
Embryo Development Stage Onset Detection by Time Lapse Monitoring
Based on Deep Learning
Wided Souid Miled
1,2
, Sana Chtourou
3
, Nozha Chakroun
3
and Khadija Kacem Berjeb
3
1
LIMTIC Laboratory, Higher Institute of Computer Science, University of Tunis El-Manar, Ariana, Tunisia
2
National Institute of Applied Science and Technology, University of Carthage, Centre Urbain Nord, Tunisia
3
University of Medicine of Tunis, Lab. of Reproductive Biology and Cytogenetic, Aziza Othmana Hospital, Tunisia
Keywords:
IVF, Pronuclei Detection, Embryo Selection, Computer Vision, Classification, Deep Learning, Sequential
Models.
Abstract:
In Vitro Fertilisation (IVF) is a procedure used to overcome a range of fertility issues, giving many couples the
chance of having a baby. Accurate selection of embryos with the highest implantation potentials is a necessary
step toward enhancing the effectiveness of IVF. The detection and determination of pronuclei number during
the early stages of embryo development in IVF treatments help embryologists with decision-making regarding
valuable embryo selection for implantation. Current manual visual assessment is prone to observer subjectivity
and is a long and difficult process. In this study, we build a CNN-LSTM deep learning model to automatically
detect pronuclear-stage in IVF embryos, based on Time-Lapse Images (TLI) of their early development stages.
The experimental results proved possible the automation of pronuclei determination as the proposed deep
learning based method achieved a high accuracy of 85% in the detection of pronuclear-stage embryo.
1 INTRODUCTION
Statistically, almost 10% to 15% of couples suffer
from infertility in the world. Multiple infertility treat-
ments have been developed over the years, collec-
tively referred to as Assisted Reproductive Technol-
ogy (ART). In Vitro Fertilization (IVF) has prevailed
as the most effective and commonly used type of
ART.
To undergo an IVF cycle, patients should have
an ovarian stimulation in order to collect multiple
oocytes which will be incubated with selected motile
sperm from a semen collection. The intra cytoplasmic
sperm injection is a more advanced technique where
every spermatozoa is injected in a mature oocyte. The
resulting embryos are kept in an incubator for three to
five days where their development is observed con-
tinuously by embryologists, on an x400 microscopic
scale, to extract their morphokinetic parameters. Mor-
phokinetics comprise the timing and morphological
changes of embryo as it grows and passes through
a series of sequential developmental stages defined
in academic guidelines (Ciray et al., 2014). Based
on these observations, embryologists decide whether
to transfer the developed embryo for implantation,
freeze it for later use, or discard it if it doesn’t show a
good implantation potential.
In recent years, new advanced IVF incubators en-
tered the market with Time Lapse Imaging (TLI) tech-
nology (Dolinko et al., 2017). These TLI incubators
make it possible to monitor embryonic development
continuously. They take photographs of each embryo
at regular intervals and compile them in a time-lapse
video, giving dynamic insight into embryonic devel-
opment in vitro without disturbing the stable culture
conditions. These incubators, often accompanied by
a dedicated annotation software, have provided both
biologists and clinicians with a new set of data re-
garding embryonic behaviour during preimplantation
development and its association with embryo quality.
As detailed in academic guidelines (Ciray et al.,
2014), the human embryo undergoes different de-
velopment stages, from a fertilized egg (zygote) to
a transferable blastocyst. The main developmental
events are polar body appearance (pPB2), pronuclei
appearance and fading (pPNa and pPNf), cleavage
or cell divisions (p2 to p9+), compaction or Morula
(phase pM), and Blastocyst formation and expansion
(pB and pEB). Figure. 1 illutrates some of these em-
bryo development phases.
Typically, the pronuclear stage occurs within
about 16-18 hours, after the sperm is combined with
368
Miled, W., Chtourou, S., Chakroun, N. and Berjeb, K.
Embryo Development Stage Onset Detection by Time Lapse Monitoring Based on Deep Learning.
DOI: 10.5220/0012390600003636
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 16th International Conference on Agents and Artificial Intelligence (ICAART 2024) - Volume 2, pages 368-375
ISBN: 978-989-758-680-4; ISSN: 2184-433X
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
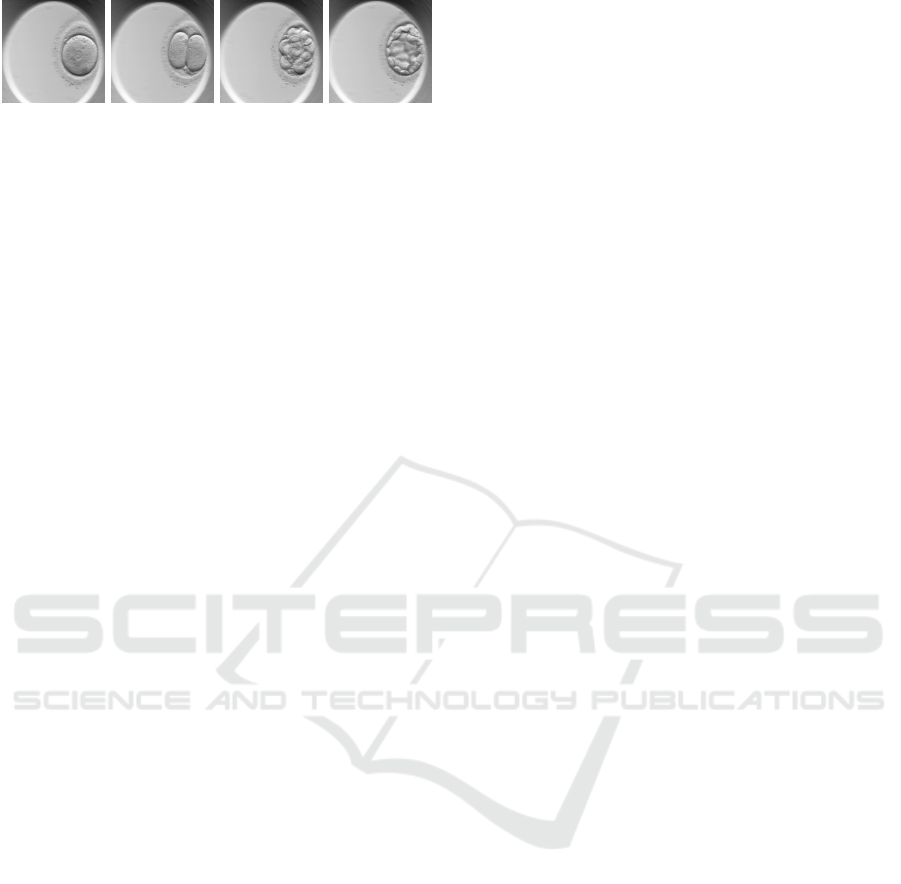
(a) (b) (c) (d)
Figure 1: Embryonic development stages. (a) Pronuclear
(b) First cleavage (c) Morula (d) Blastocyst.
the egg. At this stage, a male and female pronuclei
(2PN) appear containing the genetic material from the
sperm and the egg, respectively. The two pronuclei of
a normal fertilization are generally equal in size and
centrally located. Indeed, several studies have shown
that the morphology of the embryo at the pronuclear
stage is a valuable parameter in the process of evaluat-
ing embryo quality and developmental potential. Cur-
rently, embryologists do the assessment visually, in a
manual process, leaning on their visual experience.
This poses several challenges including: the selection
is prone to human perception error, which can lead to
the loss of promising embryos, or to failed pregnan-
cies; the process is highly subjective as it is difficult
to agree on quality assessment between embryologists
(Adolfsson and Andershed, 2018). Manual assess-
ment is also a difficult and time-consuming process.
Apart from having to take out the embryo from the in-
cubators thus disturbing its culture conditions. These
challenges suggest that an automated evaluation so-
lution leveraging computer vision and artificial intel-
ligence would provide a more reliable and accurate
solution that helps embryologists and supports their
decision-making with embryo selection.
Artificial intelligence (AI) is a field whose goal is
to create machines capable of learning and improving
themselves in an autonomous way. This technology
is proving to be useful in all intellectual tasks. The
concept of (IA) has been extended to encompass sev-
eral subfields, including image classification, which
has made considerable progress in recent years (Ya-
dav and Sawale, 2023). This progress is due to numer-
ous works in this field and to the availability of public
datasets that have allowed researchers to report the ex-
ecution of their approaches. This direction of research
has resulted in the emergence and evolution of Deep
Learning (DL), with the advent of Convolutional Neu-
ral Networks (CNN), a particular type of neural net-
work whose architecture of connections is inspired
by that of the visual cortex. In the same trend, the
use of artificial intelligence (AI) techniques is being
intensively researched in the field of IVF. Many au-
tomated systems based on artificial intelligence have
been proposed to improve IVF success rates by as-
sisting embryologists with their decision and ensuring
more consistent results. Recent AI and DL advance-
ments in the embryology laboratory are summarized
in the review of Dimitratis et al. (I. Dimitriadis and
Bormann, 2022).
In this work, we are concerned with the problem
of automatic detection of pronuclei in the early stages
of IVF embryos development. We aim to develop a
proof of concept (PoC) computer vision solution to
automatically grade the quality of pronuclei in fer-
tilized embryos, based on time-lapse images of their
early development stages.
The main contributions of this work are as follows:
• We build a supervised data collected from TLI
IVF incubators making a dataset of 250 anno-
tated time-lapse sequences of unique embryos
framed each into 20 annotated images. The an-
notations refer to critical embryo development in-
stants, namely tPB2, tPNa, tPNf, and t2. We infer
from these annotations the tPN assessment, which
confirms successful fertilization.
• We create a deep learning model based on a CNN-
LSTM network with a pre-trained VGG16 back-
bone.
• Hyperparameter selection and comparative exper-
iments are conducted to optimize and evaluate the
proposed CNN-LSTM model
• To our knowledge, this work represents the first
attempt at automatic video annotation of human
embryos from an ART center in north Africa.
2 RELATED WORK
According to the literature review by Louis et al.
(Louis et al., 2021), existing research employing
computer vision and deep learning techniques for
IVF embryo selection focuses on the following main
tasks: automatic embryo stage development anno-
tation (Gomez et al., 2022), (V. Raudonis, 2019)
cell counting and detection during cleavage (Rad and
Havelock, 2019), blastocyst quality grading accord-
ing to Gardner’s grading system (Gardner and School-
craft, 1999), (L. Lockhart and Havelock, 2019),
(G. Vaidya and Banker, 2021), (M. F. Kragh and
Karstoft, 2019) and implantation outcome prediction.
Leahy et al. (Leahy et al., 2020) created a pipeline
of five CNNs for automated measurements of key
morphological features of human embryos for IVF.
A Mask R-CNN network with a ResNet50 backbone
was proposed for pronucleus object instance segmen-
tation. The model detects pronuclei by outputting
an object mask and a confidence score from 0 to 1
for each frame of a TLI embryo sequence, cropped
Embryo Development Stage Onset Detection by Time Lapse Monitoring Based on Deep Learning
369

around the embryo region of interest. Another in-
sightful research that uses deep learning for automat-
ing assessment of human embryos in IVF treatment
is reported in (Lockhart, 2018). Three tasks were the
focus of this work: blastocyst grading, cell detection
and counting, and embryo stage classification and on-
set detection. For the latter task, the proposed model
incorporates temporal learning over the TLI sequence
and automatically detects three classes, namely cleav-
age, morula, and blastocyst stage onsets. In order to
detect stage transitions, two image sequence batches
are fed in parallel, in pairwise learning, through two
separate CNNs, which are based on VGG16 architec-
tures pre-trained on the ImageNet dataset with three
final convolution layers fine-tuned. Fully connected
layers from each classifier are concatenated and used
to predict whether the input images fed through each
branch were at the same stage. Synergic loss from this
binary output is backpropagated through both classi-
fier branches. Stage transitions predictions are then
refined using temporal context in an LSTM layer sep-
arately for each synergic branch.
Gomez et al. (Gomez et al., 2022) worked on the
automatic annotation of the 16 embryo development
phases. In addition to providing a fully annotated
dataset composed of 704 time-lapse videos, authors
applied ResNet, ResNet-LSTM and ResNet3D mod-
els to automatically annotate the stage development
phases. The evaluation results showning the superi-
ority of ResNet-LSTM and ResNet-3D over ResNet,
prove the importance of using the temporal informa-
tion in the automatic annotation process. However,
predicting the 16 classes of embryonic development
is prone to numerous challenges, primarily due to
the extensive computational requirements necessary
for training DL models on more than 300k images,
which demand high-performance GPUs. Fukunaga et
al. (Fukunaga et al., 2020) proposed an automated
pronuclei determination system based on few amount
of supervised data. In their paper, authors proposed
a framework of four stages. First, images are pre-
processed to detect and focus on the embryo area us-
ing a circular Hough mask. Then, images are passed
for main processing to two CNNs, both composed of
two convolution layers and two fully connected lay-
ers. The first model detects the outline around pronu-
clei and passes these outline images to the second
CNN, which gives a probability distribution of the
number of pronuclei (0PN, 1PN, 2PN). Finally, pre-
dictions are postprocessed through a Hidden Markov
model, while setting conditions for the change in the
number of pronuclei over time. Thus, the change of
the number of pronuclei, if occurred (the state can re-
main unchanged), is only valid from 0PN to either
1PN or 2PN and from 1PN to 2PN. This integration
of time-series information resulted in improvement of
performance in sensitivity, however the accuracy re-
mains relatively low. To the best of our knowledge,
this workb (Fukunaga et al., 2020) is the only exist-
ing reference that deals with detecting and determin-
ing pronuclei number in IVF embryos.
In this work, we aim to automate the annotation
process of the early stages of embryonic development,
from Polar Body appearance (tPB2) to just before
the first cell division (t2). We create a deep learn-
ing model that analyzes the TLI incubator’s sequences
of embryonic development and annotates tPN, de-
fined as the time at which fertilization status is con-
firmed, immediately before the time fading of pronu-
clei (tPNf) (Ciray et al., 2014).
3 METHODOLOGY
3.1 Dataset
The dataset used in this work is a collection of 352
videos of unique embryos exported from a private TLI
IVF Incubator manufactured by Esco Medical
R
. The
frames of each video are time-lapse embryo images
taken every five minutes, starting shortly after fertil-
ization. Each video contains between 600 and 1400
frames in gray scale with a resolution of 1280 × 720
pixels.
An experienced biologist notes the start and end
time of each phase of the embryo’s development.
Each image of each video has therefore a class, which
corresponds to the phase seen in the image. The an-
notations follow the same convention used by Gomez
et al. (Gomez et al., 2022) and academic guidelines
(Ciray et al., 2014). There are, in general, 16 annota-
tions corresponding to 16 different instants of embryo
evolution. Here, as we are only interested in detect-
ing two key instants, namely tPB2 and tPN, we only
consider the following phases:
• tPB2: time of appearance of second polar body
• tPNa: time of pronuclei appearance
• tPNf: time of pronuclei fading
• t2: time of first cell division marks the end of
pronuclear phase
The stage tPN, which is defined as the time at which
fertilization status is confirmed, is calculated from
tPNa and tPNf (Ciray et al., 2014). We received the
annotation in Excel sheets generated by the software
of the TLI incubator, which we had to parse to extract
useful information.
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
370
3.2 Data Preprocessing
Before feeding the sequence of images to the pro-
posed deep learning based model, we made some
preprocessing treatments. First, as we reviewed the
dataset, we observed that some videos suffered from
excessive lighting changes and motion blur. Other
images were taken from a bad angle where the em-
bryo was not entirely visible. Some other videos did
not cover some critical stages of the embryo’s devel-
opment. After discarding these unusable videos, we
obtained 250 annotated videos of unique embryos.
Then, as the embryo cell presents only a small part
of the image, we cropped them, reducing the frame
size to 360 × 360 and gaining in memory efficiency.
To achieve this, we applied the Hough transform to
detect circular shapes in the images, then we cropped
the detected circles with a fixed size of 360 ×360 pix-
els.
After choosing the frames and preprocessing
them, each frame has been labeled based on the ex-
pert’s annotations. We repeated the same process for
every video in the dataset. For each video, we ended
up with 20 images, sampled over the first 18 hours
of embryo development. The retained frames are an-
notated as 0 (neither tPB2 nor tPN occured), 1 (tPB2
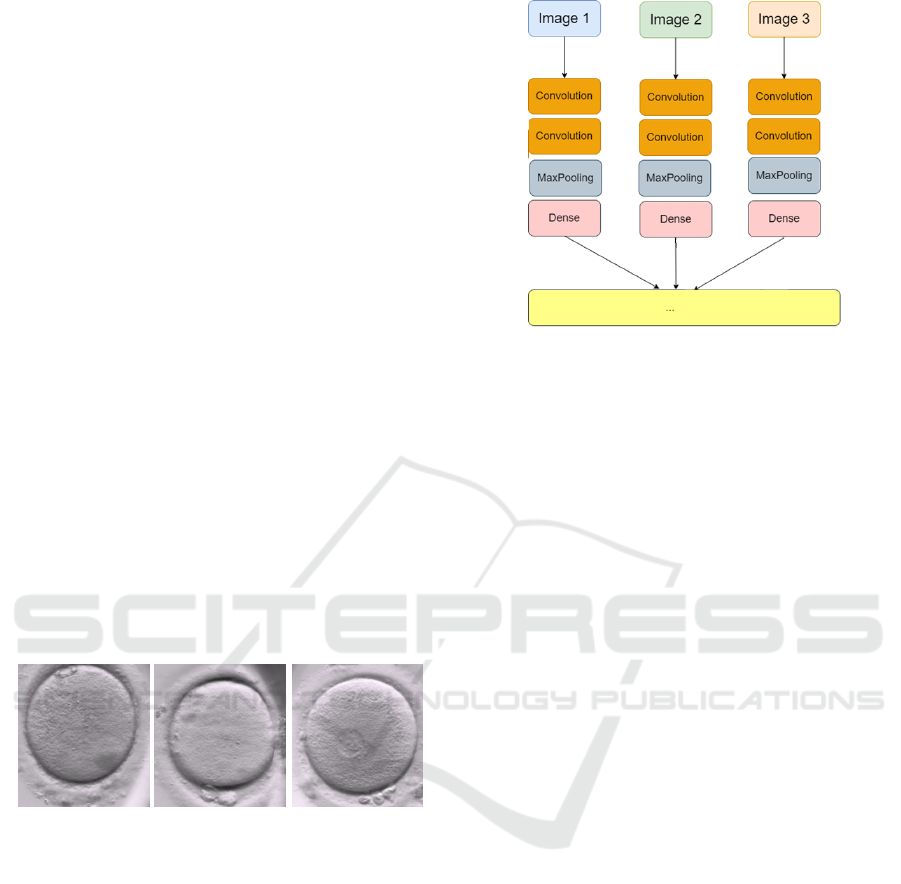
occured), 2 (tPN occured). We can see examples of
the three classes in Figure. 2.
class 0 (no event) class 1 (tPB2) class 2 (tPN)
Figure 2: Examples of labelled images from the dataset.
3.3 Proposed Model
In this work, we are concerned with a sequence clas-
sification problem, which implies that the model’s
input is not a series of independent images to be
classified as categorical targets, but rather a time-
dependent sequence of images to be predicted accord-
ing to a certain order. Sequence classification is a
challenging problem because the sequences can vary
in length, contain a very large vocabulary of input
symbols, and may require the model to learn the long-
term context or dependencies between symbols in the
input sequence. The solution to this sequentially-
classified problem is to use a combination of the two
approaches: the LSTM architecture, and the CNN ar-
Figure 3: Proposed input convolution flows.
chitecture.
It should be noted that a sequence of images must not
be fed to a single convolution. If we take a common
sequential network, each entry is connected to all the
neurons in the first layer. With multiple images as
batch entries to the CNN network, all the pixels of all
images are merged and sent to the first layer. Con-
sequently, their distinctive features and the temporal
information will be lost. To overcome this problem,
as illustrated in Figure 3, we need to share the network
layers across the video frames to reduce the number
of tensors, thus having filters for each image input,
not for the whole stack of frames.
With this adopted architecture, each image has got its
own convolution flows. If we separately train each
convolution flow, we will have several unwanted be-
haviors:
• We will need long training time because several
convolution flows need to be trained (one per in-
put image).
• Some convolution flows will not detect what other
flows could detect.
• Each convolution flow, for one sequence, can have
several different weights, and so we get different
detection features that are not linked.
In order to make sure that all the convolution flows
can extract the same features, we propose to add a
time distributed layer which applies the same convo-
lution layer to several inputs. This allows to apply
the layer operation on each timestamp. Otherwise,
when we flatten the data all the image instances will
be combined and the time dimension will be lost.
As shown in Figure 4, the proposed model has two
main parts: a CNN and an LSTM network, linked by
a time distributed layer. Each layer that is time dis-
tributed will share the same weights, saving calcula-
tion and computation time.
Embryo Development Stage Onset Detection by Time Lapse Monitoring Based on Deep Learning
371
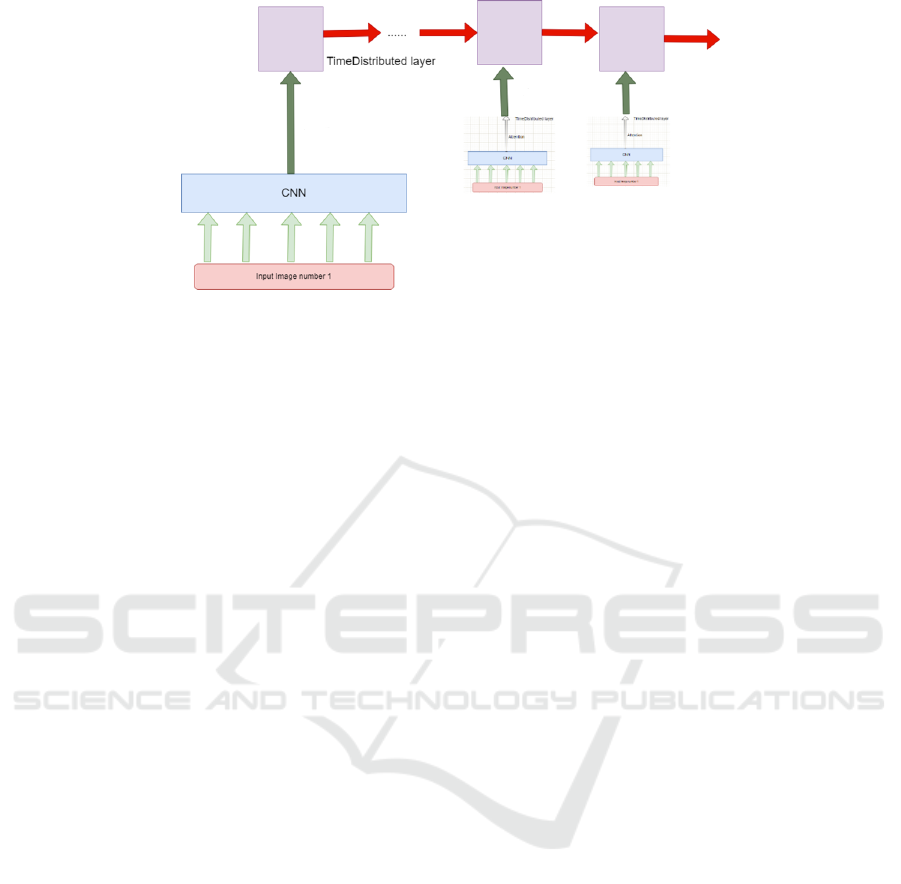
Figure 4: The proposed architecture integrating a time distributed layer.
For the CNN backbone, in the field of medical
image analysis, it is common to use a deep learn-
ing model pre-trained on a large and challenging im-
age classification task, such as the ImageNet classi-
fication competition. The research organizations that
develop models for these competitions often release
their final models under a permissive license for reuse.
These models can take days or weeks to train on mod-
ern hardware. But, we can directly use them pre-
trained employing transfer learning technique for a
target specific task. In this work, we opted for a
VGG16 model pre-trained on the ImageNet compe-
tition dataset.
4 EXPERIMENTAL RESULTS
4.1 Dataset
In this section, the performance of the proposed deep
learning model for the task of early stage human em-
bryo detection is discussed. Our dataset contains 250
annotated videos of unique embryos augmented five
times (Horizontal flip, vertical flip, transpose, and
transpose horizontal flip). We further resized the im-
ages from 360 × 360 to 180 × 180 resolution. Since
the number of frames can be very large, it is imprac-
tical to feed all of them to the model, as this would
slow the training and reduce the performance. Our
strategy was to choose 20 frames between the start
of the video and the instant tPNf (which denotes the
fading of the pronuclei) and feed them to the model,
since this range covers all the phases we are interested
in. We chose our frames in a way where the number
of frames between two consecutive chosen frames is
constant. Every sequence is therefore framed into 20
(180 × 180 × 3) images. As the VGG model requires
3-channels input images, we converted our grayscale
images into RGB. Furthermore, as each pixel value
can vary from 0 to 255, representing the color inten-
sity, feeding an image directly to the neural network
will result in complex computations and a slow train-
ing process. To address this problem, we normalize
the high numeric values to range from 0 to 1 by di-
viding all pixel values by 255. Then, we labeled the
dataset marking images in the tPB2 phase as class 1,
those attaining the stage tPN as class 2, and the re-
maining images where no event occurs into class 0.
Finally, we split the dataset, conventionally, into 80%
training data and 20% test data.
4.2 Models Implementation
Since the backbone pre-trained CNN model wasn’t
designed to annotate pronuclei stage development
phases in embryo image datasets, we have to make
it more specific to our needs, taking advantage of the
transfer learning technique and using the ImageNet
pre-tuned weights. We chose to train only the last
four layers and reduce the number of outputs using
the last pooling layer with a maximum operation ap-
plied to the convolutional values. First, we specify
the top layers by the VGG implementation, taking our
custom input 180 × 180 × 3 images. We then link the
time distributed layer with the VGG16 output layer
via a sequential mode, which will fully connects each
neuron from both sides. The next layers are the LSTM
layers, followed by five dense layers, separated with
50% dropout layers to prevent over-fitting. We use
the ReLu activation function and Softmax as a final
activation function, which will output the correspond-
ing class probabilities. As an optimization algorithm,
we opted for Adam (Adaptive Moment Estimation),
as it is straightforward to implement, is computation-
ally efficient, has little memory requirements and is
well suited for problems with large data and/or pa-
rameters (Kingma and Ba, 2014). We fix the learning
rate at a value of 0.01, to converge the learning in a
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
372
faster, more efficient way, and to avoid the problem of
vanishing gradients. We choose the categorical cross-
entropy as the loss function since we are dealing with
a multi-class dataset.
After experimenting with the transfer learning
technique, we decided to build a custom CNN model
using six convolutional layers. We also introduced
a batch normalization to reduce the inter-variance of
the layer inputs. This technique stabilizes the learn-
ing process and dramatically reduces the number of
epochs required for training. The batch normaliza-
tion momentum uses the moving average of the sam-
ple mean and variance in a mini-batch for training.
By adjusting a dynamic momentum parameter, the
noise level in the estimated mean and variance can
be well controlled. We fixed the momentum value
to 0.9. We kept the Adam optimizer and categori-
cal cross-entropy loss function. We set the learning
rate to 0.001, which is considerably lower than the
one used with the VGG16 architecture. The model is
built from scratch, so the gradients are initially ran-
domized, and to reach a similar accuracy, the weights
need to be adjusted carefully.
4.3 Evaluation
The metrics we used for the performance evaluation
of proposed DL models are accuracy and sensitivity.
Accuracy is defined as the ratio of correctly classified
instances by the total amount of instances. Sensitivity
is defined as the number of correctly classified pos-
itive samples divided by the number of all positive
samples.
We conduct a first experiment where we trained
the CNN-LSTM model based on a pre-trained
VGG16 backbone for a total of 90 epochs and a batch
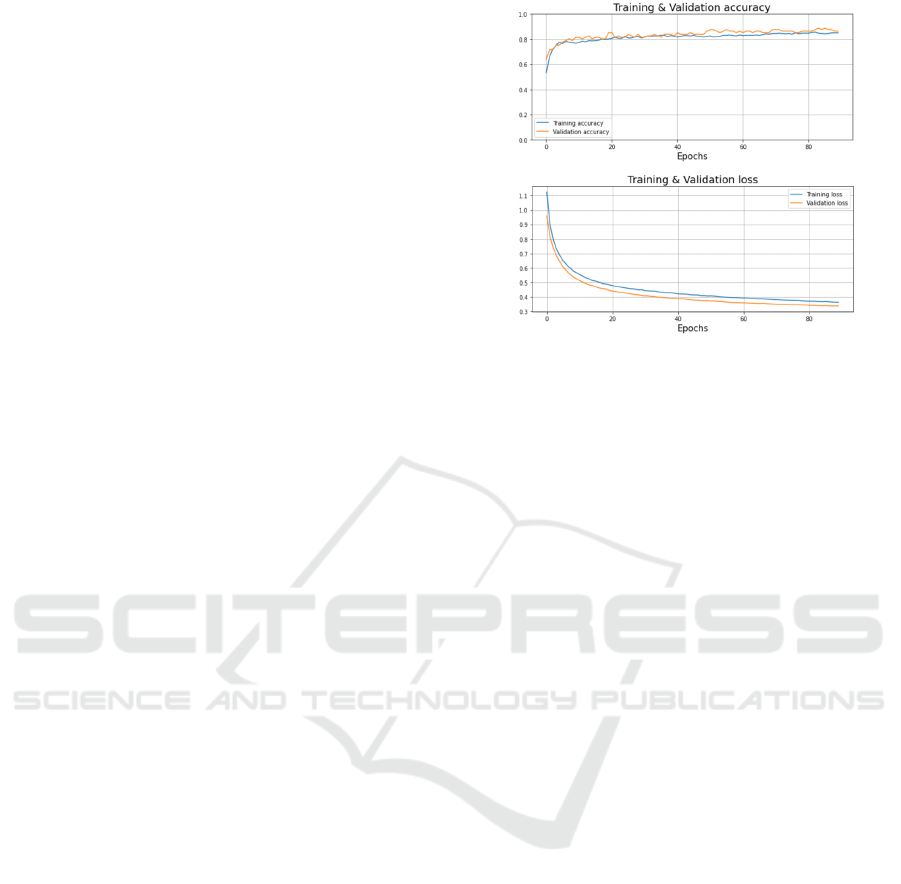
size of 16. The accuracy and loss graphs for training
and validation are shown in Figure 5. The accuracy
curve represents few variations and is up to 0.86%. In
addition, the loss curve is almost stable and the vali-
dation and training curves are almost similar, showing
that the model is well fitted.
In order to make the proposed classification model
interpretable, we implemented the Grad-Cam method
that exploits the features map from the last convolu-
tion layers to calculate the gradients of the features
map against the class score to identify the most im-
portant filters. Figure 6 shows the generated heatmaps
on the tPN stage prediction, where red pixels indicate
highest contribution towards stage prediction and no
colour represents no contribution, As seen in this fig-
ure, for tPN stage prediction, the network mainly re-
lied on the circles in the centre of the embryo, which
correspond to the two pronucleus. Thus, the Grad-
Figure 5: Accuracy and loss graphs of the CNN-LSTM pro-
posed model.
Cam method makes it possible the visualisation of the
areas that contributed the most to the prediction of the
specific tPN class.
In a second experiment, we trained the custom-
built CNN model, along with a LSTM network, for
a total of 50 epochs and a batch size of 8. We no-
ticed that this second model has taken more time, and
more failed attempts to reach the threshold accuracy.
We can visibly conclude this from all the fluctuations
in the accuracy per epoch graph in Fig. 7, where the
validation accuracy doesn’t exceed 60%. This was ex-
pected since the pre-trained model has already learned
high-level features, is assigned pre-trained weights
and only needs fine-tuning to fit the training dataset
on the target task, while the custom-built CNN model
starts with randomized weights.
4.4 Comparison with State of the Art
For state-of-the-art comparison, as there is no bench-
mark available in the literature, we reported the re-
sults of Fukunaga et al. (Fukunaga et al., 2020) and
those of Gomez et al. (Gomez et al., 2022) given
in their corresponding papers and conducted on their
own datasets. Comparative results in terms of accu-
racy and sensitivity metrics are reported in Table 1.
The common aspects between our work and the
work of Fukunaga et al. (Fukunaga et al., 2020) are
the limited amount of supervised data available, and
the classification task. However, the main difference
is the methodology of the detection systems: we pro-
posed a CNN network linked to an LSTM layer while
they developed a 2-CNN architecture, with no deploy-
ment of a sequential model that would deal with time
dependency with a deep learning technique. Their
model’s sensitivity reached 82%, but with only a 40%
accuracy rate, which makes our method more accurate
Embryo Development Stage Onset Detection by Time Lapse Monitoring Based on Deep Learning
373
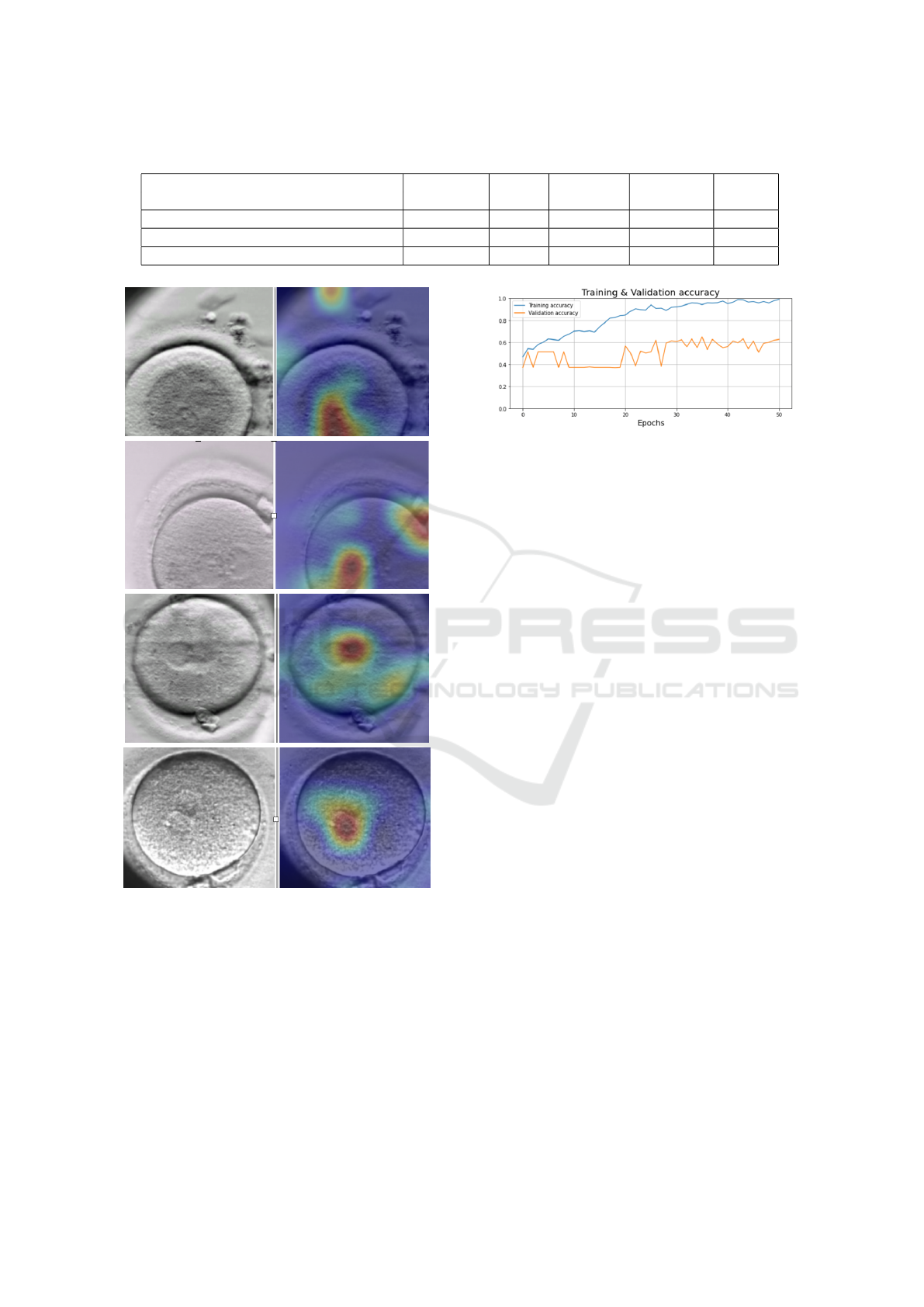
Table 1: Comparison with state-of-the-art methods.
Model Dataset LSTM Accuracy. Sensitivity Classes
usage
Proposed Model 250 videos Yes 85% 96% 3
Fukunaga et al. (Fukunaga et al., 2020) 300 videos No 40% 82% 3
Gomez et al. (Gomez et al., 2022) 873 videos Yes 73% 96% 16
Figure 6: The heatmaps generated by the Grad-CAM
method on the tPN stage.
with 85% accuracy score and 96% sensitivity score.
Regarding Gomez et al. (Gomez et al., 2022),
the used dataset is composed of 337 thousand images
from 873 annotated videos. This big ground-truth
helped apply three approaches: ResNet, LSTM, and
ResNet-3D architectures, and demonstrate that they
outperform algorithmic approaches to the automatic
annotation of embryo development phases. Further-
more, the compared models are detecting 16 classes
of 16 morphokinetic events, compared to 2 events in
Figure 7: Accuracy graph of the custom CNN proposed
model.
our case. The three models they benchmarked con-
cluded a 73% accuracy score.
5 CONCLUSION
Continuous embryo monitoring with time-lapse imag-
ing enables time based development metrics along-
side visual features to assess an embryo’s quality be-
fore transfer and provides valuable information about
its likelihood of leading to a pregnancy. In this work,
we developed a deep learning based model to classify
a sequence of time-lapse Human embryo images with
the aim of helping embryologists with embryo selec-
tion for IVF implantations. The classification task
aims to detect tPB2 and tPN key instants from an in-
put sequence of images by predicting the class of each
image among three classes; denoting the appearance
of the second polar body (tPB2), the appearance of the
pronuclei (tPN), or none of the two events. The pro-
posed model is a combination of a pre-trained VGG16
backbone, and an LSTM network. It has proven to be
powerful enough to fit the data as it achieved a high
training accuracy, In future work, our model can be
enhanced by being incorporated into a pipeline where
the second part detects the number of pronuclei as
0PN, 1PN, 2PN or more. This pipeline can then be
part of a whole automatic embryo assessment deep
learning framework, integrating the work on blasto-
cyst segmentation and cell counting.
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
374

REFERENCES
Adolfsson, E. and Andershed, A. (2018). Morphology vs
morphokinetics: A retrospective comparison of inter-
observer and intra-observer agreement between em-
bryologists on blastocysts with known implantation
outcome. JBRA assisted reproduction, 22(3):228–
237.
Ciray, H., Campbell, A., Agerholm, I., Aguilar, J.,
Chamayou, S., Esbert, M., and Sayed, S. (2014). Pro-
posed guidelines on the nomenclature and annotation
of dynamic human embryo monitoring by a time-lapse
user group. In Human Reproduction, volume 38,
pages 2650–660.
Dolinko, A. V., Farland, L. V., Kaser, D. J., and al. (2017).
National survey on use of time-lapse imaging systems
in ivf laboratories. Assisted Reproduction and Genet-
ics, 34(9):1167–1172.
Fukunaga, N., Sanami, S., Kitasaka, H., and al. (2020).
Development of an automated two pronuclei detec-
tion system on time-lapse embryo images using deep
learning techniques. Reprod Med Biol., 19(3):286–
294.
G. Vaidya, S. Chandrasekhar, R. G. N. G. D. P. and Banker,
M. (2021). Time series prediction of viable embryo
and automatic grading in ivf using deep learning. vol-
ume 15, pages 190–203.
Gardner, D. and Schoolcraft, W. (1999). In vitro culture of
human blastocyst. Towards Reproductive Certainty:
Fertility and Genetics Beyond 1999, page 378–388.
Gomez, T., Feyeux, M., and al. (2022). Towards deep
learning-powered ivf: A large public benchmark for
morphokinetic parameter prediction. https://arxiv.org/
abs/2203.00531.
I. Dimitriadis, N. Zaninovic, A. C. B. and Bormann, C. L.
(2022). Artificial intelligence in the embryology lab-
oratory: a review. volume 44, pages 435–448.
Kingma, D. P. and Ba, J. (2014). Adam: A method for
stochastic optimization. In 3rd International Confer-
ence for Learning Representations. arXiv.
L. Lockhart, P. Saeedi, J. A. and Havelock, J. (2019). Multi-
label classification for automatic human blastocyst
grading with severely imbalanced data. pages 1–6.
Leahy, B., Jang, W., Yang, H., and al. (2020). Automated
measurements of key morphological features of hu-
man embryos for ivf. CoRR, abs/2006.00067.
Lockhart, L. (2018). Automating assessment of human em-
bryo images and time-lapse sequences for ivf treat-
ment.
Louis, C., Erwin, A., Handayani, N., and al. (2021). Review
of computer vision application in in vitro fertilization:
the application of deep learning-based computer vi-
sion technology in the world of ivf. Assist Reprod
Genet., 38(3):1627–1639.
M. F. Kragh, J. Rimestad, J. B. and Karstoft, H. (2019).
Automatic grading of human blastocysts from time-
lapse imaging. volume 115, page 103494.
Rad, P. Saeedi, J. A. and Havelock, J. (2019). Cell-net:
Embryonic cell counting and centroid localization via
residual incremental atrous pyramid and progressive
upsampling convolution. volume 7, pages 81945–
81955.
V. Raudonis, A. Paulauskaite-Taraseviciene, K. S. e. a.
(2019). Towards the automation of early-stage human
embryo development detection. volume 18.
Yadav, S. and Sawale, M. D. (2023). A review on image
classification using deep learning. volume 17.
Embryo Development Stage Onset Detection by Time Lapse Monitoring Based on Deep Learning
375
