT incorrectly predicted that
axitinib does prolong the survival of pancreatic can-
cer patients, while our BioLinkBERT + Onto20Type
model gave the correct negative answer. This time,
there is no relation between axitinib and any form
of cancer listed in the KG. Therefore, our enhanced
model might have been able to rely on its injected
knowledge and deduce that there are no such connec-
tions between the entities in question.
6 CONCLUSION
This paper investigated the performance of biomed-
ical pre-trained language models when enhanced
with structured domain-specific biomedical knowl-
edge. For this purpose, we utilized two biomedi-
cal PLMs (PubMedBERT and BioLinkBERT) and ex-
ternal knowledge from two large KGs, UMLS and
OntoChem. The KGs were partitioned into smaller
subgraphs and later fused into a common knowl-
edge representation. The knowledge was injected into
the PLMs by using lightweight but powerful adapter
modules. We tested the performance on four down-
stream biomedical NLP tasks and showed that the
knowledge-enhanced models consistently improved
Diversifying Knowledge Enhancement of Biomedical Language Models Using Adapter Modules and Knowledge Graphs
383

Table 4: Examples of two instances from the BioASQ dataset (with a question, context, and verdict) where the knowledge-
enhanced model performed correctly, unlike its vanilla counterpart.
Question Context Predictions
Can Diazepam be bene-
ficial in the treatment of
traumatic brain injury?
The present experiment examined the ef-
fects of diazepam, a positive modulator
at the GABA(A) receptor, on survival
and cognitive performance in traumatically
brain-injured animals.
BioLinkBERT:
BLBERT+Onto20Type:
Gold Label:
no
yes
yes
Does axitinib prolong
the survival of pancreatic
cancer patients?
Axitinib/gemcitabine, while tolerated, did
not provide survival benefit over gemc-
itabine alone in patients with advanced
pancreatic cancer from Japan or other re-
gions [...].
BioLinkBERT:
BLBERT+Onto20Type:
Gold Label:
yes
no
no
the results, indicating a clear benefit of infusing exter-
nal structured knowledge into unstructured PLMs. By
updating the adapter weights, which are only about 1–
2% amount of PLM weights, the performance (in best
setting) increased on HoC and MedNLI for 1%, on
BioASQ 3%, and on PubMedQA 7%. This demon-
strates the power of using adapter modules to fine-
tune PLMs for domain-specific purposes. Moreover,
we have demonstrated that OntoChem is a viable al-
ternative to UMLS and other knowledge sources in
the field of biomedical knowledge enhancement.
Future Research. In future work, we would like
to further investigate the potential of the OntoChem
ontology. Besides entities and relations, every data
triple comes with the source sentence from which
it was extracted. Drawing inspiration from works
like K-Adapter (Wang et al., 2020), this linguistic
knowledge could be extracted and used in additional
adapters to enhance the models. Moreover, the idea of
merging together the data from Ontochem with sub-
graphs from MSI (Ruiz et al., 2021), UMLS (Boden-
reider, 2004), or PubChem(Kim et al., 2020) presents
a promising direction. Finally, future work could
be more human-centric and have medical profession-
als curate the KGs. This way, the resulting KELMs
would be tailored directly by those who use them.
Limitations. Our research did not come without
certain challenges and limitations. A portion of the
data from OntoChem was not usable due to incom-
plete ID mappings. As a result, only a fraction of
the available knowledge was integrated into the ex-
perimental segment of this work, which has likely
led to less thoroughly connected KGs. Additionally,
medical professionals often indicate concerns regard-
ing ethical questions and the development and use of
LLMs in bio-medicine. While our methodology and
models will likely not be used in practice without fur-
ther research and improvements, we did not specif-
ically address the medical community’s concerns in
our work. We tried to improve the overall model per-
formance and factual accuracy to reduce hallucina-
tions, but there is no way to entirely eliminate the risk
of wrong predictions and other critical issues. At the
time of writing, we are conducting a survey involv-
ing clinicians to address their concerns in our future
work.
REFERENCES
Abacha, A. B., M’rabet, Y., Zhang, Y., Shivade, C., Lan-
glotz, C., and Demner-Fushman, D. (2021). Overview
of the mediqa 2021 shared task on summarization in the
medical domain. In Proceedings of the 20th Workshop
on Biomedical Language Processing, pages 74–85.
Almazrouei, E., Alobeidli, H., Alshamsi, A., Cappelli, A.,
Cojocaru, R., Debbah, M., Goffinet, E., Heslow, D., Lau-
nay, J., Malartic, Q., Noune, B., Pannier, B., and Penedo,
G. (2023). Falcon-40B: an open large language model
with state-of-the-art performance.
Andreev, K. and R
¨
acke, H. (2004). Balanced graph par-
titioning. In Proceedings of the Sixteenth Annual ACM
Symposium on Parallelism in Algorithms and Architec-
tures, SPAA ’04, page 120–124, New York, NY, USA.
Association for Computing Machinery.
Baker, S., Ali, I., Silins, I., Pyysalo, S., Guo, Y., H
¨
ogberg,
J., Stenius, U., and Korhonen, A. (2017). Cancer Hall-
marks Analytics Tool (CHAT): a text mining approach
to organize and evaluate scientific literature on cancer.
Bioinformatics, 33(24):3973–3981.
Baker, S., Silins, I., Guo, Y., Ali, I., H
¨
ogberg, J., Stenius, U.,
and Korhonen, A. (2015). Automatic semantic classifi-
cation of scientific literature according to the hallmarks
of cancer. Bioinformatics, 32(3):432–440.
Balazevic, I., Allen, C., and Hospedales, T. (2019).
TuckER: Tensor factorization for knowledge graph com-
pletion. In Proceedings of the 2019 Conference on Em-
pirical Methods in Natural Language Processing and the
9th International Joint Conference on Natural Language
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
384

Processing (EMNLP-IJCNLP), pages 5185–5194, Hong
Kong, China. Association for Computational Linguistics.
Bapna, A. and Firat, O. (2019). Simple, scalable adap-
tation for neural machine translation. In Proceedings
of the 2019 Conference on Empirical Methods in Natu-
ral Language Processing and the 9th International Joint
Conference on Natural Language Processing (EMNLP-
IJCNLP), pages 1538–1548, Hong Kong, China. Associ-
ation for Computational Linguistics.
Beltagy, I., Lo, K., and Cohan, A. (2019). Scibert: A pre-
trained language model for scientific text. In Conference
on Empirical Methods in Natural Language Processing.
Bodenreider, O. (2004). The unified medical language sys-
tem (umls): integrating biomedical terminology. Nucleic
acids research, 32(suppl 1):D267–D270.
Bosselut, A., Rashkin, H., Sap, M., Malaviya, C., Celiky-
ilmaz, A., and Choi, Y. (2019). COMET: Common-
sense transformers for automatic knowledge graph con-
struction. In Proceedings of the 57th Annual Meeting
of the Association for Computational Linguistics, pages
4762–4779, Florence, Italy. Association for Computa-
tional Linguistics.
Chronopoulou, A., Peters, M., and Dodge, J. (2022). Effi-
cient hierarchical domain adaptation for pretrained lan-
guage models. In Proceedings of the 2022 Conference of
the North American Chapter of the Association for Com-
putational Linguistics: Human Language Technologies,
pages 1336–1351, Seattle, United States. Association for
Computational Linguistics.
Colon-Hernandez, P., Havasi, C., Alonso, J. B., Huggins,
M., and Breazeal, C. (2021). Combining pre-trained
language models and structured knowledge. ArXiv,
abs/2101.12294.
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2019). BERT: Pre-training of deep bidirectional trans-
formers for language understanding. In Proceedings of
the 2019 Conference of the North American Chapter of
the Association for Computational Linguistics: Human
Language Technologies, Volume 1 (Long and Short Pa-
pers), pages 4171–4186, Minneapolis, Minnesota. Asso-
ciation for Computational Linguistics.
Di Sciullo, A. M. (2018). Knowledge of language and
knowledge science.
Gu, Y., Tinn, R., Cheng, H., Lucas, M. R., Usuyama, N.,
Liu, X., Naumann, T., Gao, J., and Poon, H. (2020).
Domain-specific language model pretraining for biomed-
ical natural language processing. ACM Transactions on
Computing for Healthcare (HEALTH), 3:1 – 23.
He, B., Zhou, D., Xiao, J., Jiang, X., Liu, Q., Yuan, N. J.,
and Xu, T. (2020). BERT-MK: Integrating graph contex-
tualized knowledge into pre-trained language models. In
Findings of the Association for Computational Linguis-
tics: EMNLP 2020, pages 2281–2290, Online. Associa-
tion for Computational Linguistics.
Hoang, S. N., Nguyen, B., Nguyen, N. P., Luu, S. T., Phan,
H. T., and Nguyen, H. D. (2022). Enhanced task-based
knowledge for lexicon-based approach in vietnamese
hate speech detection. In 2022 14th International Con-
ference on Knowledge and Systems Engineering (KSE),
pages 1–5.
Houlsby, N., Giurgiu, A., Jastrzebski, S., Morrone, B.,
De Laroussilhe, Q., Gesmundo, A., Attariyan, M., and
Gelly, S. (2019). Parameter-efficient transfer learning for
nlp. In International Conference on Machine Learning,
pages 2790–2799. PMLR.
Hu, E. J., yelong shen, Wallis, P., Allen-Zhu, Z., Li, Y.,
Wang, S., Wang, L., and Chen, W. (2022). LoRA: Low-
rank adaptation of large language models. In Interna-
tional Conference on Learning Representations.
Hu, L., Liu, Z., Zhao, Z., Hou, L., Nie, L., and Li, J. (2023).
A survey of knowledge enhanced pre-trained language
models. IEEE Transactions on Knowledge and Data En-
gineering.
Irmer, M., Bobach, C., B
¨
ohme, T., P
¨
uschel, A., and Weber,
L. (2013). Using a chemical ontology for detecting and
classifying chemical terms mentioned in texts. Proceed-
ings of Bio-Ontologies 2013.
Ji, S., Pan, S., Cambria, E., Marttinen, P., and Philip, S. Y.
(2021). A survey on knowledge graphs: Representation,
acquisition, and applications. IEEE transactions on neu-
ral networks and learning systems, 33(2):494–514.
Jin, Q., Dhingra, B., Liu, Z., Cohen, W., and Lu, X. (2019).
PubMedQA: A dataset for biomedical research question
answering. In Proceedings of the 2019 Conference on
Empirical Methods in Natural Language Processing and
the 9th International Joint Conference on Natural Lan-
guage Processing (EMNLP-IJCNLP), pages 2567–2577,
Hong Kong, China. Association for Computational Lin-
guistics.
Johnson, A. E., Pollard, T. J., Shen, L., Lehman, L.-w. H.,
Feng, M., Ghassemi, M., Moody, B., Szolovits, P., An-
thony Celi, L., and Mark, R. G. (2016). Mimic-iii, a
freely accessible critical care database. Scientific data,
3(1):1–9.
Karypis, G. and Kumar, V. (1997). Metis: A software
package for partitioning unstructured graphs, partition-
ing meshes, and computing fill-reducing orderings of
sparse matrices.
Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S.,
Li, Q., Shoemaker, B., Thiessen, P., Yu, B., Zaslavsky,
L., Zhang, J., and Bolton, E. (2020). Pubchem in 2021:
New data content and improved web interfaces. Nucleic
Acids Research, 49.
Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He,
S., Li, Q., Shoemaker, B. A., Thiessen, P. A., Yu, B.,
et al. (2019). Pubchem 2019 update: improved access to
chemical data. Nucleic acids research, 47(D1):D1102–
D1109.
Lai, T. M., Zhai, C., and Ji, H. (2023). Keblm: Knowledge-
enhanced biomedical language models. Journal of
Biomedical Informatics, 143:104392.
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., and
Kang, J. (2019). BioBERT: a pre-trained biomedical lan-
guage representation model for biomedical text mining.
Bioinformatics, 36(4):1234–1240.
Lu, Q., Dou, D., and Nguyen, T. H. (2021). Parameter-
efficient domain knowledge integration from multiple
sources for biomedical pre-trained language models. In
Findings of the Association for Computational Linguis-
tics: EMNLP 2021, pages 3855–3865. Association for
Computational Linguistics.
Diversifying Knowledge Enhancement of Biomedical Language Models Using Adapter Modules and Knowledge Graphs
385

Luo, M., Mitra, A., Gokhale, T., and Baral, C. (2022). Im-
proving biomedical information retrieval with neural re-
trievers. In Proceedings of the AAAI Conference on Arti-
ficial Intelligence, volume 36, pages 11038–11046.
Meng, Z., Liu, F., Clark, T., Shareghi, E., and Collier, N.
(2021). Mixture-of-partitions: Infusing large biomedi-
cal knowledge graphs into BERT. In Proceedings of the
2021 Conference on Empirical Methods in Natural Lan-
guage Processing, pages 4672–4681, Online and Punta
Cana, Dominican Republic. Association for Computa-
tional Linguistics.
Nentidis, A., Bougiatiotis, K., Krithara, A., and Paliouras,
G. (2020). Results of the seventh edition of the bioasq
challenge. In Cellier, P. and Driessens, K., editors, Ma-
chine Learning and Knowledge Discovery in Databases,
pages 553–568, Cham. Springer International Publish-
ing.
Peters, M. E., Neumann, M., RobertL.Logan, I., Schwartz,
R., Joshi, V., Singh, S., and Smith, N. A. (2019). Knowl-
edge enhanced contextual word representations. In Con-
ference on Empirical Methods in Natural Language Pro-
cessing.
Pfeiffer, J., Kamath, A., R
¨
uckl
´
e, A., Cho, K., and Gurevych,
I. (2020a). Adapterfusion: Non-destructive task compo-
sition for transfer learning. ArXiv, abs/2005.00247.
Pfeiffer, J., R
¨
uckl
´
e, A., Poth, C., Kamath, A., Vuli
´
c, I.,
Ruder, S., Cho, K., and Gurevych, I. (2020b). Adapter-
Hub: A framework for adapting transformers. In Pro-
ceedings of the 2020 Conference on Empirical Methods
in Natural Language Processing: System Demonstra-
tions, pages 46–54, Online. Association for Computa-
tional Linguistics.
Ravichander, A., Hovy, E., Suleman, K., Trischler, A., and
Cheung, J. C. K. (2020). On the systematicity of prob-
ing contextualized word representations: The case of hy-
pernymy in BERT. In Proceedings of the Ninth Joint
Conference on Lexical and Computational Semantics,
pages 88–102, Barcelona, Spain (Online). Association
for Computational Linguistics.
Rogers, A., Kovaleva, O., and Rumshisky, A. (2021). A
primer in bertology: What we know about how bert
works. Transactions of the Association for Computa-
tional Linguistics, 8:842–866.
Romanov, A. and Shivade, C. (2018). Lessons from natural
language inference in the clinical domain. In Proceed-
ings of the 2018 Conference on Empirical Methods in
Natural Language Processing, pages 1586–1596, Brus-
sels, Belgium. Association for Computational Linguis-
tics.
Ruiz, C., Zitnik, M., and Leskovec, J. (2021). Identification
of disease treatment mechanisms through the multiscale
interactome. Nature Communications, 12:1–15.
Schneider, P., Schopf, T., Vladika, J., Galkin, M., Sim-
perl, E., and Matthes, F. (2022). A decade of knowledge
graphs in natural language processing: A survey. In He,
Y., Ji, H., Li, S., Liu, Y., and Chang, C.-H., editors, Pro-
ceedings of the 2nd Conference of the Asia-Pacific Chap-
ter of the Association for Computational Linguistics and
the 12th International Joint Conference on Natural Lan-
guage Processing (Volume 1: Long Papers), pages 601–
614, Online only. Association for Computational Lin-
guistics.
Speer, R., Chin, J., and Havasi, C. (2017). Conceptnet 5.5:
An open multilingual graph of general knowledge. In
Proceedings of the AAAI conference on artificial intelli-
gence, volume 31.
Sung, M., Jeong, M., Choi, Y., Kim, D., Lee, J., and Kang,
J. (2022). Bern2: an advanced neural biomedical named
entity recognition and normalization tool. Bioinformat-
ics, 38(20):4837–4839.
Tsatsaronis, G., Balikas, G., Malakasiotis, P., Partalas, I.,
Zschunke, M., Alvers, M. R., Weissenborn, D., Krithara,
A., Petridis, S., Polychronopoulos, D., et al. (2015). An
overview of the bioasq large-scale biomedical seman-
tic indexing and question answering competition. BMC
bioinformatics, 16(1):1–28.
Vladika, J. and Matthes, F. (2023). Scientific fact-checking:
A survey of resources and approaches. In Rogers, A.,
Boyd-Graber, J., and Okazaki, N., editors, Findings
of the Association for Computational Linguistics: ACL
2023, pages 6215–6230, Toronto, Canada. Association
for Computational Linguistics.
Vladika, J., Schneider, P., and Matthes, F. (2023). Healthfc:
A dataset of health claims for evidence-based medical
fact-checking.
Wang, A., Singh, A., Michael, J., Hill, F., Levy, O., and
Bowman, S. (2018). GLUE: A multi-task benchmark
and analysis platform for natural language understand-
ing. In Proceedings of the 2018 EMNLP Workshop
BlackboxNLP: Analyzing and Interpreting Neural Net-
works for NLP, pages 353–355, Brussels, Belgium. As-
sociation for Computational Linguistics.
Wang, R., Tang, D., Duan, N., Wei, Z., Huang, X., Ji, J.,
Cao, G., Jiang, D., and Zhou, M. (2020). K-adapter: In-
fusing knowledge into pre-trained models with adapters.
In Findings.
Wang, X., Tsvetkov, Y., Ruder, S., and Neubig, G. (2021).
Efficient test time adapter ensembling for low-resource
language varieties. In Conference on Empirical Methods
in Natural Language Processing.
Wang, Z., Zhang, J., Feng, J., and Chen, Z. (2014). Knowl-
edge graph embedding by translating on hyperplanes. In
Proceedings of the AAAI conference on artificial intelli-
gence, volume 28.
Wei, X., Wang, S., Zhang, D., Bhatia, P., and Arnold, A. O.
(2021). Knowledge enhanced pretrained language mod-
els: A compreshensive survey. ArXiv, abs/2110.08455.
White, J. (2020). Pubmed 2.0. Medical reference services
quarterly, 39(4):382–387.
Yasunaga, M., Leskovec, J., and Liang, P. (2022). Linkbert:
Pretraining language models with document links. In An-
nual Meeting of the Association for Computational Lin-
guistics.
Yuan, Z., Liu, Y., Tan, C., Huang, S., and Huang, F. (2021).
Improving biomedical pretrained language models with
knowledge. In Proceedings of the 20th Workshop on
Biomedical Language Processing, pages 180–190, On-
line. Association for Computational Linguistics.
Zhang, Z. (2018). Improved adam optimizer for deep neural
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
386

networks. In 2018 IEEE/ACM 26th international sympo-
sium on quality of service (IWQoS), pages 1–2. Ieee.
Zheng, D., Ma, C., Wang, M., Zhou, J., Su, Q., Song, X.,
Gan, Q., Zhang, Z., and Karypis, G. (2020). Distdgl:
Distributed graph neural network training for billion-
scale graphs. In 2020 IEEE/ACM 10th Workshop on
Irregular Applications: Architectures and Algorithms
(IA3), pages 36–44, Los Alamitos, CA, USA. IEEE
Computer Society.
Diversifying Knowledge Enhancement of Biomedical Language Models Using Adapter Modules and Knowledge Graphs
387
