
Efficient and Accurate Hyperspectral Image Demosaicing with Neural
Network Architectures
Eric L. Wisotzky
1,2 a
, Lara Wallburg
1
, Anna Hilsmann
1 b
, Peter Eisert
1,2 c
,
Thomas Wittenberg
3,4
and Stephan G
¨
ob
3,4 d
1
Computer Vision & Graphics, Fraunhofer HHI, Einsteinufer 37, 10587 Berlin, Germany
2
Department of Informatics, Humboldt University, Berlin, Germany
3
Fraunhofer IIS, Erlangen, Germany
4
Chair of Visual Computing, Friedrich-Alexander-Universit
¨
at Erlangen-N
¨
urnberg, Erlangen, Germany
Keywords:
Sensor Array and Multichannel Signal Processing, Deep Learning, Biomedical Imaging Techniques, Image
Analysis, Image Upsamling.
Abstract:
Neural network architectures for image demosaicing have been become more and more complex. This results
in long training periods of such deep networks and the size of the networks is huge. These two factors prevent
practical implementation and usage of the networks in real-time platforms, which generally only have limited
resources. This study investigates the effectiveness of neural network architectures in hyperspectral image
demosaicing. We introduce a range of network models and modifications, and compare them with classical
interpolation methods and existing reference network approaches. The aim is to identify robust and efficient
performing network architectures. Our evaluation is conducted on two datasets, ”SimpleData” and ”SimReal-
Data,” representing different degrees of realism in multispectral filter array (MSFA) data. The results indicate
that our networks outperform or match reference models in both datasets demonstrating exceptional perfor-
mance. Notably, our approach focuses on achieving correct spectral reconstruction rather than just visual
appeal, and this emphasis is supported by quantitative and qualitative assessments. Furthermore, our findings
suggest that efficient demosaicing solutions, which require fewer parameters, are essential for practical appli-
cations. This research contributes valuable insights into hyperspectral imaging and its potential applications
in various fields, including medical imaging.
1 INTRODUCTION
The use of multispectral images (MSIs) or hyperspec-
tral images (HSIs), which encompass a wide range
of different spectral channels across various wave-
lengths both within and beyond the visible spectrum,
has gained increasing prominence in recent years.
These types of images find broad applications in var-
ious fields such as healthcare (Calin et al., 2014; Lu
and Fei, 2014; Zhang et al., 2017), industrial appli-
cations (Shafri et al., 2012), and agriculture (Jung
et al., 2006; Moghadam et al., 2017). However, con-
ventional acquisition methods and devices are associ-
ated with significant drawbacks, including high costs
a
https://orcid.org/0000-0001-5731-7058
b
https://orcid.org/0000-0002-2086-0951
c
https://orcid.org/0000-0001-8378-4805
d
https://orcid.org/0000-0002-1206-7478
and lengthy acquisition times (Wisotzky et al., 2018;
M
¨
uhle et al., 2021).
In recent times, alternative approaches have been
developed to address these challenges. One promis-
ing technique is based on the use of spectral mask-
ing at the pixel level, utilizing only a single sensor
plane. This concept is known as Multi-Spectral Fil-
ter Arrays (MSFAs). Unlike RGB images, which are
composed of three color values (red, green and blue),
MSFAs map the spectrum in more than three spec-
tral bands, e.g., nine, 16 or 25 bands (Hershey and
Zhang, 2008). A data cube is formed from the deter-
mined data. Its edges represent the image dimensions
in x- and y-direction and the determined wavelengths
in λ-direction. In contrast to MSIs, HSIs use several
hundred spectral bands to capture the spectrum of a
source.
When image data is acquired by a multispectral
camera using the principle of MSFA, not all image
Wisotzky, E., Wallburg, L., Hilsmann, A., Eisert, P., Wittenberg, T. and Göb, S.
Efficient and Accurate Hyperspectral Image Demosaicing with Neural Network Architectures.
DOI: 10.5220/0012392300003660
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 19th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2024) - Volume 3: VISAPP, pages
541-550
ISBN: 978-989-758-679-8; ISSN: 2184-4321
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
541

information of a data cube can be acquired. An in-
crease in spectral resolution is accompanied by a loss
of spatial resolution. This missing information needs
a successive interpolation using image processing or
image analysis techniques. Common image process-
ing techniques for spectral reconstruction include bi-
linear and nonlinear filters, referred to as debayering
or demosaicing.
While MSFAs offer the advantage of real-time im-
plementation and general applicability, accurate in-
terpolation of missing spectral or spatial information
is challenging. This, however, is crucial for precise
spatial localization of entities such as cancer cells in
healthcare, damage in plants in agriculture, or objects
in industrial applications. To enhance existing meth-
ods, neural networks have recently been proposed as
promising approaches.
Figure 1: Description of the processing pipeline and demo-
saicing of multispectral cameras.
The contribution of this work is as follows. We
present a pipeline to achieve real-world MSFA data
from different hyperspectral datasets. Further, we
propose different new and extended demosaicing net-
works and compare these using classical public avail-
able datasets and a dataset processed with our intro-
duced pipeline.
The analysis is performed from the perspective
that the networks should provide good reconstruction
quality and be able to compute it quickly, i.e., con-
tain as few parameters as possible for potential near-
sensor processing.
The remainder of this paper is as follows. The next
chapter gives an overview on related publications rel-
evant for this work. Chapter 3 describes the proposed
network architectures, before chapter 4 explains data
processing and presents training and evaluation pa-
rameters. Chapter 5 describes experiments and re-
sults, followed by a thorough discussion and conclu-
sion.
2 RELATED WORK
The most commonly used algorithms for demosaicing
in imaging are analytical ones such as nearest neigh-
bor, bilinear interpolation or cubic interpolation. The
multispectral debayering relies on these existing al-
gorithms used in RGB imaging. The basic approach
involves classic interpolation techniques. However,
more complex methods take into account not only the
information from the nearest pixel of the same chan-
nel but also the next of other channels or even fur-
ther away pixels. There are many different variations
of demosaicing algorithms available for RGB imag-
ing (Malvar et al., 2004). However, implementing
these algorithms for multispectral filter structures is
challenging due to the different number of channels.
An approach based on the weighted bilinear interpo-
lation (Brauers and Aach, 2006) is the advancement
of linear interpolation using MSFAs, called Intensity
Difference (ID interpolation) (Mihoubi et al., 2015).
Further, interpolation methods based on image fu-
sion have been proposed (Eismann and Hardie, 2005;
Bendoumi et al., 2014; Zhang, 2014). Fusion based
methods usually require the availability of a guiding
image with higher spatial resolution, which is difficult
to obtain in many scenarios.
Demosaicing by interpolation based techniques,
both traditional as well as fusion-based, is easy to
achieve, however, these methods suffer from color ar-
tifacts and lead to lower spatial resolution. Especially
at edges, they do not take into account the spectral
correlations between adjacent bands as well as due to
crosstalk. This results in spectral distortions in the
demosaiced image, especially for increasing mosaic
filter size.
Alternatively, deep neural networks can be trained
to account for scene information as well as correla-
tions between individual spectral bands.
2.1 Network-Based Demosaicing
Demosaicing using convolutional neural networks
(CNN) was first proposed for images with 2×2 Bayer
pattern (Wang, 2014; Gharbi et al., 2016). In recent
years, CNN based color image super resolution (SR)
has gained popularity. Examples of such networks
include SRCNN (Dong et al., 2014), DCSCN (Ya-
manaka et al., 2017) and EDSR (Lim et al., 2017).
Due to their success, these networks have been ex-
tended to HSI super resolution (Li et al., 2018). The
underlining aspect of all CNN based HSI demosaic-
ing networks is the utilization of spatial and spectral
context from the data during training.
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
542

Nevertheless there is only a small number of pub-
lications where CNNs are used for patterns > 2 × 2.
In the NTIRE 2022 Spectral Demosaicing Challange
several deep learning-based demosaicing algorithms
were introduced (Arad et al., 2022b). The leading
methods all involved very large and complex net-
work structures, such as an enhanced HAN (Niu et al.,
2020), NLRAN and a Res2-Unet (Song et al., 2022)
based method. An interesting approach for the recon-
struction of MSI uses leaner network structures of five
residual blocks (Shinoda et al., 2018). Bilinear inter-
polated data are used as input for this refinement ap-
proach. Furthermore, there are already approaches to
replace the bilinear interpolation by the ID interpo-
lation (G
¨
ob et al., 2021). Dijkstra et al. (2019) pro-
posed a similarity maximization network for HSI de-
mosaicing, inspired by single image SR. This network
learns to reconstruct a downsampled HSI by upscal-
ing via deconvolutional layers. The network results
are presented for a 4 × 4 mosaic pattern and the de-
mosaiced HSI showed high spatial and spectral reso-
lution. Habtegebrial et al. (2019) used residual blocks
to learn the mapping between low and high resolu-
tion HSI, inspired by the HCNN+ architecture (Shi
et al., 2018). These two networks use 2D convolu-
tions in order to learn the spectral-spatial correlations.
An important characteristic of HSI is the correlation
between adjacent spectral bands which are not taken
into account when using 2D convolutional based net-
works. These correlations can be incorporated by us-
ing 3D convolutional networks. Mei et al. (2017) pro-
posed one of the first 3D CNNs for hyperspectral im-
age SR addressing both the spatial context between
neighboring pixels as well as the spectral correlation
between adjacent bands of the image. A more effec-
tive way to learn the spatial and spectral correlations
is through a mixed 2D/3D convolutional network (Li
et al., 2020).
Further, deep learning (DL) have been employed
in order to predict HSI data from MSI or even classic
RGB image data (Arad et al., 2022a, 2020). However,
the main problem here is the lack of training data and
the large dependence of the method on the application
and training data. This means that the spectral behav-
ior of individual scenes is learned by the DL model for
interpolation and thus there is a dependency between
the planned application and training data, which can
lead to poor results and incorrect hyperspectral data if
the training data is improperly selected, of poor qual-
ity, or too small in scope (Wang et al., 2021). Thus,
the need of high quality ground truth data is essential.
In such a dataset, each pixel should contain the entire
spectral information, which is difficult to acquire in a
natural environment.
2.2 Datasets
In order to be able to train and evaluate networks, both
fully completed data cubes and the corresponding raw
data are required. For this purpose, a number of dif-
ferent databases containing are available. One major
challenge for the task of snapshot mosaic HSI demo-
saicing using neural networks is the lack of real world
ground truth data.
The only databases available are those that use
either the pushbroom method or spatio-spectral-line
scans, usually realized by a liquid crystal. Hence, the
data has different characteristics than snapshot mosaic
data (e.g., missing cross-talk) and therefore a trained
network cannot adequately represent reality. How-
ever, we are not aware of any data sets that include
MSFAs. All HSI databases used in this work are pre-
sented in Tab. 1 and include a large number of differ-
ent colored objects.
One alternative to include real MSFA data into
training is to downsample captured snapshot mosaic
data as presented in Dijkstra et al. (2019). How-
ever, simple downsampling leads to differences in dis-
tances of adjacent pixels, which affects the network
results in an unknown manner. Therefore, this ap-
proach is not been followed in this work.
3 NETWORK ARCHITECTURE
In the following, three different architecture types are
presented. All are based on different works recently
published and have been modified in order to achieve
improved demosaicing results.
3.1 ResNet-Based Architecture
In the initial phase, we aimed to enhance the network
proposed by Shinoda et al. (2018). The modifications
can be classified into two components: the prepro-
cessing of input data and the CNN itself. Due to its
superior effectiveness, we use ID interpolation as in-
put of the network.
The alterations made to the CNN were intended
to reduce the number of parameters without compro-
mising the quality and are visualized in Fig. 2. Ini-
tially, the network comprised of five residual blocks.
The first two blocks contained two conv-blocks in the
main-path and one conv-block in the skip-connection,
while the next three residual blocks consisted of three
conv-blocks in the main-path and one conv-block
in the skip-connection. The number of filters in-
creased from the first to the fifth residual block with
8/16/32/64/64, and were formed by a 3D kernel-size
Efficient and Accurate Hyperspectral Image Demosaicing with Neural Network Architectures
543
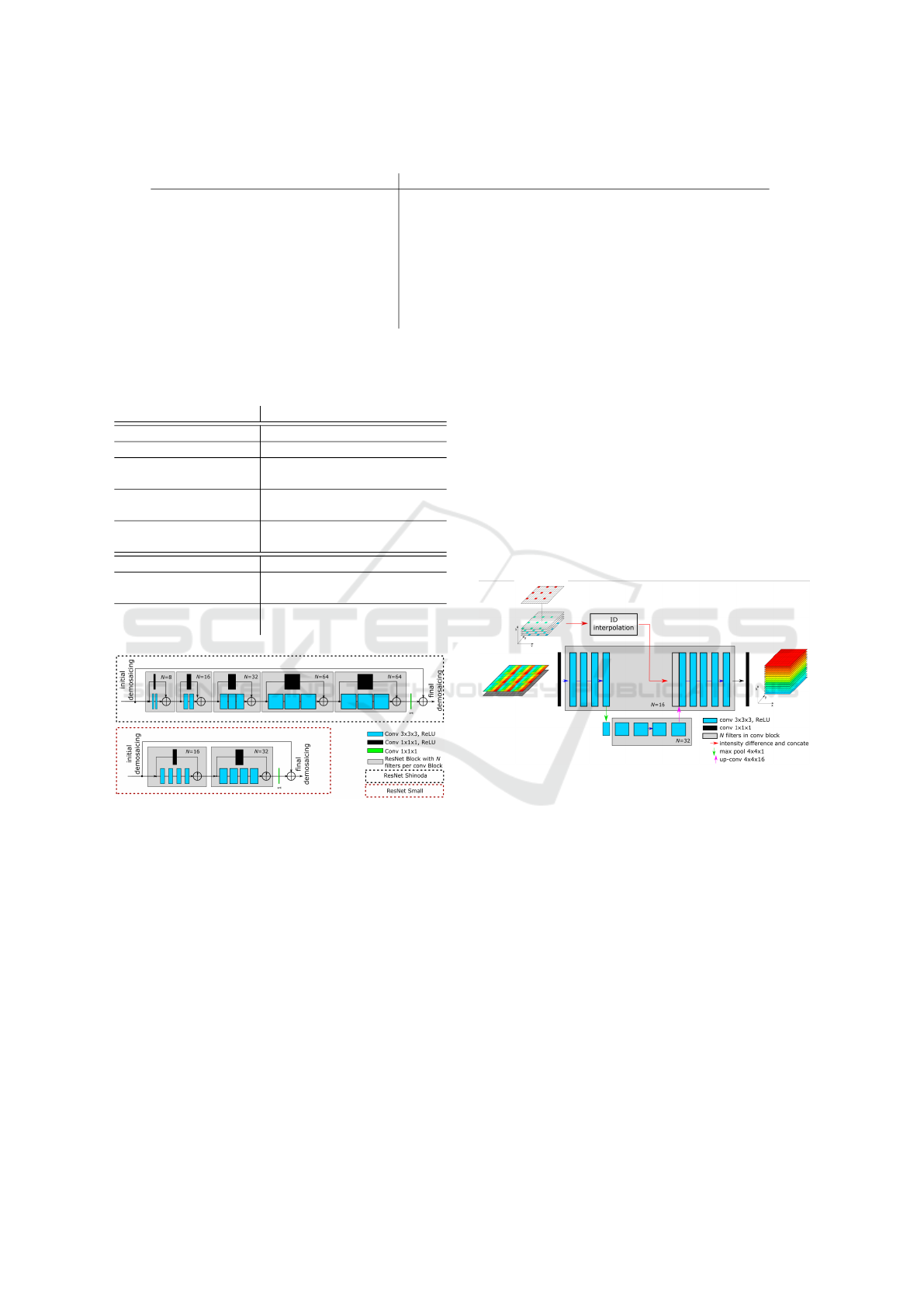
Table 1: The used hyperspectral data collections.
Dataset # Images Size [px] Spectrum [nm] Bands Range
CAVE (Yasuma et al., 2010) 32 512x512 400-700 31 1:65536
HyTexiLa (Khan et al., 2018) 112 1024x1024 405-996 186 0:1
TokyoTech31 (Monno et al., 2015) 30 500x500 420-720 31 0:1
TokyoTech59 (Monno et al., 2018) 16 512x512 420-1000 59 0:1
SIDRI-v10 (Mirhashemi, 2019) 5 640x480 400-1000 121 0:1
SIDRI-vis (Mirhashemi, 2019) 1 640x480 400-720 31 0:1
ODSI-DB Nuance (Hyttinen et al., 2020) 139 1392x1040 450-950 51 1:65536
ODSI-DB Specim (Hyttinen et al., 2020) 171 Various 400-1000 204 1:65536
Google 670 480x1312 400-1000 396 1:4096
Table 2: The number of parameters and sizes of the input
images of the different networks. If two input sizes are
stated, the network structure needs different input.
Network Parameter Input Size
ID-ResNet-L 697k [16 × 100 × 100]
ID-ResNet-S 118k [16 × 100 × 100]
ID-UNet 128k
[16 × 100 × 100]
[1 × 100 × 100]
Parallel-S 331k
[16 × 100 × 100]
[1 × 100 × 100]
Parallel-L 382k
[16 × 25 × 25]
[1 × 100 × 100]
UNet 227k [1 × 100 × 100]
ResNet
697k [16 × 100 × 100]
(Shinoda et al., 2018)
Parallel
281k
[16 × 25 × 25]
(Wisotzky et al., 2022) [1 × 100 × 100]
Figure 2: The ResNet-based architectures. Top: ResNet
according to Shinoda et al. (2018). Bottom: our modified
version.
of 3 × 3 × 3. Since our data input shows improved
quality, we optimize the architecture by reducing the
number of residual blocks to two. To achieve the de-
sired quality of the CNN, we modified the number of
conv-blocks to four in the main-path and one in the
skip-connection, with 16 filters in the first residual
block and 32 in the second. These modifications re-
duced the overall number of parameters presented in
Tab. 2.
3.2 U-Net-Based Architecture
Originally introduced for medical image segmenta-
tion, the U-Net is a classical approach to reconstruct
images (Ronneberger et al., 2015). Specifically, the
downsampling path of the U-Net captures the con-
text of the image, while the upsampling path performs
the complete reconstruction. To transfer information
from the downsampling path to the upsampling path,
skip connections are used. In this work, we modified
the skip connections to insert external information for
reconstruction improvements. We utilize a fully re-
constructed image obtained by classical demosaicing,
i.e., ID interpolation, which is then inserted into the
upsampling path via the skip connection. A detailed
schematic of this approach is provided in Fig. 3.
Figure 3: The U-Net-based architecture. A small U-Net
structure is used, while instead of the skip connection, we
insert the results from ID interpolation in the network.
The input to the U-Net is the 2D MSFA. To en-
sure a small, simple, and effective network architec-
ture, we limit the downsampling layer to a max-pool
4 ×4× 1 operation and reconstruct the image in its 16
spectral regions using a 4 × 4 × 16 kernel in the up-
sampling path. The conv-blocks in the network con-
sist of 16 filters in the upper layers and 32 filters in the
lower layers and are provided with 3 × 3 × 3 kernels.
3.3 Parallel Architecture
As third, we use a CNN architecture with paral-
lel building blocks to reconstruct the correct spatial-
spectral distribution in the image. We elaborated
two general types based on literature (Dijkstra et al.,
2019; Shinoda et al., 2018; Habtegebrial et al., 2019;
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
544

Wisotzky et al., 2022). First, two parallel feature
extracting layers using a mosaic to cube converter
(M2C) on one side and ResNet blocks on the other
side are used followed by a feature adding and two
deconvolution (deconv) layers to upsample the spatial
dimensions of the image. The second implementation
combines two effective approaches introduced by Di-
jkstra et al. (2019) and Shinoda et al. (2018)/Habtege-
brial et al. (2019), which are added and refined to
form the demosaiced output. Both implementations
use 3D kernels and are presented in Fig. 4.
Figure 4: The parallel CNN architectures. Top: network
Parallel-L is an extension of Wisotzky et al. (2022) using
the small ResNet-structure (cf. Fig. 2). Bottom: network
Parallel-S is a combination of two effective approaches.
4 DATA AND TRAINING
During training, we used image patches of size [1 ×
100 × 100] (representing the mosaic sensor output) as
input for the network. The output and ground truth
data had a size of [16 × 100 × 100]. Depending on
the model, the input is either left unchanged or was
transformed in the following way:
• Transformation to a sparse 3D cube where empty
values are filled with mean; size: [16 ×100× 100]
• Transformation to a 3D cube with low resolution
and smaller spatial size; size: [16 × 25 × 25]
• Transformation to a 3D cube by ID interpolation;
size: [16 × 100 × 100].
On each side of the images, we removed four pixels
resulting in a shape of 92×92 px, because ID interpo-
lation distorts those outermost pixels making it more
difficult for the models to predict the correct interpo-
lation.
In order to train and validate the networks pro-
posed in this work, a large HSI data set was built
from different data collections introduced earlier. To
allow comparability between the individual data col-
lections, the value range of all HSI data are normal-
ized to [0, 1]. The data set was split into 75% training,
15% validation and 10% test data. As described, the
data does not represent real snapshot mosaic behavior
including cross-talk and characteristic filter responses
for each mosaic pixel. Therefore, an MSFA had to be
simulated. First, we just selected 16 wavelengths in
the range of 450 − 630 nm, interpolated these bands
from the HSI data and built an MSFA as widely been
done in literature (Arad et al., 2020, 2022a; G
¨
ob et al.,
2021; Habtegebrial et al., 2019). We refer to this data
set as SimpleData.
In addition, we built a data set better represent-
ing real captured MSFAs by transforming the stated
reflectances of the data r(λ) into real MSI snapshot
camera measurements r
b
at band b according to
r
b
=
R
λ
max
λ
min
T (λ)I(λ) f
b
(λ)r(λ)dλ
R
λ
max
λ
min
T (λ)I(λ) f
b
(λ)dλ
, (1)
where T is the optical transmission profile of the op-
tical components of our used hardware setup, f
b
char-
acterize the optical filter responses of each of the b
spectral bands of the used camera (M
¨
uhle et al., 2021;
Wisotzky et al., 2020), and I is the relative irradiance
of the light source. All profiles are well known and
allow to transform HSI data into real MSI snapshot
camera outputs. We refer to this data set as SimReal-
Data.
For training, we used the ADAM optimizer with
an adaptive learning rate strategy and an initial learn-
ing rate of 0.0002. At every 10th epoch, the learn-
ing rate is reduced by the factor 0.9. We trained each
model for 100 epochs. The batch size was 20. The
loss function for calculating the difference between
the ground truth and the predicted full-spectrum hy-
perspectral cube is defined by the mean squared error
(MSE)
MSE(o, p) =
1
N
N
∑
i=0
|o
i
− p
i
|
2
, (2)
where o is the ground truth and p is the predicted
value. To evaluate model performance, we calculated
the structural similarity index (SSIM), peak signal-to-
noise ratio (PSNR) and spectral angle mapper (SAM).
5 RESULTS AND DISCUSSION
In the following, we present the evaluation results
of the predictions of our presented network archi-
tectures. For this purpose, they are examined and
compared in a quantitative and qualitative manner.
Efficient and Accurate Hyperspectral Image Demosaicing with Neural Network Architectures
545

We compare our proposed networks with classical
interpolation approaches, bilinear and ID interpola-
tion, as well as with different reference network ap-
proaches introduced in Sec. 2: ResNet (Shinoda et al.,
2018), HSUp (Dijkstra et al., 2019), U-Net and Paral-
lel (Wisotzky et al., 2022).
Further, we analyze the resulting images visually
and, to show the usability of our work, we visually an-
alyze intraoperative snapshot images acquired during
a surgery of parotidectomy.
5.1 Quantitative Results
All networks, our proposed networks as well as the
different references, were trained on the two cre-
ated datasets SimpleData and SimRealData. For both
dataset modifications, the networks learned to predict
a full spectral cube of dimension [16 × 100 × 100]
from the given input images. Our networks outper-
formed or were in the same range as the reference
networks for both datasets.
5.1.1 SimpleData
For the SimpleData without cross-talk all modified
and newly introduced networks perform better that
the state-of-the-art methods, see Tab. 3. The large
modified ResNet-based model using ID interpolation
as input performed significantly best. The larger the
network in terms of trainable parameter the better the
results for all three evaluation measures, but also the
slower the network performance. In comparison with
the reference ResNet model (Shinoda et al., 2018),
the quality of the input image, i.e., the quality of the
initial demosaicing, is of high relevance. Using bet-
ter input quality (ID instead of bilinear interpolation),
the ResNet is able to achieve an relatively higher in-
creases in reconstruction accuracy. It can be assumed
that with even better initial demosaiced data by us-
ing network-based demosaicing methods, the perfor-
Table 3: Demosaicing results on SimpleData patches. Best
result is bold, second best is bold-italic and third rank is
italic.
Network SSIM PSNR [dB] SAM
ID-ResNet-L 0.9891 52.3846 3.48e-02
ID-ResNet-S 0.9856 51.1377 3.90e-02
ID-UNet 0.9868 51.3331 3.85e-02
Parallel-S 0.9865 51.4654 3.78e-02
Parallel-L 0.9857 51.1386 3.93e-02
UNet 0.9839 50.5408 4.25e-02
ResNet 0.9792 48.9982 4.07e-02
Parallel 0.9863 51.1316 3.96e-02
HSUp 0.9846 50.8835 4.05e-02
Bilinear 0.9235 37.5282 5.90e-02
ID 0.9671 39.7182 5.27e-02
mance is further improved. However, this would also
increase the number of parameters and thus reduce
performance.
The network Parallel-S follows the ID-ResNet-L
model in reconstruction accuracy. It performed as
second best in PSNR and SAM metrics and third in
SSIM. The performance order is switched with the
ID-UNet, which is third in PSNR and SAM, and sec-
ond in SSIM. The Parallel-L network, which includes
the smaller ID-ResNet-S, shows an improvement in
comparision to ID-ResNet-S. Both methods are fourth
and fifth in terms of the analyzed metrics. Thus, all
proposed methods and modifided networks perform-
ing better than the compared modalities from litera-
ture. This also shows that it is possible to greatly
reduce the network complexity (by up to six times:
118k network parameters for ID-ResNet-S instead
of 697k parameters for ID-ResNet-L) with only mi-
nor loss of quality compared to the best performing
model.
Interestingly, also with respect to other recent
work (Arad et al., 2022b), it appears that the complex-
ity of the models, i.e., the number of model paramter,
has a great impact on the quality of the demosaicing
results on simple, rather unrealistic, data sets. Be-
cause after all, the best results in this study are also
achieved with the most complex models.
5.1.2 SimRealData
For the SimRealData, which much better represent
real captured data, the results of all networks, pro-
posed as well as reference networks, are very much
improved in comparison to SimpleData, see Tab. 4.
Further, the difference between the reconstruction re-
sults of all networks is reduced. Again the largest net-
work ID-ResNet-L performed best, but closely fol-
lowed by the parallel networks. Interestingly, the
state-of-the-art parallel model is performing second
best, closely followed by Parallel-S. The other net-
works, expect for the standard U-Net, are following
closely. Thus, networks with only 40% or 50% of
the parameters (Parallel or Parallel-S, respectively)
Table 4: Demosaicing results on SimRealData. Best result
is bold, second best is bold-italic and third rank is italic.
Network SSIM PSNR SAM
ID-ResNet-L 0.9989 62.0690 1.29e-02
ID-ResNet-S 0.9984 60.6000 1.55e-02
ID-UNet 0.9985 60.7327 1.58e-02
Parallel-S 0.9983 60.9021 1.44e-02
Parallel-L 0.9982 60.5968 1.52e-02
UNet 0.9973 58.3865 2.28e-02
Parallel 0.9986 61.2197 1.41e-02
HSUp 0.9983 60.6398 1.50e-02
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
546
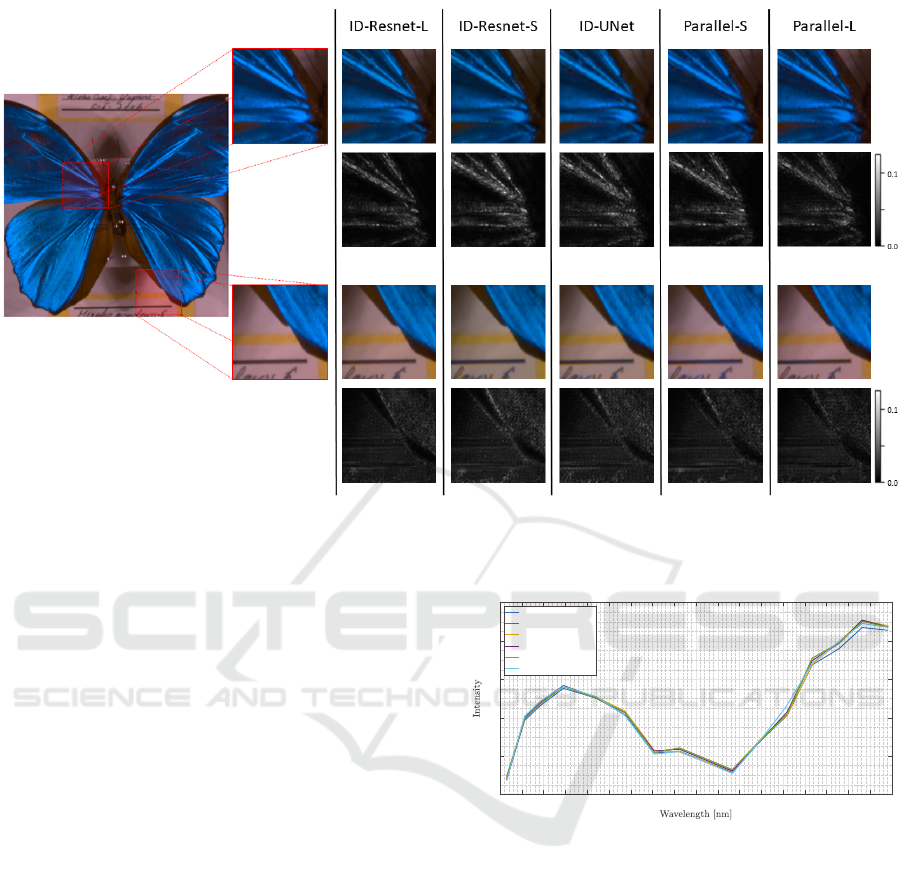
Figure 5: Visual results and error images. Spectral data are represented in RGB and error images of two region of interest
(ROI) are build using l
1
-Norm. The maximum errors in the top ROI are 0.0598, 0.0994, 0.1001, 0.0805, 0.0667 and in the
bottom ROI are 0.0401, 0.0597, 0.0614, 0.0520, 0.0366 in order of appearance of the models from left to right.
achieve similarly good reconstruction results.
Obviously, the complexity of the networks is not
decisive for the reconstruction results, but rather the
quality of the input data and the general structure.
The networks can draw information about all spectral
bands from the complex spectral behavior of the in-
dividual pixels on an MSFA. The effect of cross-talk
contains essential data, which are useful to all net-
works for a more precise spectral interpolation. This
allows using simpler network structures while main-
taining similar high reconstruction quality.
5.2 Qualitative Results
An analysis of the individual spectral channels did not
reveal any discernible deviations in the quality char-
acteristics between the spectral channels. Thus, no
channel stands out as particularly defective during the
demosaicing process. Therefore, the qualitative anal-
ysis of the results is made on RGB-calculated images.
As can be seen in Fig. 5, all reconstruction results
appear in similar quality at first glance. On closer in-
spection, minor differences are evident at strong im-
age edges, e.g., at the bottom right wing. These small
differences can be quantitatively represented in a dif-
ference image (Fig. 5) or in a spectral plot, Fig. 6.
In addition, we have demosaiced intraoperative
images, see Fig. 7. In terms of quality, these im-
460 470 480 490 500 510 520 530 540 550 560 570 580 590 600 610 620 630 640
0
0.2
0.4
0.6
0.8
1
Ground Truth Data
Parallel-S
Parallel-L
ID-ResNet-L
ID-ResNet-S
Unet
Figure 6: Spectral plot of a central left point in the left top
wing of the butterfly in Fig. 5. The SAM of the spectra in
comparison to the ground truth are 0.0133, 0.0157, 0.0158,
0.0265, 0.0276 for the models ID-Resnet-L, Parallel-S,
Parallel-L, ID-UNet and ID-Resnet-S, respectively.
ages show a high resolution as small details as well
as edges are clearly visible. It is also noticeable that
artifacts such as color fringes are reduced.
6 CONCLUSION
In conclusion, our results clearly show that signif-
icant improvements in demosaicing algorithms can
be achieved by using well-designed neural network
architectures. The networks and modifications we
have introduced feature excellent reconstruction of
the ground truth data while reducing or at least hold-
Efficient and Accurate Hyperspectral Image Demosaicing with Neural Network Architectures
547
Figure 7: Demosaiced medical image showing qualitatively
high image quality.
ing the model parameters constant. The results were
tested both quantitatively and qualitatively, showing
convincing results over traditional as well as CNN-
based demosaicing methods.
Due to comparatively fewer network parame-
ter, our networks result in more efficient computa-
tion proving the capability for real-time application,
e.g., for intraoperative hyperspectral image applica-
tion. Especially in comparison to current work (Arad
et al., 2022b), which uses very complex networks and
achieves similar results, this is a gain. Importantly,
our focus is on correct spectral reconstruction rather
than visual attractiveness, which is of high importance
for the mentioned applications and supported by the
quantitative results confirmed by our qualitative eval-
uation.
Moreover, the differences between the results of
different datasets, in agreement with the existing liter-
ature Dijkstra et al. (2019), provide valuable insights
for the demosaicing of real camera data. This un-
derscores the importance of developing demosaicing
solutions that train on data being as close as possi-
ble to real MSFA data. Our results demonstrated that
the use of synthetic representatives of real MSFA data
are suitable for training and networks trained on these
data perform well despite fewer training parameters,
thereby enabling fast processing. This pursuit of ef-
ficient solutions is critical for practical applications
in various fields, including medical imaging and re-
mote sensing, allowing to integrate compact acquisi-
tion concepts like snapshot mosaic imaging into such
processes.
ACKNOWLEDGMENT
This work was funded by the German Federal
Ministry for Economic Affairs and Climate Action
(BMWK) under Grant No. 01MK21003 (NaLamKI).
Only tissue that has been exposed during normal sur-
gical treatment has been scanned additionally with
our described camera. This procedure has been ap-
proved by Charit
´
e–Universit
¨
atsmedizin Berlin, Ger-
many.
REFERENCES
Arad, B., Timofte, R., Ben-Shahar, O., Lin, Y.-T., and Fin-
layson, G. D. (2020). Ntire 2020 challenge on spectral
reconstruction from an rgb image. In Proceedings of
the IEEE/CVF Conference on Computer Vision and
Pattern Recognition (CVPR) Workshops.
Arad, B., Timofte, R., Yahel, R., Morag, N., Bernat, A.,
Cai, Y., Lin, J., Lin, Z., Wang, H., Zhang, Y., Pfis-
ter, H., Van Gool, L., Liu, S., Li, Y., Feng, C., Lei,
L., Li, J., Du, S., Wu, C., Leng, Y., Song, R., Zhang,
M., Song, C., Zhao, S., Lang, Z., Wei, W., Zhang, L.,
Dian, R., Shan, T., Guo, A., Feng, C., Liu, J., Agarla,
M., Bianco, S., Buzzelli, M., Celona, L., Schettini,
R., He, J., Xiao, Y., Xiao, J., Yuan, Q., Li, J., Zhang,
L., Kwon, T., Ryu, D., Bae, H., Yang, H.-H., Chang,
H.-E., Huang, Z.-K., Chen, W.-T., Kuo, S.-Y., Chen,
J., Li, H., Liu, S., Sabarinathan, S., Uma, K., Bama,
B. S., and Roomi, S. M. M. (2022a). Ntire 2022
spectral recovery challenge and data set. In 2022
IEEE/CVF Conference on Computer Vision and Pat-
tern Recognition Workshops (CVPRW), pages 862–
880.
Arad, B., Timofte, R., Yahel, R., Morag, N., Bernat, A.,
Wu, Y., Wu, X., Fan, Z., Xia, C., Zhang, F., Liu, S.,
Li, Y., Feng, C., Lei, L., Zhang, M., Feng, K., Zhang,
X., Yao, J., Zhao, Y., Ma, S., He, F., Dong, Y., Yu,
S., Qiu, D., Liu, J., Bi, M., Song, B., Sun, W., Zheng,
J., Zhao, B., Cao, Y., Yang, J., Cao, Y., Kong, X., Yu,
J., Xue, Y., and Xie, Z. (2022b). Ntire 2022 spectral
demosaicing challenge and data set. In Proceedings
of the IEEE/CVF Conference on Computer Vision and
Pattern Recognition, pages 882–896.
Bendoumi, M. A., He, M., and Mei, S. (2014). Hyper-
spectral image resolution enhancement using high-
resolution multispectral image based on spectral un-
mixing. IEEE Transactions on Geoscience and Re-
mote Sensing, 52(10):6574–6583.
Brauers, J. and Aach, T. (2006). A color filter array based
multispectral camera. In 12. Workshop Farbbildverar-
beitung, pages 5–6. Ilmenau.
Calin, M. A., Parasca, S. V., Savastru, D., and Manea, D.
(2014). Hyperspectral imaging in the medical field:
present and future. Applied Spectroscopy Reviews,
49(6):435–447.
Dijkstra, K., van de Loosdrecht, J., Schomaker, L., and
Wiering, M. A. (2019). Hyperspectral demosaicking
and crosstalk correction using deep learning. Machine
Vision and Applications, 30(1):1–21.
Dong, C., Loy, C. C., He, K., and Tang, X. (2014). Learn-
ing a deep convolutional network for image super-
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
548

resolution. In European conference on computer vi-
sion, pages 184–199. Springer.
Eismann, M. T. and Hardie, R. C. (2005). Hyperspectral
resolution enhancement using high-resolution multi-
spectral imagery with arbitrary response functions.
IEEE Transactions on Geoscience and Remote Sens-
ing, 43(3):455–465.
Gharbi, M., Chaurasia, G., Paris, S., and Durand, F. (2016).
Deep joint demosaicking and denoising. ACM Trans-
actions on Graphics (TOG), 35(6):1–12.
G
¨
ob, S., G
¨
otz, T. I., and Wittenberg, T. (2021). Multispec-
tral single chip reconstruction using dnns with appli-
cation to open neurosurgery. Current Directions in
Biomedical Engineering, 7(2):37–40.
Habtegebrial, T. A., Reis, G., and Stricker, D. (2019). Deep
convolutional networks for snapshot hypercpectral de-
mosaicking. In 2019 10th Workshop on Hyperspectral
Imaging and Signal Processing: Evolution in Remote
Sensing (WHISPERS), pages 1–5. IEEE.
Hershey, J. and Zhang, Z. (2008). Multispectral digital
camera employing both visible light and non-visible
light sensing on a single image sensor. US Patent
7,460,160.
Hyttinen, J., F
¨
alt, P., J
¨
asberg, H., Kullaa, A., and Hauta-
Kasari, M. (2020). Oral and dental spectral image
database—odsi-db. Applied Sciences, 10(20):7246.
Jung, A., Kardev
´
an, P., and T
˝
okei, L. (2006). Hyperspectral
technology in vegetation analysis. Progress in Agri-
cultural Engineering Sciences, 2(1):95–117.
Khan, H. A., Mihoubi, S., Mathon, B., Thomas, J.-B., and
Hardeberg, J. Y. (2018). Hytexila: High resolution
visible and near infrared hyperspectral texture images.
Sensors, 18(7):2045.
Li, Q., Wang, Q., and Li, X. (2020). Mixed 2d/3d con-
volutional network for hyperspectral image super-
resolution. Remote Sensing, 12(10):1660.
Li, Y., Zhang, L., Dingl, C., Wei, W., and Zhang, Y.
(2018). Single hyperspectral image super-resolution
with grouped deep recursive residual network. In 2018
IEEE Fourth International Conference on Multimedia
Big Data (BigMM), pages 1–4. IEEE.
Lim, B., Son, S., Kim, H., Nah, S., and Mu Lee, K. (2017).
Enhanced deep residual networks for single image
super-resolution. In Proceedings of the IEEE con-
ference on computer vision and pattern recognition
workshops, pages 136–144.
Lu, G. and Fei, B. (2014). Medical hyperspectral imaging:
a review. Journal of Biomedical Optics, 19(1):10901.
Malvar, H. S., He, L.-w., and Cutler, R. (2004). High-
quality linear interpolation for demosaicing of bayer-
patterned color images. In 2004 IEEE International
Conference on Acoustics, Speech, and Signal Process-
ing, volume 3, pages iii–485. IEEE.
Mei, S., Yuan, X., Ji, J., Zhang, Y., Wan, S., and Du, Q.
(2017). Hyperspectral image spatial super-resolution
via 3d full convolutional neural network. Remote
Sensing, 9(11):1139.
Mihoubi, S., Losson, O., Mathon, B., and Macaire, L.
(2015). Multispectral demosaicing using intensity-
based spectral correlation. In International Confer-
ence on Image Processing Theory, Tools and Applica-
tions (IPTA), volume 5, pages 461–466. IEEE.
Mirhashemi, A. (2019). Configuration and registration of
multi-camera spectral image database of icon paint-
ings. Computation, 7(3):47.
Moghadam, P., Ward, D., Goan, E., Jayawardena, S., Sikka,
P., and Hernandez, E. (2017). Plant disease detection
using hyperspectral imaging. In 2017 International
Conference on Digital Image Computing: Techniques
and Applications (DICTA), pages 1–8. IEEE.
Monno, Y., Kikuchi, S., Tanaka, M., and Okutomi, M.
(2015). A practical one-shot multispectral imaging
system using a single image sensor. IEEE Transac-
tions on Image Processing, 24(10):3048–3059.
Monno, Y., Teranaka, H., Yoshizaki, K., Tanaka, M., and
Okutomi, M. (2018). Single-sensor rgb-nir imaging:
High-quality system design and prototype implemen-
tation. IEEE Sensors Journal, 19(2):497–507.
M
¨
uhle, R., Markgraf, W., Hilsmann, A., Malberg, H., Eis-
ert, P., and Wisotzky, E. L. (2021). Comparison
of different spectral cameras for image-guided or-
gan transplantation. Journal of Biomedical Optics,
26(7):076007.
Niu, B., Wen, W., Ren, W., Zhang, X., Yang, L., Wang,
S., Zhang, K., Cao, X., and Shen, H. (2020). Sin-
gle image super-resolution via a holistic attention net-
work. In Computer Vision–ECCV 2020: 16th Euro-
pean Conference, Glasgow, UK, August 23–28, 2020,
Proceedings, Part XII 16, pages 191–207. Springer.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-
net: Convolutional networks for biomedical image
segmentation. In Medical Image Computing and
Computer-Assisted Intervention–MICCAI 2015: 18th
International Conference, Munich, Germany, October
5-9, 2015, Proceedings, Part III 18, pages 234–241.
Springer.
Shafri, H. Z., Taherzadeh, E., Mansor, S., and Ashurov, R.
(2012). Hyperspectral remote sensing of urban areas:
an overview of techniques and applications. Research
Journal of Applied Sciences, Engineering and Tech-
nology, 4(11):1557–1565.
Shi, Z., Chen, C., Xiong, Z., Liu, D., and Wu, F. (2018).
Hscnn+: Advanced cnn-based hyperspectral recovery
from rgb images. In Proceedings of the IEEE Con-
ference on Computer Vision and Pattern Recognition
Workshops, pages 939–947.
Shinoda, K., Yoshiba, S., and Hasegawa, M. (2018). Deep
demosaicking for multispectral filter arrays. arXiv
preprint arXiv:1808.08021.
Song, B., Ma, S., He, F., and Sun, W. (2022). Hyperspec-
tral reconstruction from rgb images based on res2-unet
deep learning network. Opt Precis Eng, 30(13):1606.
Wang, Y., Cao, R., Guan, Y., Liu, T., and Yu, Z. (2021).
A deep survey in the applications of demosaick-
ing. In 2021 3rd International Academic Exchange
Conference on Science and Technology Innovation
(IAECST), pages 596–602. IEEE.
Wang, Y.-Q. (2014). A multilayer neural network for image
demosaicking. In 2014 IEEE International Confer-
Efficient and Accurate Hyperspectral Image Demosaicing with Neural Network Architectures
549

ence on Image Processing (ICIP), pages 1852–1856.
IEEE.
Wisotzky, E. L., Daudkane, C., Hilsmann, A., and Eisert, P.
(2022). Hyperspectral demosaicing of snapshot cam-
era images using deep learning. In DAGM German
Conference on Pattern Recognition, pages 198–212.
Springer.
Wisotzky, E. L., Kossack, B., Uecker, F. C., Arens, P., Hils-
mann, A., and Eisert, P. (2020). Validation of two
techniques for intraoperative hyperspectral human tis-
sue determination. Journal of Medical Imaging, 7(6).
Wisotzky, E. L., Uecker, F. C., Arens, P., Dommerich, S.,
Hilsmann, A., and Eisert, P. (2018). Intraoperative hy-
perspectral determination of human tissue properties.
Journal of Biomedical Optics, 23(9):1–8.
Yamanaka, J., Kuwashima, S., and Kurita, T. (2017). Fast
and accurate image super resolution by deep cnn with
skip connection and network in network. In Interna-
tional Conference on Neural Information Processing,
pages 217–225. Springer.
Yasuma, F., Mitsunaga, T., Iso, D., and Nayar, S. K. (2010).
Generalized assorted pixel camera: postcapture con-
trol of resolution, dynamic range, and spectrum. IEEE
transactions on image processing, 19(9):2241–2253.
Zhang, Y. (2014). Spatial resolution enhancement of hy-
perspectral image based on the combination of spec-
tral mixing model and observation model. In Image
and Signal Processing for Remote Sensing XX, vol-
ume 9244, page 924405. International Society for Op-
tics and Photonics.
Zhang, Y., Wirkert, S. J., Iszatt, J., Kenngott, H., Wag-
ner, M., Mayer, B., Stock, C., Clancy, N. T., Elson,
D. S., and Maier-Hein, L. (2017). Tissue classification
for laparoscopic image understanding based on multi-
spectral texture analysis. Journal of Medical Imaging,
4(1):015001–015001.
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
550
