
Covid-19 Impact on Standard Coding Systems Update
Elena Cardillo
1a
, Maria Teresa Chiaravalloti
1b
and Erika Pasceri
2c
1
Institute of Informatics and Telematics, National Research Council, Rende, Italy
2
Department of Culture, Education and Society, University of Calabria, Rende, Italy
Keywords: Coding Systems, Covid-19, Standards Updates, Knowledge Organization Systems, Healthcare
Interoperability.
Abstract: The outbreak of Covid-19 pandemic has sped up many healthcare processes and practices. Both stakeholders
and standard organizations and authorities had to quickly implement new guidelines and codes to uniquely
identify the disease and all the related healthcare data. The object of this work is to study the impact of the
Covid-19 pandemic on clinical coding systems, in terms of updates and introduction of new specific codes
for the identification of the SARS-CoV-2 virus, with the aim of allowing a better description of the disease
and interoperability of the clinical data. The analysis is focused on ICD, SNOMED CT, LOINC, ATC as
coding systems either included into the Italian EHR regulation or widely used internationally. Results show
that coding systems that created a plenty of new codes for Covid-19 have: i) a flexible structure; ii) a speed
process for updates; iii) a large user community for inputs. Others instead demonstrated in this circumstance
that they are limited by hierarchical structures or excessively cumbersome updating processes, which conflict
with the flexibility required to standards to represent the evolution of clinical knowledge. This is especially
true in exceptional situation like the pandemic one.
1 INTRODUCTION
Systems for organising information and knowledge
are essential to reduce possible semantic conflicts
(ambiguities) and issues related to the specialization
of a domain terminology. They are commonly known
as Knowledge Organization Systems (KOSs).
“Knowledge is the lifeblood of modern society, but
without organization knowledge is dead. We could
even say that without organization knowledge is not
knowledge at all. Knowledge must be organized in
order to be used, be it by people or by machines”
(Dagobert, 2009). To this end, KOS in clinical
domain are used to classify, represent and encode
diseases and other types of data (symptoms, medical
procedures, drugs, etc.) in a unique way, with the
purposes of indexing and retrieving information of
interest, supporting epidemiological studies and
decision-making, and ensuring semantic
interoperability during data exchange between
different information systems. In addition, they
a
https://orcid.org/0000-0001-5003-205X
b
https://orcid.org/0000-0003-4695-2026
c
https://orcid.org/0000-0001-9917-2184
support the digitization process of clinical settings
and enable physicians to access and process relevant
data to support diagnosis, define risk profiles, and
facilitate statistical and epidemiological studies.
Some clinical KOSs are reference standards for the
semantic area they represent. They differ mainly in
structure, relationships with other standards, and
updating process.
The pandemic was an exceptional testing ground
in many respects, as it gave the opportunity to test the
functioning of systems under conditions of stress and
urgency. Globally, the Coronavirus Disease 2019
(Covid-19) had a significant impact not only on
public health, but also on the economy and society,
involving millions of people and causing serious
health crises, forcing many countries to implement
restrictive measures to reduce contagion, such as
lockdown, social distancing and the use of masks.
The Covid-19 confirmed cases growth put pressure
on healthcare systems around the world, with
hospitals and intensive care units often overcrowded.
490
Cardillo, E., Chiaravalloti, M. and Pasceri, E.
Covid-19 Impact on Standard Coding Systems Update.
DOI: 10.5220/0012387600003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 2, pages 490-497
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

Additionally, Covid-19 left many patients with
serious long-term symptoms, known as “long-
haulers”. From the point of view of clinical coding
systems, this meant keeping up with the expressive
needs of the Covid-19 and everything related to it
(e.g. biological studies) to correctly record the
pandemic data, share them without ambiguity, be able
to aggregate them for surveillance and forecasts.
Therefore, this paper aims to carry out an analysis of
how much and how some clinical KOSs have updated
their contents in relation to the Covid-19 disease to
verify whether and how timely they have been and if
they have covered the semantic areas of the new
concepts, determined by the contingent situation. The
analysis was carried out on KOSs which are either
included in the Italian regulations to manage EHR
data or widely used internationally and with a well-
known effort made to adapt to the pandemic
emergency.
The paper is structured as follows: Section 2 gives
an overview of the Background related to the
pandemic and its impact on healthcare services;
Section 3 focuses on the analyzed coding systems and
their updates related to Covid-19; Section 4 is
dedicated to Discussion and Conclusions,
highlighting future perspectives of the study.
2 BACKGROUND AND
SIGNIFICANCE
Covid-19 is an infectious disease caused by the
SARS-CoV-2 virus, belonging to the family of
coronavirus, firstly identified in Wuhan, China, in
December 2019 and quickly becoming a pandemic.
The outbreak of the Covid-19 pandemic was
impactful from many points of view as it introduced
a new way of thinking and acting for previously
unknown aspects and, at the same time, required a
review and readjustment of the known ones. The role
of telehealth, for example, was enhanced and became
crucial to handle ordinary medical activities, when
hospitals’ departments and outpatient care facilities
were “closed” because of the infections. The
provision of medical care remotely allowed – in many
cases - to manage the pandemic more safely and
efficiently, by reducing the risk of virus transmission
and ensuring safe access to care for patients who
would otherwise have struggled during lockdown or
isolation. Furthermore, telemedicine allowed remote
monitoring of patients with Covid-19 and other
chronic conditions, facilitating collection of data and
treatment management.
Although Covid-19 had an unprecedented impact on
the world with disastrous health and economic
consequences and highlighting significant gaps in
healthcare systems all over the world, it led to a series
of advances in terms of scientific and technological
improvements. This posed new challenges and
opportunities for innovation in healthcare, like the
new vaccine development approach (it has been done
at unprecedented speed, with the usual sequential
steps done in parallel), recognized now as being
successful (Buchy et al., 2021).
Throughout the pandemic, many clinical
terminologies / classification and coding systems
were quickly updated to introduce new specific codes
for the SARS-CoV-2 virus, with the aim of allowing
a better understanding and description of the disease.
The intent was also to uniquely identify it ab origine
to distinguish it from existing diseases of the
respiratory system and correctly classify it. It allowed
a more detailed registration and monitoring of
epidemiological data, guaranteeing improved
analyses and researches related to the Covid-19
pandemic. As an example, the code U07.1 (Covid-19,
Virus Identified) was added among the ICD-10 codes
shortly after the pandemic began. During the
pandemic it was particularly difficult for health
systems to keep up with evolving diagnostic and
procedural coding recommendations, but the new
codes had been extremely decisive in strengthening
observational studies, characterising the disease
phenotype, and for responding to other important
epidemiological questions (such as disease
prevalence) (Marwaha et al., 2021).
Among the
various types of clinical data, diagnostic tests for
Covid-19 played a crucial role as the main tool for
identifying confirmed cases of Covid-19. To quickly
respond to the pandemic urgency, the LOINC
Committee released a set of nearly 1,300 new codes
to identify new clinical and laboratory observations
related to the Covid-19 pandemic to guide the
uniform coding of these concepts (Dong et al., 2020).
A similar study was previously conducted by
(Zeng et al., 2020), but while recognizing the
fundamental role played by KOSs in critical moments
when the correct identification of information
becomes crucial, it has the limitation of having been
published in May 2020. Therefore, it begins to trace
the changes to coding systems in relation to the
pandemic but cannot provide full recognition of them
for obvious temporal reasons.
Covid-19 Impact on Standard Coding Systems Update
491

3 CLINICAL STANDARDS’
UPDATES RELATED TO
COVID-19
During the Covid-19 pandemic, medical coding
systems played a crucial role in the standardization
process, organization of clinical information and for
the definition of pandemic epidemiological aspects,
being able to uniquely identify the type of pathogen
involved and other relevant connected data (e.g., its
transmissibility, incubation times and duration of the
disease). They had also been used for several
purposes, including identifying at-risk groups,
developing care plans, supporting the institutional
authorities' planning of both primary and secondary
prevention measures, such as containment strategies
and the need for the supply of protective equipment.
The rapid and unexpected nature of the pandemic
required careful monitoring of the progress of
infections and enshrined the importance of the Health
Surveillance as «the continuous, systematic
collection, analysis and interpretation of health-
related data», as defined by the WHO. According to
a study published in Journal of Hospital Infection
(Lin et al., 2020), thanks to a health surveillance
algorithm, based on the data codified and collected in
electronic medical records, it was possible to identify
patients admitted to hospital whose “pneumonia” did
not show a clear improvement with antibiotic
treatment. This had produced daily alerts addressed to
general practitioners. The surveillance algorithm,
thanks to the data registered within the EHR,
demonstrated the utility of the information
technology to facilitate infection control.
To achieve the objectives of this study, a double
survey was carried out regarding the contents related
to Covid-19 in the coding systems considered. For
each of them, the official web page, guidelines and
other resources provided by the respective Standards
Development Organizations (SDOs) were
investigated, and an additional check through the
UMLS Metathesaurus Terminology Services (UTS)
allowed to evaluate a possible expansion of the
coding systems / value sets to be considered. The
specific keywords used, in this case, were: “Covid-
19”, “SARS-CoV-2”, “Covid-19 Vaccine”, “SARS-
CoV-2 vaccine”, “Covid19 (disease)”, “Suspected
Covid-19”, while the semantic groups applied were:
“Disorders”, “Chemical & Drugs, “Living beings”;
the semantic types applied were: Disease or
Syndrome, Pharmacologic Substance Immunologic
Factor, Laboratory Procedure.
3.1 Covid-19 in ICD
Because of the advent of the new pathogen SARS-
COV-2, WHO introduced in February 2020 two
emergency codes in the ICD-10 classification. These
codes are: i) U07.1 – Covid-19, virus identified (for
confirmed cases), ii) U07.2 – Covid-19, virus not
identified (for suspected cases). Initially, U07.1 was
referred to as “acute respiratory disease 2019-nCoV”,
and this code was not made available for use until
April 2020. Subsequently, it was labelled as “Covid-
19, virus identified” (De Lissovoy, 2020). The codes
for Covid-19 disease, defined between 2020 and
2021, have been integrated into “Chapter XXII -
Special purpose codes”, block “U00-U49 Assignment
provisional of new diseases of uncertain aetiology or
for emergency use” of the 2019 edition of ICD-10.
Although it was generally suggested to assign U07.1
to cases with a lab-confirmed diagnosis and U07.2 to
cases with a clinical diagnosis, rules for recognizing
Covid-19 diagnoses and thus for using these codes
were not that easy. In fact, in US some flowcharts
were provided and used to guide the proper
assignment of diagnostic codes related to Covid-19
encounters (Varghees, 2020). The introduction of
new codes was based on requests of Member States
to ensure accurate reporting of Covid-19 related
conditions. The need to distinguish between acute
disease, long-term effects, and complications led to
the adoption of the neutral term “post-Covid”. This
term implies no specific etiologic relationship,
allowing any condition to be linked to a previous
acute Covid-19 infection. The term “post-Covid”
refers to persistent, recurrent or new symptoms (e.g.,
fatigue, short of breath, insomnia, etc.) and other
health effects that occur after the acute phase of
Covid-19 infection. Despite the early detection of this
condition, a specific code was not made available for
clinical use until 2021 in ICD-10 (the suggestion was
to use U09.9, “Unspecified post-Covid-19
condition”). From January 2021, new codes had been
introduced on immunization to prevent Covid-19 and
adverse reactions to vaccines against Covid-19.
These codes are: U11.9 “Need for vaccination against
Covid-19, not specified” and U12.9 “Vaccines
against Covid-19 that cause adverse effects in
therapeutic use, unspecified”. Figure 1. summarises
the Covid-19 coding flowchart using ICD,
considering both ICD-10 and ICD-11, the last
revision of the classification, and distributing the new
codes according to the purpose (e.g., for diagnoses,
prevention).
HEALTHINF 2024 - 17th International Conference on Health Informatics
492

Figure 1: Use of ICD codes for Covid-19 during the disease
outbreak.
The promptness in the introduction of the new
ICD codes has been extremely helpful in accurately
recording and documenting the diagnoses and
conditions of patients with Covid-19, for monitoring
and surveillance on the diffusion of the pandemic and
for collecting epidemiological data that help in
identifying clusters of cases, monitoring the spread of
the virus and assess the impact of control measures.
3.2 Covid-19 in LOINC
LOINC is the most widely used coding system for
laboratory and clinical observations (McDonald et al.,
2003). After the pandemic outbreak, a specific
LOINC subset was created for facing the emergency,
during that time, new term requests related to Covid-
19 from worldwide users had a preferential route in
the submission process. Furthermore, it was created a
specific webpage on the standard website to give a
quick and direct access to LOINC content related to
the disease. The mentioned subset contains 562 terms.
Some of them belong to pre-pandemic LOINC
versions (existing codes), therefore they were not
created for the specific purpose, but their semantics
adapt to it, while others are codes created starting
from LOINC version 2.68 (June 2020), so specifically
required to respond to the needs posed by Covid-19
outbreak. They are further divided according to their
purpose of use in the following subcategories:
▪ SARS-CoV-2 lab tests: all the 160 codes of this
category are newly created after pandemic
outbreak;
▪ SARS-CoV-2 AOE questions: 11 codes,
including 4 existing codes;
▪ Convalescent plasma: 2 newly added codes;
▪ LOINC terms related to public health case
reporting: 110 codes, and 44 of them released
before June 2020;
▪ Covid-19 and Telehealth documents: 120 codes
(48 belonging to previous LOINC versions, and
72 created after the Covid-19 outbreak);
▪ Covid-19 Survey terms: 159 codes, including 1
already existing LOINC code.
The Covid-19 pandemic produced effects also on
the types of clinical documents produced by the
healthcare stakeholders, because new ones had been
introduced and the existing ones needed to be better
organized. Clearly identifying clinical document
types is a non-trivial task because often behind
identical names there is not the same content and,
viceversa, there are plenty of different names for
documents related to the same semantic area. With
the aim of standardizing this field, in the first 2000s
the joint effort of the LOINC committee and HL7
created the LOINC Document Ontology (Frazier et
al., 2001). This section of the standard models the six
main LOINC axes to uniquely identify clinical
document types based on different metadata (e.g.,
role, subject, type of service, etc.).
LOINC document type (DT) codes are one of the
metadata used in the registries of the Italian EHR
infrastructure to index clinical documents. Admitted
values are specified into the Affinity Domain (AD)
document, which regulates interoperability services
among regional EHR systems. Usually AD specifies
LOINC DT which have a corresponding CDA2 HL7
Implementation Guide (IG) already defined or in case
of special needs, as those posed by the pandemic. This
latter was the case of the two new LOINC codes
proposed by LOINC Italy to address the urgency to
have these kinds of documents indexed into the
patients’ EHR:
▪ LOINC 97500-3 Proof of Covid-19
immunization or negative status certificate,
which is the so-called Green Pass, a document
in digital or paper format that certifies
vaccination against the Covid-19 disease or,
alternatively, the recent negative test result or
recent recovery from the infection;
▪ LOINC 97499-8 Proof of Covid-19 recovery
certificate, which is a document certifying the
recovery from the disease and can be around
others. It is issued by the GP.
LOINC DT codes related to Covid-19 are 120: 48
belonging to previous LOINC versions, and 72 new
codes created ad hoc for the scope. Many of the new
codes have the value Telehealth in the “System” axis,
reflecting the fact that the pandemic was really a
boost for remote healthcare services. There is no
prevalence of new codes assigned to a specific
medical specialty, demonstrating how the impact has
been truly global and has had, despite the very high
costs in terms of human lives, also important positive
implications for the progress of the healthcare sector
Covid-19 Impact on Standard Coding Systems Update
493
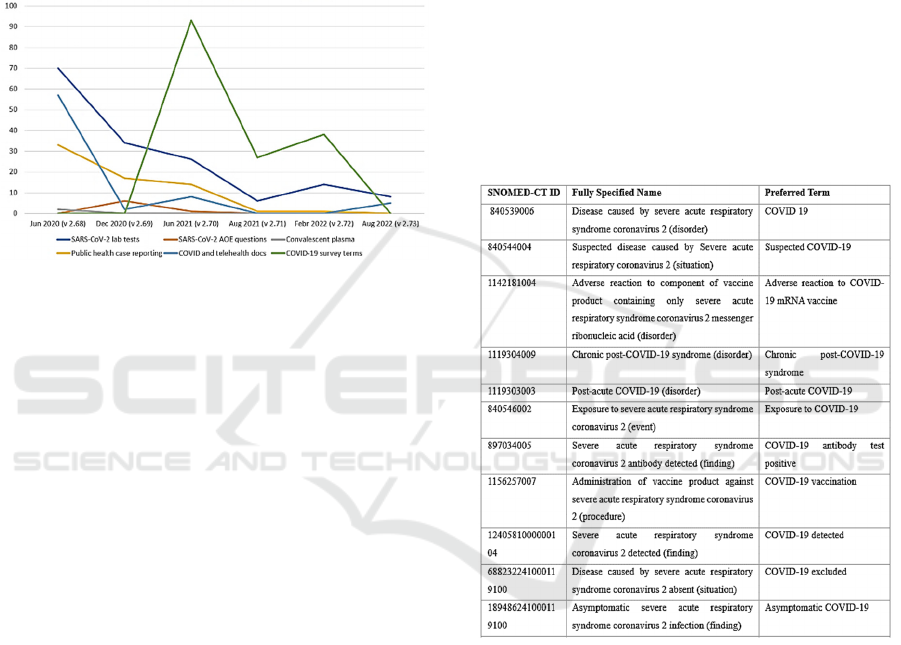
towards full digitalization. Figure 2 shows the trend
of LOINC Covid-19 related codes introduced over the
last three years.
Another aspect to consider is the need highlighted
by the pandemic to have aggregable and immediately
available comparable data, both for reporting and for
clinical / scientific analyses and studies, which in
those months underwent an extraordinary
acceleration to contain the virus and propose
solutions (e.g., vaccines).
Figure 2: Covid-19 related LOINC terms over the years.
To this end, the LOINC codes of the types of
clinical documents served to trace the flow of
documents in the federated EHRs, as in the case of
the Italian one (namely FSE), especially about
laboratory reports and vaccination cards. We have
above mentioned the two new types of documents
introduced during the pandemic, but it was also
required a further specification of the existing ones so
that their tracking in the FSE can immediately lead
back to Covid-19. To give an example, to track the
Covid swabs carried out daily in Italy, values for the
EventCodeList metadata have been inserted in AD to
specify that a LOINC DT Laboratory report contains
the result of the execution of an antigen or molecular
Covid-19 swab test or the outcome of a serological
Covid-19 test. They are LP418019-8 Covid-19
antigen swab test; LP417541-2 Covid-19 molecular
swab test; 96118-5 Qualitative Serological Test and
94503-0 Quantitative Serological Test. Similarly, the
DT identifying the Single vaccination card could be
combined with the values of the metadata
EventCodeList to specify the ATC code of the
administered vaccine type (J07BN Covid-19 vaccines
for the specific pandemic purpose, but admissible
values for this metadata include those belonging to
the WHO Anatomical Therapeutic Chemical
classification (ATC) coding system). This combined
use of the metadata of the AD made it possible to have
daily reports on the progress of the Country's
immunization process.
3.3 SNOMED CT Updates
As the global terminology for health, SNOMED CT
can serve as a common language for recording,
sharing, integrating and analyzing Covid-19 related
data elements, such as symptoms, risk factors and test
results. Following the advent of the pandemic,
SNOMED International quickly published a
comprehensive version of the Covid-19 concepts
including updated descriptions and mappings from
SNOMED CT to ICD-10. This version provided
physicians, researchers and administrators the most
up-to-date terminology needed to properly code,
analyse and address Covid-19 (Sutton, 2020). Some
of these concepts are illustrated in Table 1.
Table 1: Examples of SNOMED-CT Covid-19 related
codes.
A guide presenting concrete examples of subsets
of SNOMED CT codes that can be used to code
different types of Covid-19 data, including
symptoms, risk factors and test results, has been
published on the official SNOMED International
website. These subsets can be adapted according to
the specific needs of different health communities to
ensure health service delivery, pandemic monitoring,
international cooperation and retrospective data
analysis, as seen for the other coding systems.
SNOMED CT subsets have been organized into a
number of categories, based on groupings of data that
can be recorded together, for example the “Provider
and Facility Details” (i.e., Health profession, place of
care, personal protective equipment), “Clinical
Assessments”, “Test and Investigations”, etc.
HEALTHINF 2024 - 17th International Conference on Health Informatics
494

During the last three years also specific SNOMED
CT value sets related to Covid-19 have been created
by different organizations to be used in clinical
documents and for interoperability purposes (i.e., use
in EHRs, HL7 CDA documents). These includes the
“LIVD SARS-CoV2 Test Result Codes value set” or
the “COVID_19 (Antibody Substance in Lab
Results)” created respectively in May 2020 and in
May 2021 as Extensions. From a terminological point
of view, it is important to make clear the definition
and thus the purpose of use of these subsets /value
sets. As stated in (Rossander et. al., 2021) a subset is
“a collection of components from a terminology.
SNOMED CT subsets presented in RF2-format are
simple reference sets”. These subsets can be called
value sets in some use cases. A SNOMED CT subset
can include either SNOMED CT concepts, which can
be represented by any of the descriptions linked to
them, or specified descriptions. The use of SNOMED
CT Covid-19 related subsets and value sets was
important in the context of data exchange so in HL7
messages, where we can see the complementary use
with LOINC codes, LOINC used for coding the
testing method, and SNOMED CT used for coding
non-numeric answers. For example, in the case of the
LOINC code 94500-6 “SARS-CoV-2 (COVID-19)
RNA [Presence] in Respiratory system specimen by
NAA with probe detection”, SNOMED CT answer
codes indicated are 10828004 “positive”, 260385009
“negative”, 455371000124106 “invalid result”). This
use is motivated by the fact that both standards are
already commonly in place within laboratory
information systems.
3.4 ATC Updates
The Anatomical Therapeutic Chemical (ATC)
Classification System, a classification system that
classifies the active ingredients of drugs according to
the organ or system on which they act and their
therapeutic, pharmacological and chemical
properties, has the purpose to help monitoring drug
use and improving quality medication use. The ATC
is updated twice a year. With the advent of the Covid-
19 pandemic, ATC updates were provided for new
drugs and therapeutic indications associated with the
treatment of the virus. For example, codes were added
for Covid-19 vaccines, as well as for antiviral drugs
and other treatments used in the management of the
disease (it is the case of the code J07BN02 “Covid-
19, viral vector, non-replicating”.
Since the purpose of ATC is to codify the name
of the drug molecule, its additions relating to Covid-
19 have consequently had repercussions on specific
coding systems for the identification of drug packages
placed on the market (such as, for example, the AIC -
Autorizzazione all’Immissione in Commercio -
coding system for drugs on the Italian market or
FDA's National Drug Code for the USA). The
antivirals used for Covid-19 in ATC are: J05AE30
“nirmatrelvir and ritonavir” and J05AB16
“remdesivir”. The prescription and the use of
antivirals for the treatment of COVID-19 is subject to
close monitoring, thus enabling a rapid identification
of new possible safety information. Healthcare
professionals are required to report any adverse
reaction using the National Pharmacovigilance
Network.
4 DISCUSSION AND
CONCLUSIONS
Data coding plays a crucial role in managing and
analyzing complex information, especially in health
emergency situations such as those posed by the
Covid-19 pandemic outbreak. Data collection has
been essential to monitor the spread of the virus, track
cases, identify affected areas and evaluate the
effectiveness of control measures. Sometimes,
however, the effectiveness of the data depends on
multiple elements, including the completeness of the
information recorded and the accuracy with which it
was acquired. The analyzed material and the results
of the study, reported in Section 3, on one hand, made
immediately evident the considerable work done and
effort spent by the different working groups of each
SDO in the update process, above all in the case of
LOINC and SNOMED CT, and the contribution
given with the introduction of new codes to ensure an
accurate registration of the health conditions and a
continuity in tracking and monitoring relevant
information useful during the pandemic as well as for
their regular use in clinical documents.
On the other hand, results showed a very
heterogeneous trend of updates applied over the last
three years among the considered systems. It can be
seen, in fact, that classification systems with a
hierarchical structure (ICD ones and ATC) had a
slower and more complex updating process (implying
the introduction of new classes/codes) mainly for two
reasons: i) the branches of the classification have a
limited extensive possibility, so it is often difficult to
find further space in structured notational systems
(for example those who use numeric digits up to 9);
ii) in the event that notations are available, the
location of insertion of a new concept must be
Covid-19 Impact on Standard Coding Systems Update
495
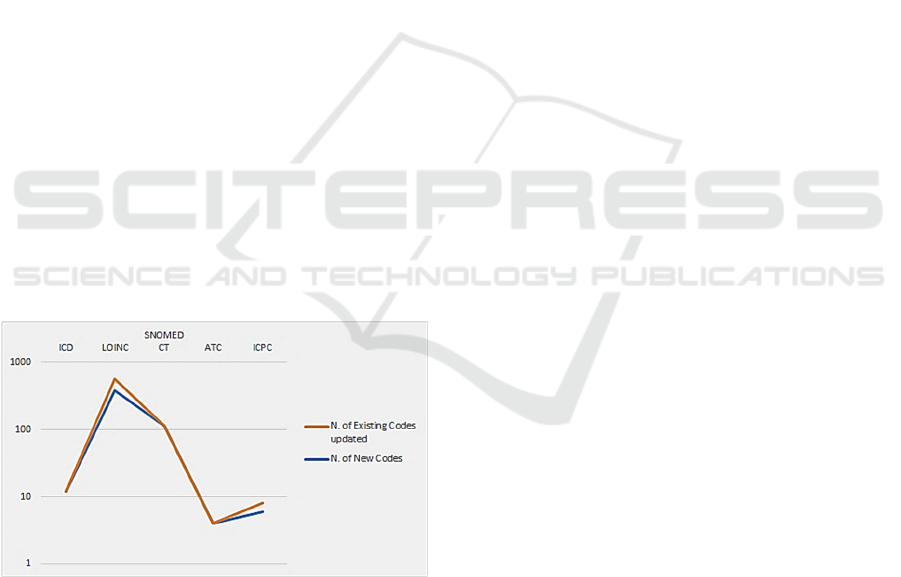
carefully evaluated by the appointed commissions, in
order to respect the hierarchical ordering of the
concepts and preserve the association of the new
concept/class to the semantic category covered by
chapters/ranges; iii) it is important to avoid semantic
inconsistencies considering also the inclusions and
exclusions criteria. On the contrary, in the case of
coding systems which present a linear structure and
are not based on decimal system for code notation,
i.e., SNOMED CT and LOINC it was easier to
introduce new codes. To confirm these observations,
Cimino, listing the desiderata for controlled medical
vocabularies (Cimino, 1998), and (Harrison, 2021
and Awaysheh, 2018) highlighted how flexibility is
the requirement that most supports the diffusion and
use of a coding system.
As shown in Figure 3, it was however common
for some of the systems to update existing codes by
proposing an adaptation of them in terms of
integrating so-called 'unspecified' classes and
modifying inclusion criteria. The analysis confirmed
that to make data truly useful, it is essential that they
are managed according to shared norms and
standards. Among KOSs, clinical coding systems are
used to structure data and uniquely identify pieces of
information. Nonetheless, the analysis of coded data
may encounter obstacles and potential alterations due
to the improper application of these systems.
Regarding this issue, the study confirmed that the
choice of coding systems must be well thought out
according to the context and purposes for which they
have to be used.
Figure 3: Covid-19 related updates trend.
For example, for hospital discharge letters, Italy
is still adopting, by regulations, ICD in its ninth
revision. Giving this, the national guidelines for
coding Covid-19 related discharge letters (Italian
Ministry of Health, 2020) indicate to use this version
to identify pathological conditions associated with the
virus and possible complications related to it, while
the use of ICD-10 is preferred for expressing the
cause of the death in death certificates. The Italian
Institute of Statistics in the COVID-19: interim report
on definition, certification and classification of
causes of death. (ISS et al., 2021), highlighted some
errors in the classification of the causes of death: i)
Lack of specificity: the initial cause of death should
be specific, and for example, a viral infection may be
an initial cause, but specifying the infectious agent
and type subsequent disease (e.g., "Covid-19
pneumonia"); ii) Intermediate causes: for example,
“pneumonia” could be an intermediate cause of death
because it can be caused by different infectious agents
or by inhaling a liquid or chemical substance; iii)
Illogical sequences: e.g., “chronic obstructive
pulmonary disease” cannot cause an infection,
although it may increase its criticality, so Covid-19
should be reported as the initial cause and the disease
as a consequence (ISS, 2021).
The study allowed also to check some preliminary
alignment among the considered coding systems with
respect to some generic semantic categories. In
particular, searching in UMLS among all the Covid-
19 related subsets and codes, we found exact
mappings for just 4 concepts: 1) Covid-19 (in
SNOMED CT “840539006” and ICD-10-CM
“U07.1”); 2) Post-acute Covid-19 condition (in
SNOMED CT “1119303003” and ICD-10-CM
“U09.9”), 3) Covid-19 vaccine (in SNOMED CT
“28531000087107” and ATC “J07BN”); and finally
Unvaccinated for Covid-19 / Vaccination not done (in
SNOMED CT “591191000124106” and ICD-10-CM
“Z28.310”. Although more mappings were expected
between ICD-10 and SNOMED CT, for example, it
was confirmed that the joint initiatives taken by the
SDOs to face with the Covid-19 coding and
identification emergency promoted the
complementary use of the coding systems and thus of
most of the new codes. Despite these observations,
the approach has shown some limitations in
comparing Covid-19 related subsets and value sets,
requiring a more accurate analysis and the use of
additional tools (e.g., the VSAC comparison tool of
the NLM) to improve results, including the flow.
In the healthcare domain, the use of coding and
classification systems sometimes has limitations due
to several factors, often related to a lack of
information about the systems themselves.
Healthcare professionals responsible for filling out
medical records are not able to take full advantage of
these systems lacking the proper training. Sometimes
physicians fill out clinical documents having no
access to complete information about patients’
medical history, including the incidence of Covid-19
on their clinical course. Accuracy in differencing
HEALTHINF 2024 - 17th International Conference on Health Informatics
496

deaths caused by Covid-19 from those caused by
other diseases with the ongoing Covid-19 infection
can be complex and subject to many variables. This
is especially true in a period with high prevalence of
death causing infections.
In conclusion, the main results of this study
highlighted the need for proper information collection
and management to have data structured according to
shared rules to be quickly accessible to the
appropriate government agencies. Despite advances
in the digital health domain there are still gaps in
semantic interoperability of these tools. One of the
lessons learnt from the study is that no matter how
good and widely used were coding systems in the
past, if there isn’t the required flexibility, any new
outbreak will, inevitably, bring different challenges
for their update, so the systems will never be perfect.
An improvement of this study will be the
improvement of the comparative analysis of the
versioning of the considered systems and the
adaptation of mappings between standards.
ACKNOWLEDGEMENTS
This study moved from the thesis of Dr. Maria
Francesca Littera (University of Calabria, Master’s
Degree in Management and Preservation of Digital
Documents), tutored by the authors. A special thank
is due for providing some materials and analyses.
REFERENCES
Awaysheh A, Wilcke J, Elvinger F, Rees L, Fan W,
Zimmerman K. (2018) A review of medical
terminology standards and structured reporting. J Vet
Diagn Invest. Jan;30(1):17-25. doi:
10.1177/1040638717738276.
Buchy, P., Buisson, Y., Cintra, O., Dwyer, DE., Nissen, M.,
Ortiz de Lejarazu, R., Petersen, E. (2021) Covid-19
pandemic: lessons learned from more than a century of
pandemics and current vaccine development for
pandemic control. Int J Infect Dis. Nov (112):300-317.
doi: 10.1016/j.ijid.2021.09.045.
Cimino JJ. (1998) Desiderata for controlled medical
vocabularies in the twenty-first century. Methods Inf
Med. Nov;37(4-5):394-403.
Dagobert, S. (2009). “Knowledge Organization Systems.
Overview”. P.2.
De Lissovoy G. (2020). Codes, Coding, and Covid-19.
Medical care, Volume 58, Issue 12, pp.1035–1036.
Dong X., Li J., Soysal E., Bian J., DuVall SL., Hanchrow
E., Liu H, Lynch KE., Matheny M., Natarajan K., Ohno
Ohno-Machado L., Pakhomov S., Reeves RM., Sitapati
AM., Abhyankar S., Cullen T., Deckard J, Jiang X,
Murphy R, Xu H. (2020) “COVID Covid-19 TestNorm:
A tool to normalize COVID Covid-19 testing names to
LOINC codes”. J Am Med Inform Assoc., 27(9):1437-
1442. doi:10.1093/jamia/ocaa145.
Frazier, P., Rossi-Mori, A., Dolin, R.H., Alschuler, L.,
Huff, S.M. (2001). The creation of an ontology of
clinical document names. In Stud Health Technol
Inform. 84(Pt 1):94-8.
Harrison JE, Weber S, Jakob R, Chute CG. (2021) ICD-11:
an international classification of diseases for the
twenty-first century. BMC Med Inform Decis Mak.
Nov 9;21(Suppl 6):206. doi: 10.1186/s12911-021-
01534-6.
Istituto Superiore di Sanità - ISS (2021). “Covid-19: interim
report on definition, certification and classification of
causes of death”. Updating Rapporto ISS Covid-19 n.
49/2020. Version of April 26, 2021. Working groups
ISS Cause di morte Covid-19 and Sovrintendenza
sanitaria centrale – INAIL, ISTAT 2021, ii, 16 p.
Rapporto ISS Covid-19 n. 10/2021 (in Italian).
Italian Ministry of Health (2020), General Directorate for
Health Planning, Linee Guida per la codifica della SDO
per casi affetti da malattia da SARS–COV–2 (COVID–
19), published on March 20, 2020.
Lin C-Y., Cheng C C-H., Lu P P-L., Shih D D-C., Hung C
C-T., Lo H H-H., Tsai M M-J., Hung J J-Y.. (2020).
Active surveillance for suspected Covid-19 cases in
inpatients with information technology. In Journal of
Hospital Infection, Volume 105, Issue 2, pp.197-199.
doi: 10.1016/j.jhin.2020.03.027.
Marwaha, J. S., Halamka, J.D., Brat, G.A, Gordon, W.J.
(2021). “Repurposing Billing and Administrative
Terminologies as Instruments of Public Health:
Lessons From The Covid-19 Pandemic”, in Health
Affairs Blog.
McDonald CJ, Huff SM, Suico JG, Hill G, Leavelle D,
Aller R, Forrey A, Mercer K, DeMoor G, Hook J,
Williams W, Case J, Maloney P. (2003) LOINC, a
universal standard for identifying laboratory
observations: a 5-year update. Clin Chem.
Apr;49(4):624-33. doi: 10.1373/49.4.624.
Rossander A, Lindsköld L, Ranerup A, Karlsson D. (2021).
A State-of-the Art Review of SNOMED CT
Terminology Binding and Recommendations for
Practice and Research. In Methods Inf Med. Dec;60(S
02):e76-e88. doi: 10.1055/s-0041-1735167.
Sutton K. (2020). Keeping up with Covid-19. In Wolkers
Kluwer, Health, March 25..
Varghees S. and Magoon V. (2020). Covid-19 diagnosis
coding explained in a flowchart, in FPM Journal,
Quick Tips, Jul 05.
Marcia Lei Zeng, Yi Hong, Julaine Clunis, Shaoyi He, L.P.
Coladangelo. (2020) Implications of Knowledge
Organization Systems for Health Information Exchange
and Communication during the COVID-19 Pandemic,
Data and Information Management, 4(3):148-170,
doi.org/10.2478/dim-2020-0009.
Covid-19 Impact on Standard Coding Systems Update
497
