
Combining Datasets with Different Label Sets for Improved Nucleus
Segmentation and Classification
Amruta Parulekar
1,∗
, Utkarsh Kanwat
1,∗
, Ravi Kant Gupta
1
, Medha Chippa
1
, Thomas Jacob
1
,
Tripti Bameta
2
, Swapnil Rane
2
and Amit Sethi
1
1
Indian Institute of Technology, Bombay, Mumbai, India
2
Tata Memorial Centre-ACTREC (HBNI), Mumbai, India
{medha6271, tripti.bameta, raneswapnil82}@gmail.com
Keywords:
Cell Nuclei, Classification, Histopathology, Segmentation.
Abstract:
Segmentation and classification of cell nuclei using deep neural networks (DNNs) can save pathologists’
time for diagnosing various diseases, including cancers. The accuracy of DNNs increases with the sizes
of annotated datasets available for training. The available public datasets with nuclear annotations and labels
differ in their class label sets. We propose a method to train DNNs on multiple datasets where the set of classes
across the datasets are related but not the same. Our method is designed to utilize class hierarchies, where the
set of classes in a dataset can be at any level of the hierarchy. Our results demonstrate that segmentation and
classification metrics for the class set used by the test split of a dataset can improve by pre-training on another
dataset that may even have a different set of classes due to the expansion of the training set enabled by our
method. Furthermore, generalization to previously unseen datasets also improves by combining multiple other
datasets with different sets of classes for training. The improvement is both qualitative and quantitative. The
proposed method can be adapted for various loss functions, DNN architectures, and application domains.
1 INTRODUCTION
Histopathology is practice of preparation and obser-
vation of tissue slides to visually identify signs and
grades of various diseases, including cancers. The vi-
sual features include nucleus to cytoplasm ratio, nu-
clear pleomorphism, and counts of various types of
cells. Usually histopathological examination relies on
nuclear details for estimating these features as the cell
(cytoplasmic) boundaries are not easy to identify in
hematoxylin and eosin (H&E) stained samples, which
is a staple of histolpathology. Automating instance
segmentation and classification of nuclei using deep
neural networks (DNNs), such as HoVerNet (Graham
et al., 2019) and StarDist (Schmidt et al., 2018) on
whole slide images (WSIs) acquired using scanners,
can bring efficiencies and objectivity to several types
of histological diagnostic and prognostic tasks.
Because DNNs can be scaled to generalize bet-
ter with more diverse and larger datasets, it is neces-
sary to accurately annotate and label multiple large
datasets for their training. In the last few years,
*
These authors contributed equally to this work
several annotated histological datasets have been re-
leased that differ in the sets of nuclear class la-
bels, magnification, source hospitals, scanning equip-
ment, organs, and diseases. For instance, while
the PanNuke dataset covers 19 organs with semi-
automated annotation of five nuclear classes – neo-
plastic, non-neoplastic epithelial, inflammatory, con-
nective, dead (Gamper et al., 2020); the MoNuSAC
covers four organs with the following four nuclear
classes – epithelial, lymphocytes, macrophages, and
neutrophils (Verma et al., 2021). While most of
this input diversity is beneficial to train generalized
DNNs, combined training across datasets with differ-
ent sets of class labels remains a challenge. Existing
methods to train DNNs on multiple datasets that dif-
fer class label sets are not satisfactory. For instance,
transfer (Yosinski et al., 2014) and multi-task learn-
ing (Zhang et al., 2014), do not train the last (few)
layer(s) of a DNN on more than one dataset.
We propose a method to train DNNs for instance
segmentation and classification over multiple related
datasets for the same types of objects that have differ-
ent class label sets. Specifically, we make the follow-
ing contributions. (1) We propose a method to mod-
Parulekar, A., Kanwat, U., Gupta, R., Chippa, M., Jacob, T., Bameta, T., Rane, S. and Sethi, A.
Combining Datasets with Different Label Sets for Improved Nucleus Segmentation and Classification.
DOI: 10.5220/0012380800003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 1, pages 281-288
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
281
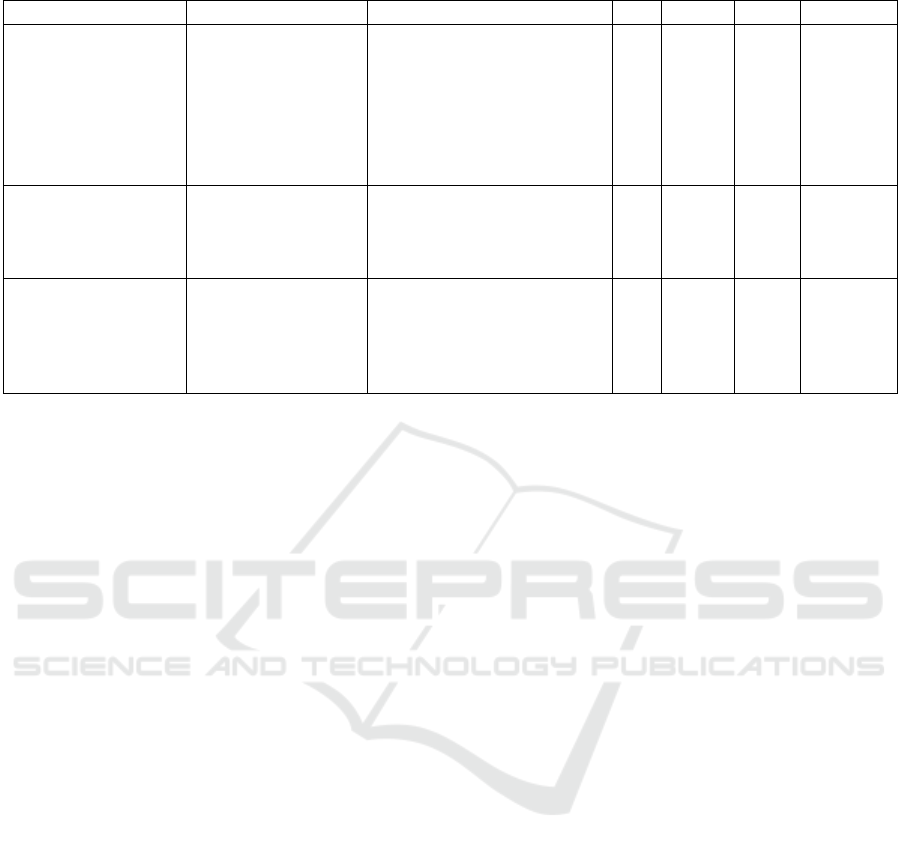
Table 1: Characteristics of notable nucleus segmentation and classification datasets.
Dataset Classes Organs Mag. Nuclei Images Img. Size
PanNuke
(Gamper et al., 2020)
5: Inflammatory,
Neoplastic,
Dead, Connective,
Non-neoplastic
Epithelial
19: Bladder, Ovary, Pancreas,
Thyroid, Liver, Testis,
Prostrate, Stomach, Kidney,
Adrenal gland, Skin,
Head & Neck, Cervix,
Lung, Uterus, Esophagus,
Bile-duct, Colon, Breast
40x 216,345 481 224x224
MoNuSAC
(Verma et al., 2021)
4: Epithelial,
lymphocytes,
macrophages,
neutrophils
4: Breast, Kidney,
Liver, prostrate
40x
46,909 310
82x35
to
1422x2162
CoNSeP
(Graham et al., 2019)
7: Healthy Epithelial,
Inflammatory,
Muscle, Fibroblast,
Malignant Epithelial,
Endothelial,Other
1: Colon
40x
24,319 41 1000x1000
ify a wide variety of loss functions used for segmen-
tation and classification. (2) The method is applica-
ble whenever the class label sets across the datasets
can be expressed as a part of a common coarse-to-
fine class hierarchy tree. That is, the method can
jointly utilize multiple datasets of the same types of
objects wherein some datasets may have labels for
finer sub-classes while others may have labels for
coarser super-classes, or a mix of these, from the same
class hierarchy tree. Apart from this type of relation
among datasets, the method has no other constraints.
That is, it can be used to train a wide variety of DNNs
for instance segmentation and classification for var-
ious types of objects of interest, although we used
the segmentation of nuclei in histopathology using
StarDist (Schmidt et al., 2018) as a case study. (3)
We demonstrate quantitative and qualitative improve-
ments in nuclear segmentation and classification test
accuracy using the proposed method to train on multi-
ple datasets with different class label sets. (4) We also
show that thus using multiple datasets also improves
domain generalization on a previously unseen dataset.
2 RELATED WORKS
In this section, we review nucleus segmentation
datasets and methods, and previous attempts to com-
bine knowledge from multiple datasets.
Over the last few years, several datasets with care-
ful annotations and labeling of cell nuclei have been
released to the public to enable research on better in-
stance segmentation and classification models. Some
notable datasets are shown in Table 1. These datasets
meet our goals as they contain images with nuclear
details at 40x magnification and labels for nuclei from
multiple classes, unlike, for example, MoNuSeg (Ku-
mar et al., 2017) or CryoNuSeg (Mahbod et al., 2021).
Research on nucleus segmentation and classifi-
cation methods has recently seen the development
of combination architectures (fusion of multiple net-
works) and specialized architectures. For instance,
HoVerNet (Graham et al., 2019) was proposed to
predict whether a pixel location is inside a nu-
cleus and its horizontal and vertical distances from
the nuclear boundary. This concept has been gen-
eralized to predict multi-directional distance using
StarDist (Schmidt et al., 2018). These architectures
are specifically designed for histological images with
overcrowded nuclei and have demonstrated state-of-
the-art results compared to previous methods.
In order to combine knowledge from multiple
datasets, transfer and multi-task learning have been
proposed for natural and medical images. For in-
stance, (Reis and Turk, 2023) proposes a transfer
learning technique using the MedCLNet database.
DNNs were pre-trained through the proposed method
and were used to perform classification on the
colorectal histology MNIST dataset. The GSN-
HVNET(Zhao et al., 2023) was proposed with an
encoder-decoder structure for simultaneous segmen-
tation and classification of nuclei, and was pre-trained
on the PanNuke dataset.
Although coarse-to-fine class structure has been
exploited for knowledge transfer in other domains (Li
et al., 2019), it has not been used in medical datasets
for increasing the data for training or domain general-
ization.
All the methods described so far have only dealt
BIOIMAGING 2024 - 11th International Conference on Bioimaging
282
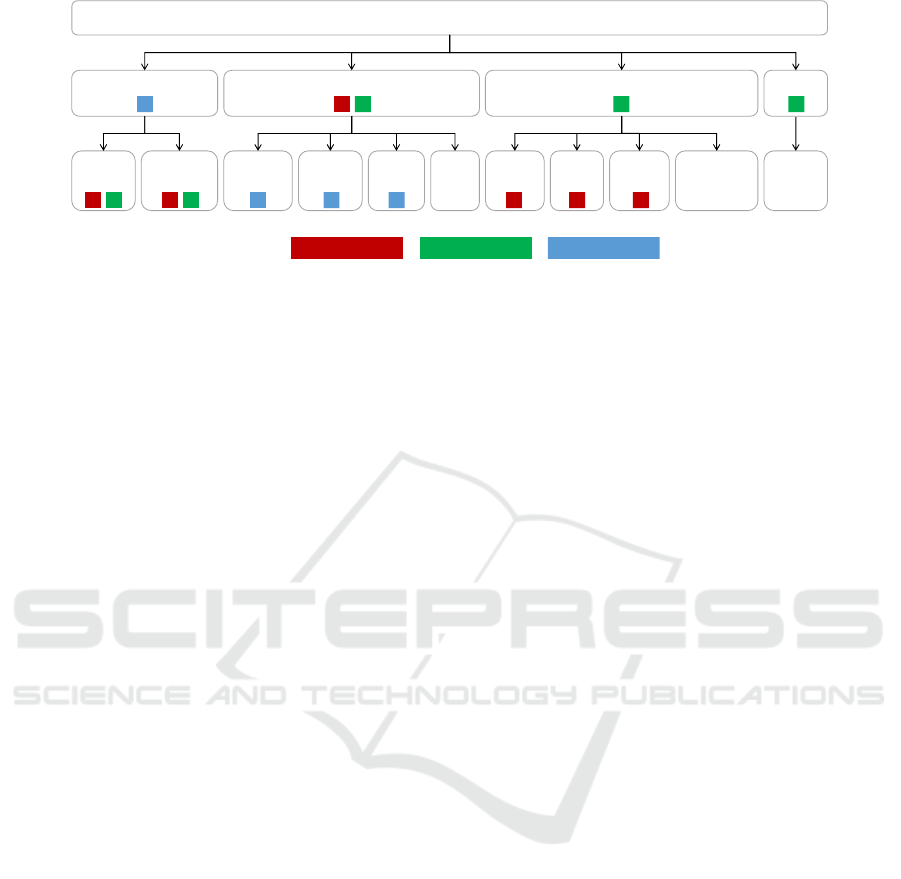
Cell Nuclei
Epithelial / Glandular Immune / Inflammatory Stromal / Connective Other
Normal
epith.
Tumoral /
Neoplastic
Lympho-
cyte
Macro-
phage
Other
Immu.
Necrotic
Muscle
Fibro-
blast
Endo-
thelial
Other
connective
Neutro-
phil
CoNSeP classMoNuSAC class PanNuke classLegend
Figure 1: Hierarchy of nucleus classes and their correspondence to the three label sets of the datasets used in this study.
with the scenario of carrying out segmentation and
classification by splitting the same dataset into train-
ing and testing, or using the same set of classes across
training and testing. At best, transfer learning was
carried out where only the lower pretrained layers
were retained and new upper layers were randomly
initialized and trained on target datasets. There are
no loss functions or training methods that can train
an all layers of a DNN on multiple datasets for cross-
domain (dataset) generalization for segmentation and
classification.
3 PROPOSED METHOD
We propose a method to train DNNs for segmenta-
tion and classification on multiple datasets with re-
lated but potentially different class label sets. We as-
sume that the class label sets across the datasets are
different cuts of the same class hierarchy tree. Within
each dataset, the class labels are mutually exclusive,
but need not be collectively exhaustive. An example
of a class hierarchy tree with different cuts for labels
for three different datasets is given in Figure 1, where
nuclei can be divided into four super-classes, which in
turn can be divided into 11 sub-classes. Deeper and
wider hierarchies can also be used. Class label sets
that are not a part of a common class hierarchy tree
are out of the scope of this work.
Our key idea is to modify a class of loss functions
whose computation involves sums over predicted and
ground truth class probability terms in conjunction
with sums over instances or pixels. This descrip-
tion covers a wide array of loss functions, includ-
ing cross entropy, Dice loss (Sudre et al., 2017), fo-
cal loss (Lin et al., 2017), Tversky loss, focal Tver-
sky loss (Abraham and Khan, 2018). We propose to
sum the predicted probabilities of fine-grained sub-
classes when the class label is only available at their
coarser super-class level. The set of finer sub-classes
to be combined using this method of loss computation
can even dynamically change from dataset-to-dataset,
epoch-to-epoch, batch-to-batch, or even instance-to-
instance. To keep things simple, we first train the
model on one dataset for a few epochs, and then train
it on a second dataset for the remaining epochs.
This method is also applicable to any DNN archi-
tecture or application domain (e.g., natural images)
that can be trained using these losses. As a case
study, we use it to modify cross entropy and focal
Tversky loss functions (Abdolhoseini et al., 2019)
to train a UNet-based StarDist DNN (Schmidt et al.,
2018) for H&E-stained histopathology nucleus seg-
mentation and classification on MoNuSAC (Verma
et al., 2021), PanNuke(Gamper et al., 2020), and
CoNSeP(Graham et al., 2019) datasets.
Although this method can be extended to multiple
levels, for simplicity of explanation we will assume
that a class label can be at one of the two levels –
a super-class or a sub-class. We design a neural ar-
chitecture that makes predictions at the finest level of
the hierarchy, which is the set of all sub-classes (plus
background) in this case. When the label for a training
instance is available at the super-class level, we add
the predicted probabilities of its sub-classes, and up-
date their weights with an equal gradient, as should be
done backwards of a sum node. This way, the weights
leading to the prediction of all sub-classes are trained
even when only the super-class label is available. The
gradient and output obtained from this approach is at
the finest (sub-class) level, but we interpret the results
for a dataset only for its corresponding training la-
bel set. That is, we do not assess sub-class level per-
formance when only super-class labels are available,
even though we train the DNN to predict at the sub-
class level. On the other hand, when we come across
a training instance where a sub-class label is avail-
able, we skip the sum-based merging of probability
masses. In this case, class-specific weight update and
the interpretation of predictions proceeds in the usual
Combining Datasets with Different Label Sets for Improved Nucleus Segmentation and Classification
283

fashion.
Consider the cross entropy loss for a fixed set of
class labels:
L
CE
= −
n
∑
i=1
c
∑
j=1
t
i j
log(y
i j
), (1)
where n is the number of training instances, c is the
number of classes, t
i j
are one-hot labels, and y
i j
are
the predicted class probabilities such that ∀i
∑
j
t
i j
=
1,
∑
j
y
i j
= 1. In case a subset of classes belong to
a super-class k denoted by j ∈ S
k
, then we modify
Equation 1 as follows:
L
MCE
= −
n
∑
i=1
m
∑
k=1
t
ik
logy
ik
= −
n
∑
i=1
m
∑
k=1
t
ik
log
∑
j∈S
k
y
i j
!
,
(2)
where t
ik
is a binary indicator label for the super-class
k, and m is the size of the class label set. That is,
t
ik
=
∑
j∈S
k
t
i j
, but the individual terms t
i j
may not
be known in the given labels. The class probability
predictions y
i j
can remain at the finest level across
datasets, while the labels t
ik
can be at the finest or
a coarser level. Just as it was the case for the la-
bels, the predicted probabilities also naturally satisfy
the relation y
ik
=
∑
j∈S
k
y
i j
. As is clear from Equa-
tion 2, that although for notational simplicity, the sum
over classes runs at the super-classes enumerated by
k at the same level, the modification applies indepen-
dently to each branch of the class hierarchy tree (see
Figure 1 for an example), as was done in our im-
plementation. Additionally, it is also clear that the
method can be extended to deeper and wider hierar-
chy trees with label sets that are arbitrary cuts of the
tree.
We next consider a slightly more complex loss –
the focal Tversky loss (Abraham and Khan, 2018):
L
FT
=
n
∑
i=1
1 −
∑
c
j=1
(t
i j
y
i j
+ ε)
α
∑
c
j=1
t
i j
+ (1 − α)
∑
c
j=1
y
i j
+ ε
!
γ
,
(3)
where ε is a small constant to prevent division by 0,
and α > 0, γ > 0 are hyper-parameters. Following the
same principles as used to propose the loss in Equa-
tion 2, we now propose a modified focal Tversky loss:
L
MFT
=
n
∑
i=1
1 −
∑
m
k=1
(t
ik
∑
j∈S
k
y
i j
+ ε)
α
∑
m
k=1
t
ik
+ (1 − α)
∑
m
k=1
∑
j∈S
k
y
i j
+ ε
γ
.
(4)
Once again, it is clear that L
MFT
can also be modi-
fied to be applied independently to each branch and
sub-branch of a class hierarchy tree, including la-
bels at different levels of the tree that are in different
branches.
In our implementation of nuclear instance seg-
mentation and classification, we used a positive com-
bination of L
MCE
and L
MFT
.
4 EXPERIMENTS AND RESULTS
We tested two hypotheses in our experiments. Firstly,
we hypothesized that using the proposed method, pre-
training on a related source dataset A with class la-
bels derived from the same class hierarchy tree as that
of a target dataset B can improve the instance seg-
mentation and classification metrics on the held-out
test cases of dataset B compared to training only on
dataset B. Secondly, we hypothesized that using the
proposed method, domain generalization to a previ-
ously unseen dataset C can improve when trained on
datasets A and B, as compared to training only on
dataset B, where the label sets for the three datasets
may be different but are derived from the same class
hierarchy tree. For experiments to confirm either hy-
potheses, we did not discard the last (few) layer(s) af-
ter training on dataset A, as is done in transfer learn-
ing and multi-task learning. We trained, retained, and
re-trained the same last layer by using the proposed
adaptive loss functions.
Due to their large size, 40x magnification with
clear nuclear details, and a minimal overlap in nuclear
classes, we selected three datasets for our experiments
– the Multi-organ Nuclei Segmentation And Classifi-
cation (MoNuSAC) (Verma et al., 2021) dataset, the
PanNuke dataset (Gamper et al., 2020), and the Col-
orectal Nuclear Segmentation and Phenotypes (CoN-
SeP) dataset (Graham et al., 2019). More details
about these datasets can be found in Section 2.
Due to its integrated evaluation of instance seg-
mentation and classification, we used panoptic quality
(PQ) (Kirillov et al., 2019) to assess our results.
This metric is now widely used as the primary
metric in papers such as (Weigert and Schmidt, 2022)
for assessing nucleus segmentation and classification.
(Kirillov et al., 2019) shows rigorous experimen-
tal evaluation that demonstrates the variation of PQ,
along with its comparison to other metrics like inter-
section over union (IoU) and average precision(AP)
We used an instance segmentation and classifica-
tion architecture used in (Weigert and Schmidt, 2022)
(which is a modification of the StarDist (Schmidt
et al., 2018) model) because it has specific train-
ing procedures and post-processing steps for H&E-
stained histology images. It also gives enhanced ob-
ject localization, leading to higher precision in seg-
mentation, especially of overlapping or closely lo-
cated nuclei.
BIOIMAGING 2024 - 11th International Conference on Bioimaging
284
Patches of size 256x256 were extracted from each
dataset. Smaller images were appropriately padded.
Some patches were overlapping while others were
cut-off to fit within 256x256. Images had three chan-
nels corresponding to RGB. The ground truth masks
had two channels – the first was the instance segmen-
tation map ranging from 0 to number of nuclei and the
second was the classification map ranging from zero
to number of classes in the dataset’s class label set.
To combat staining variability, random brightness,
hue, and saturation augmentations were performed
on the images. To combat class imbalance, geo-
metric augmentations (90 degree rotations and flips)
and elastic augmentations were performed more fre-
quently on the less populated classes.
The optimizer used was Adam. We monitored
the validation loss for early stopping. Once we fin-
ished training the model on one dataset (dataset A)
using one instantiation of the modified loss function
for a few epochs, we further trained (finetuned) the
same model - without adding or removing any lay-
ers or weights - on the second dataset (dataset B) for
a few more epochs by adapting the loss to the sec-
ond day. The method is flexible enough to take train-
ing instances from multiple datasets down to batch-
level, but we simplified the procedure to keep the
training consistent at an episode (group of epochs)
level, where only one dataset was used for training
per episode.
4.1 Test Dataset Results
Table 2 summarizes the results of testing the first
hypothesis that the test results can improve by
pre-training on another dataset using the proposed
method. Pre-training on another dataset and then fine-
tuning for a small number of epochs (Eps) on our tar-
get dataset consistently gave better results for all three
target datasets as compared to training only on the tar-
get dataset. Additionally, these results compare favor-
ably with the state-of-the-art for training and testing
on various splits of a single dataset (Schmidt et al.,
2018).
Table 3 shows that we have achieved state-of-the-
art panoptic quality value on the CoNSeP dataset with
our proposed method, pretrained on PaNNuke and
fine-tuned on CoNSeP. For MoNuSAC, we find that
our method surpasses the Unet (Ronneberger et al.,
2015), DeepLabV3 (Chen et al., 2017) and PSPNet
(Zhao et al., 2017) models, which have PQ values of
0.350, 0.396, and 0.387, respectively.
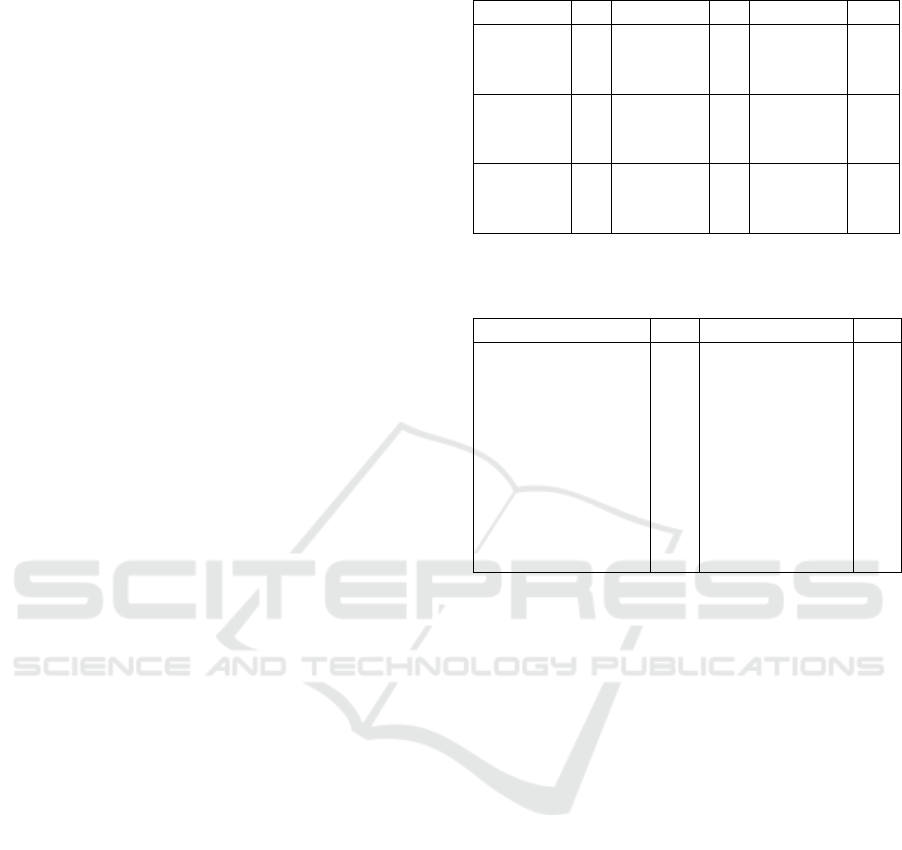
A sample of qualitative results shown in Figure 2
also shows better overlap between predicted nuclei
and annotations for test images when multiple train-
Table 2: Quantitative results on test splits.
Pre-Train Eps Fine-tune Eps Test PQ
CoNSeP 100 - - CoNSeP 0.540
MoNuSAC 175 CoNSeP 75 CoNSeP 0.555
PanNuke 250 CoNSeP 75 CoNSeP 0.571
MoNuSAC 175 - - MoNuSAC 0.579
CoNSeP 100 MoNuSAC 130 MoNuSAC 0.587
PanNuke 250 MoNuSAC 130 MoNuSAC 0.602
PanNuke 250 - - PanNuke 0.610
CoNSeP 100 PanNuke 187 PanNuke 0.605
MoNuSAC 175 PanNuke 187 PanNuke 0.610
Table 3: Comparison of results with existing models for
CoNSeP dataset. Best result is highlighted as bold.
Model PQ Model PQ
UNet++
(Zhou et al., 2018)
0.405
DSCANet
(Ye et al., 2024)
0.426
DiffMix
(Oh and Jeong, 2023)
0.505
GradMix
(Doan et al., 2022)
0.504
Hovernet
(Graham et al., 2019)
0.532
SMILE
(Pan et al., 2023)
0.530
DRCANet
(Dogar et al., 2023)
0.546
DeepLabV3+
(Chen et al., 2017)
0.373
Stardist
(Schmidt et al., 2018)
0.540 Our Method 0.571
ing datasets are used for training using our method as
compared to training on a single dataset.
It is worth noting that the improvement is more
pronounced when the pretraining dataset is more gen-
eralized and has a super-set of classes and organs as
compared to the target dataset. For example, the Pan-
Nuke dataset has most of the cell classes present in it.
Thus, pre-training on PanNuke and then fine-tuning
on other more specialized datasets gives significant
improvement in the predictions on those datasets.
Based on this observation and reasoning, the most
general dataset in terms of labels can be chosen for
pre-training by surveying the classes of the available
open source datasets.
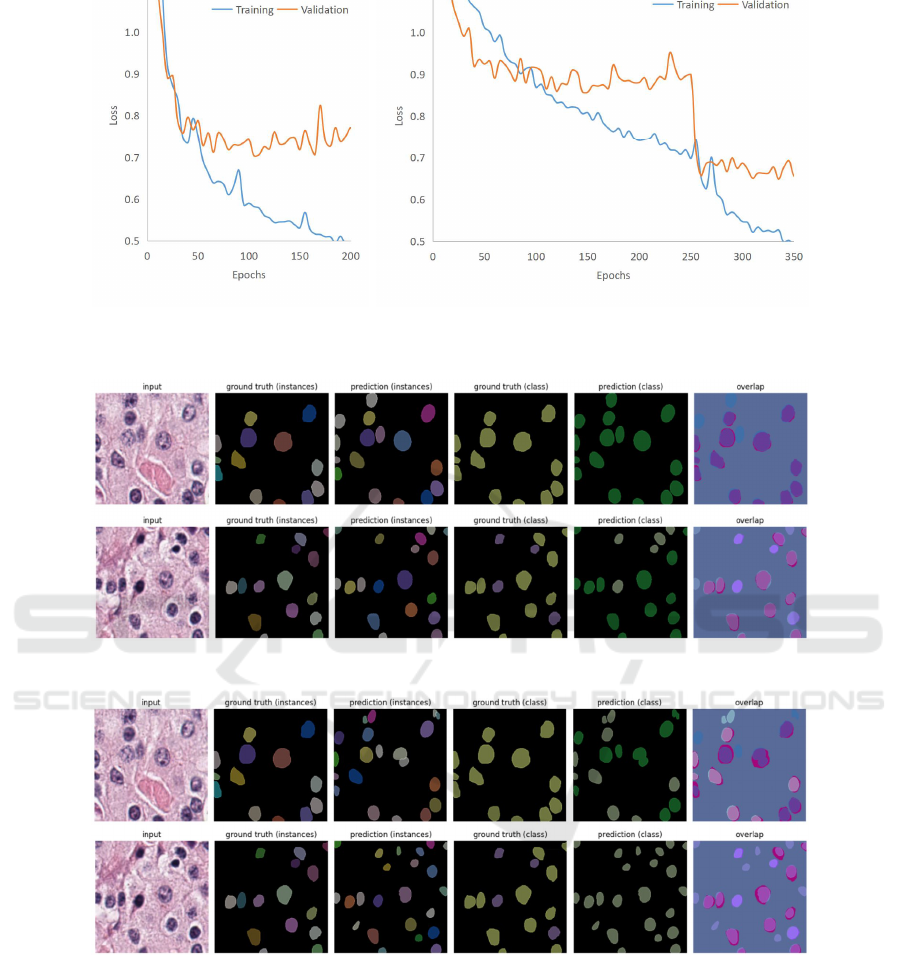
4.2 Evolution of Loss
Figure 3 shows an example of evolution of the
losses as the training progressed for the MoNuSAC
dataset as the target dataset. When trained only on
MoNuSAC (case (a)), the model starts to overfit as
it can be seen that the validation loss starts to in-
crease. However, when pretrained on PanNuke (case
(b)), the validation loss shows a marked further drop
when the dataset is switched to the training subset of
MoNuSAC as compared to that of case (a).
Combining Datasets with Different Label Sets for Improved Nucleus Segmentation and Classification
285
(a) Predictions on MoNuSAC of the model pretrained on PanNuke for 250 epochs followed by fine-
tuning on MoNuSAC for 130 epochs.
(b) Predictions on MoNuSAC of the model trained on MoNuSAC for 175 epochs before overfitting
starts to occur.
Figure 2: A qualitative sample of test split results.
4.3 Domain Generalization
To test that domain generalization can improve by
training on multiple datasets, we trained the model
on the first dataset while monitoring its validation
loss to prevent overfitting. After this, we fine-tuned
the model on a second dataset. Then we tested on a
third dataset, which did not contribute to the training
at all. Table 4 summarizes the results of this exper-
iment. Pre-training on a dataset and then fine-tuning
for a small number of epochs (Eps) on another dataset
gives better results on an unseen dataset as compared
to training only on the first dataset. Thus, our model
is able to consolidate the knowledge of two datasets.
Table 4: Quantitative results for domain generalization.
Pre-Train Eps Fine-tune Eps Test PQ
CoNSeP 100 - - MoNuSAC 0.433
CoNSeP 100 PanNuke 62 MoNuSAC 0.563
CoNSeP 100 - - PanNuke 0.433
CoNSeP 100 MoNuSAC 43 PanNuke 0.434
MoNuSAC 175 - - CoNSeP 0.344
MoNuSAC 175 PanNuke 62 CoNSeP 0.449
MoNuSAC 175 - - PanNuke 0.396
MoNuSAC 175 CoNSeP 25 PanNuke 0.405
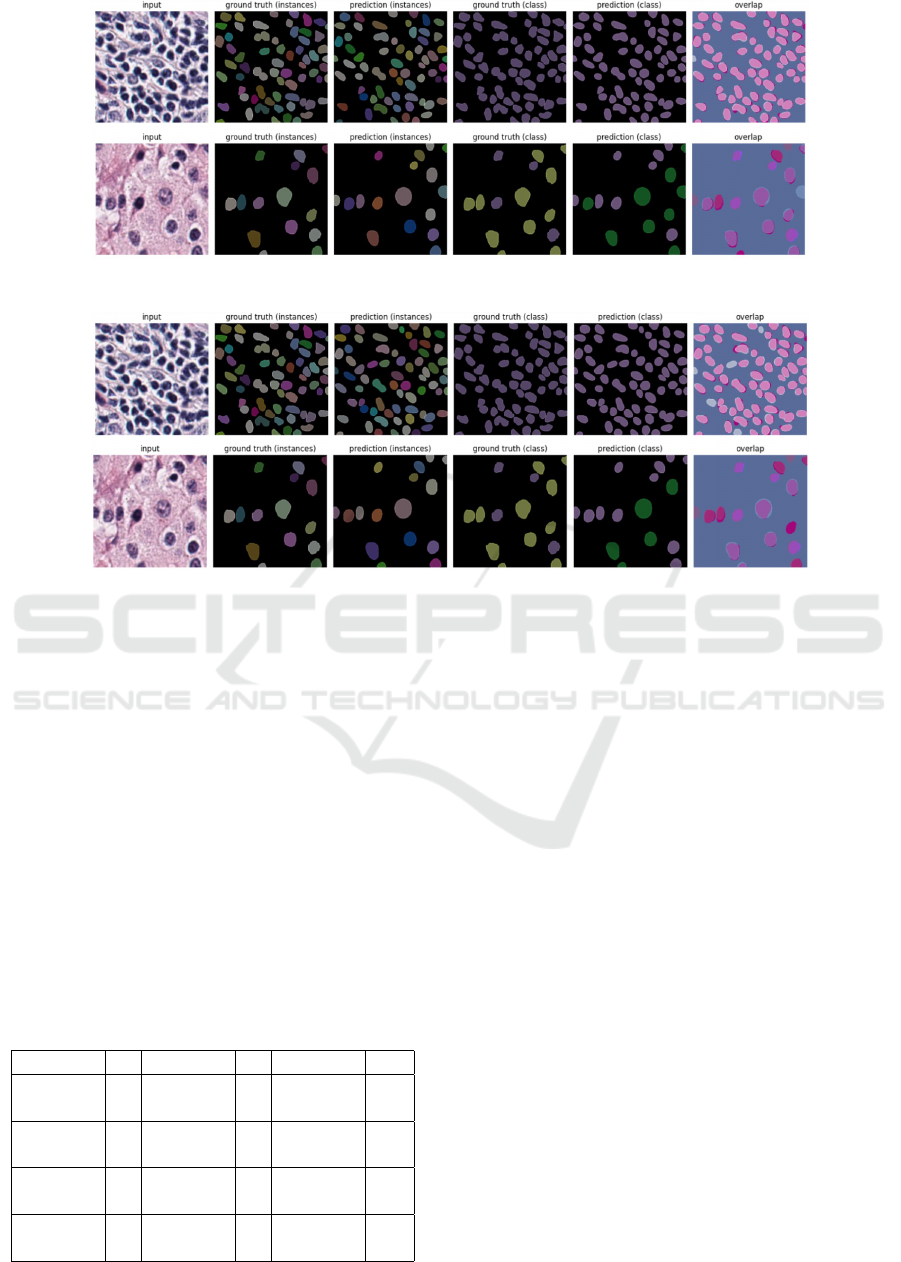
A sample of qualitative results shown in Figure 4
also shows better overlap between predicted nuclei
and annotations for images from an unseen dataset
when multiple training datasets are used for training
using our method as compared to training on a single
dataset.
We can observe that a more pronounced improve-
ment occurs when the fine-tuning dataset is more gen-
eralized and has a super-set of classes and organs as
compared to the other datasets. We must take care
not to use the most generalized dataset (with a su-
perset of classes) for pretraining because on finetun-
ing with a more specialized dataset, the model loses
its accuracy on the unseen dataset instead of benefit-
ting from the fine-tuning. For example, CoNSeP and
MoNuSAC are more specialized datasets with classes
that have less overlap, but their classes are both sub-
sets of the classes present in PanNuke. In this case,
using CoNSeP to finetune the model that was pre-
trained on PanNuke will lead to decreased perfor-
mance on MoNuSAC. Now the most general dataset
in terms of labels can be chosen by surveying the
classes of the available open source datasets.
BIOIMAGING 2024 - 11th International Conference on Bioimaging
286
(a) Training on MoNuSAC, testing on MoNuSAC (b) Pretraining on PanNuke, finetuning on MoNuSAC, testing on MoNuSAC
Figure 3: Evolution of training and validation losses for testing on MoNuSAC when (a) trained only on MoNuSAC leading
to overfitting, and (b) when pretrained on PanNuke followed by finetuning on MoNuSAC after 250 epochs.
(a) Predictions on MoNuSAC of the model pre-trained on CoNSeP for 100 epochs followed by fine-
tuning on PanNuke for 62 epochs.
(b) Predictions on MoNuSAC of the model trained on CoNSeP for 100 epochs, before overfitting
starts to occur.
Figure 4: A qualitative sample of domain generalization results.
5 DISCUSSION
We have proposed a method to train a neural network
on multiple datasets with different class labels for seg-
mentation and classification. Unlike transfer or multi-
task learning, even the last layer is shared between
datasets. We achieved this by first creating a hierar-
chical class label tree to relate the class label sets of
different datasets to each other as various cuts of the
same tree. We then devised a way to combine the
losses of the sub-classes, allowing us to train models
sequentially on multiple datasets even when the labels
are available at a coarser super-class level for some of
the classes and datasets. We show improved results
Combining Datasets with Different Label Sets for Improved Nucleus Segmentation and Classification
287

on test splits and unseen datasets. Our technique can
easily be applied to other DNN architectures, appli-
cation domains and tasks (such as, object detection);
more importantly, it can also be adapted to other loss
functions.
REFERENCES
Abdolhoseini, M. et al. (2019). Segmentation of heavily
clustered nuclei from histopathological images. Sci-
entific reports, 9(1):4551.
Abraham, N. and Khan, N. M. (2018). A novel focal tversky
loss function with improved attention u-net for lesion
segmentation.
Chen, L.-C. et al. (2017). Rethinking atrous convolution for
semantic image segmentation.
Doan, T. N. N. et al. (2022). Gradmix for nuclei segmenta-
tion and classification in imbalanced pathology image
datasets.
Dogar, G. M. et al. (2023). Attention augmented distance
regression and classification network for nuclei in-
stance segmentation and type classification in histol-
ogy images. Biomedical Signal Processing and Con-
trol, 79:104199.
Gamper, J. et al. (2020). Pannuke dataset extension, insights
and baselines.
Graham, S. et al. (2019). Hover-net: Simultaneous seg-
mentation and classification of nuclei in multi-tissue
histology images.
Kirillov, A. et al. (2019). Panoptic segmentation. In Pro-
ceedings of the IEEE/CVF conference on computer vi-
sion and pattern recognition, pages 9404–9413.
Kumar, N. et al. (2017). A dataset and a technique for
generalized nuclear segmentation for computational
pathology. IEEE transactions on medical imaging,
36(7):1550–1560.
Li, Z. et al. (2019). Exploiting coarse-to-fine task transfer
for aspect-level sentiment classification. In Proceed-
ings of the AAAI conference on artificial intelligence,
volume 33, pages 4253–4260.
Lin, T.-Y. et al. (2017). Focal loss for dense object detec-
tion. In Proceedings of the IEEE International Con-
ference on Computer Vision (ICCV).
Mahbod, A. et al. (2021). Cryonuseg: A dataset for
nuclei instance segmentation of cryosectioned h&e-
stained histological images. Computers in biology and
medicine, 132:104349.
Oh, H.-J. and Jeong, W.-K. (2023). Diffmix: Diffu-
sion model-based data synthesis for nuclei segmenta-
tion and classification in imbalanced pathology image
datasets.
Pan, X. et al. (2023). Smile: Cost-sensitive multi-task
learning for nuclear segmentation and classification
with imbalanced annotations. Medical Image Anal-
ysis, 88:102867.
Reis, H. C. and Turk, V. (2023). Transfer learning approach
and nucleus segmentation with medclnet colon cancer
database. Journal of Digital Imaging, 36(1):306–325.
Ronneberger, O. et al. (2015). U-net: Convolutional
networks for biomedical image segmentation. In
Medical Image Computing and Computer-Assisted
Intervention–MICCAI 2015: 18th International Con-
ference, Munich, Germany, October 5-9, 2015, Pro-
ceedings, Part III 18, pages 234–241. Springer.
Schmidt, U., Weigert, M., et al. (2018). Cell detection with
star-convex polygons. In Medical Image Computing
and Computer Assisted Intervention – MICCAI 2018,
pages 265–273. Springer International Publishing.
Sudre, C. H. et al. (2017). Generalised dice overlap
as a deep learning loss function for highly unbal-
anced segmentations. In Deep Learning in Medical
Image Analysis and Multimodal Learning for Clini-
cal Decision Support: Third International Workshop,
DLMIA 2017, and 7th International Workshop, ML-
CDS 2017, Held in Conjunction with MICCAI 2017,
Qu
´
ebec City, QC, Canada, September 14, Proceed-
ings 3, pages 240–248. Springer.
Verma, R. et al. (2021). Monusac2020: A multi-organ nu-
clei segmentation and classification challenge. IEEE
Transactions on Medical Imaging, 40(12):3413–3423.
Weigert, M. and Schmidt, U. (2022). Nuclei instance seg-
mentation and classification in histopathology images
with stardist. In 2022 IEEE International Symposium
on Biomedical Imaging Challenges (ISBIC). IEEE.
Ye, Z. et al. (2024). Dsca-net: Double-stage codec attention
network for automatic nuclear segmentation. Biomed-
ical Signal Processing and Control, 88:105569.
Yosinski, J. et al. (2014). How transferable are features in
deep neural networks? Advances in neural informa-
tion processing systems, 27.
Zhang, Z. et al. (2014). Facial landmark detection by
deep multi-task learning. In Computer Vision–ECCV
2014: 13th European Conference, Zurich, Switzer-
land, September 6-12, 2014, Proceedings, Part VI 13,
pages 94–108. Springer.
Zhao, H. et al. (2017). Pyramid scene parsing network. In
Proceedings of the IEEE conference on computer vi-
sion and pattern recognition, pages 2881–2890.
Zhao, T. et al. (2023). Gsn-hvnet: A lightweight, multi-task
deep learning framework for nuclei segmentation and
classification. Bioengineering, 10(3):393.
Zhou, Z. et al. (2018). Unet++: A nested u-net architec-
ture for medical image segmentation. In Deep Learn-
ing in Medical Image Analysis and Multimodal Learn-
ing for Clinical Decision Support: 4th International
Workshop, DLMIA 2018, and 8th International Work-
shop, ML-CDS 2018, Held in Conjunction with MIC-
CAI 2018, Granada, Spain, September 20, 2018, Pro-
ceedings 4, pages 3–11. Springer.
BIOIMAGING 2024 - 11th International Conference on Bioimaging
288
