
ResNet-101 Empowered Deep Learning for Breast Cancer Ultrasound
Image Classification
Agnesh Chandra Yadav
1 a
, Maheshkumar H. Kolekar
1 b
and Mukesh Kumar Zope
2
1
Department of Electrical Engineering, IIT Patna, Patna, India
2
Department Medical Physics, IGIMS Patna, Patna, India
Keywords:
Breast Cancer Classification, Ultrasound Images, Deep Learning, ResNet-101.
Abstract:
In the modern era, accurate breast cancer classification plays a crucial role in early detection and treatment
planning. This article introduces a modified ResNet-101 architecture tailored specifically for classifying breast
cancer using ultrasound images. The ultrasound images undergo pre-processing before passing through our
adapted ResNet-101 model, which includes the integration of shortcut connections to enhance gradient stabil-
ity and deep structure adaptability for effective learning and classification. The dataset comprises 780 images
categorized into normal, benign, and malignant cases. To address class imbalance, data augmentation tech-
niques are employed, enriching diversity and enhancing modeling precision. The proposed model achieves
exceptional performance, boasting precision, recall, F1-score, and accuracy values of 0.9855, 0.9677, 0.9756,
and 0.9743, respectively. The comparative analysis highlights the superiority of our model over existing tech-
niques. Furthermore, we explore its potential for clinical application using real-world datasets. Our findings
indicate significant promise in revolutionizing breast cancer detection, offering a robust tool for early and ac-
curate diagnosis with the potential to impact patient outcomes greatly.
1 INTRODUCTION
As per the American Cancer Society’s projections
for 2022, there is an anticipated surge of approx-
imately 1,918,030 new cancer cases, leading to an
estimated 609,360 deaths within the United States
alone(Ferlay et al., 2018), (Loizidou et al., 2023).
Among these, breast cancer, an exceptionally preva-
lent and potentially life-threatening ailment affecting
women on a global scale, has risen to become the pri-
mary cause of mortality in nearly every nation. This
multifaceted disease, which accounts for about 30%
of all female cancers, necessitates timely identifica-
tion and detection, as underscored by recent stud-
ies, to facilitate effective treatment and improve pa-
tient outcomes(Aavula et al., 2019)-(Chaurasia et al.,
2018). Cancer progresses through discernible stages,
and detecting it in an advanced phase presents con-
siderable risks. In the realm of medical image analy-
sis, particularly in the realm of breast cancer diagno-
sis, deep learning techniques have demonstrated sub-
stantial promise in enabling the accurate identification
and classification of breast cancer(Kaushik and Kaur,
2016). Over the past years, deep learning, a subset
a
https://orcid.org/0009-0007-8707-5142
b
https://orcid.org/0000-0002-4272-3528
of artificial intelligence, has emerged as a highly aus-
picious methodology in various medical domains, in-
cluding the detection of breast cancer(Rabiei et al.,
2022). Consequently, according to the World Health
Organization’s report in 2019, precise and early detec-
tion plays a pivotal role in advancing diagnosis and el-
evating the survival rate of breast cancer patients from
20% to 60%. With approximately 1.5 million women
receiving a diagnosis each year and half a million suc-
cumbing to the disease, breast cancer stands as a sig-
nificant health challenge(Lotter et al., 2021).
Furthermore, the application of deep learning
methodologies in breast cancer detection holds the
promise of advancing personalized medicine through
its ability to offer insights into the subtype classifica-
tion of breast cancer. This information stands as a piv-
otal factor in tailoring treatment regimens to individ-
ual patients, ultimately resulting in more precise ther-
apeutic interventions and enhanced prognostic out-
comes. Given the recent strides and encouraging out-
comes witnessed in the realm of deep learning-driven
breast cancer detection, there is a burgeoning impe-
tus among researchers to delve deeper into refining
and expanding these methodologies. Through the ju-
dicious utilization of extensive datasets, fine-tuning of
network architectures, and integration of multi-modal
imaging modalities, deep learning stands poised to re-
Yadav, A., Kolekar, M. and Zope, M.
ResNet-101 Empowered Deep Learning for Breast Cancer Ultrasound Image Classification.
DOI: 10.5220/0012377800003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 1, pages 763-769
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
763

define the landscape of breast cancer diagnosis, im-
parting a substantial influence on patient care. This
impetus is substantiated by recent scholarly works,
including those by Ramadan et al. (Ramadan et al.,
2020), Hakin et al. (Hakim et al., 2021), Zeiser et
al. (Zeiser et al., 2020), and Rehman et al. (Rehman
et al., 2021), who have proffered CNN-based models
for breast cancer classification. These investigations
underscore the vast potential of deep learning in the
realm of breast cancer detection and categorization,
laying a robust groundwork for continued exploration
and progress in this pivotal domain of research.
J. W. Li et al. (Li et al., 2022) undertook an in-
depth analysis of ultrasound images aimed at predict-
ing the behavior of breast invasive ductal carcinoma.
This endeavor offers a noninvasive means of assess-
ing, quantifying tumor characteristics, and tailoring
treatment decisions on an individualized basis. How-
ever, it is imperative to acknowledge and surmount
challenges such as data variability, optimal feature se-
lection, mitigating overfitting, conducting robust ex-
ternal validation, and ensuring clinical applicability
to facilitate the successful translation of these findings
into clinical practice.
In a separate study, Sharma et al. (Sharma et al.,
2020) engaged in a comprehensive multi-class classi-
fication analysis to compare the performance of di-
verse classifiers, including decision trees, k-nearest
neighbor (kNN), support vector machine (SVM), and
ensemble classifiers. Their primary objective was
the early prediction and detection of dementia. No-
tably, it is crucial to highlight that these classifiers
were not specifically employed for early breast can-
cer detection in their research. The focus of their in-
vestigation was squarely on dementia prediction, and
the aforementioned classifiers were rigorously eval-
uated within the confines of that specific context.
Aboutalib et al. (Aboutalib et al., 2018) conducted
a study delving into the application of groundbreak-
ing deep learning techniques to discern recalled yet
benign mammography images from negative exam-
inations and those displaying malignancy. Through
this approach, they achieved remarkable outcomes in
terms of accuracy, sensitivity, and specificity. By har-
nessing the potential of deep learning, the authors
showcase the possibility of enhancing the early de-
tection of breast cancer by precisely identifying and
distinguishing among various types of mammography
images. On a related note, S. Mishra et al. (Misra
et al., 2021) proposed an ensemble transfer learning
methodology that incorporates elastography and B-
mode breast ultrasound images, aiming to enhance di-
agnostic precision and generalization capabilities.
In 2022, several methodologies were developed
to advance breast cancer detection and classification.
(Ueda et al., 2022) Developed and validated a deep
learning model for breast cancer detection in mam-
mography and a clinical decision support system us-
ing ultrasound images, with a limitation in obtain-
ing a diverse annotated dataset for robust training
and generalization. (Ragab et al., 2022) Introduced
an Ensemble Deep-Learning-Enabled Clinical Deci-
sion Support System utilizing VGG-16, VGG-19, and
SqueezeNet for feature extraction from ultrasound
images, highlighting enhanced performance but rais-
ing concerns about the computational resources re-
quired. (Jabeen et al., 2022) Proposed a breast cancer
classification framework involving modification of a
DarkNet-53 model, transfer learning, and optimiza-
tion algorithms, with potential challenges in the in-
terpretability of selected features. (Althobaiti et al.,
2022) Presented a deep transfer learning-based model
for breast cancer detection using photoacoustic mul-
timodal imaging, demonstrating promise but raising
questions about resilience to variations in imaging
conditions. (Jabeen et al., 2022) introduced an auto-
mated model for breast cancer diagnosis using digital
mammograms, incorporating pre-processing and hy-
perparameter tuning, with potential sensitivity to tun-
ing processes. However, it should be noted that this
approach comes with escalated computational com-
plexity and entails challenges in determining suitable
combination strategies and ensuring model diversity.
The aim of this current study was to employ a
range of varied deep-learning methodologies and in-
tegrate multiple influencing factors into the modeling
process for the prediction of breast cancer. The fol-
lowing key contributions underscore the significance
of this work:
1. We have modified the ResNet-101 architecture
specifically for breast cancer ultrasound image
classification. In this model, we have employed
Shortcut Connections for Gradient Stability to ad-
dress the vanishing gradient problem with ’Bot-
tleneck’ blocks.
2. In our proposed model, we have implemented a
mechanism for Deep Structure Adaptability, al-
lowing the network to effectively handle intricate
architectures, facilitating enhanced learning and
classification capabilities.
3. We have conducted a comparative analysis be-
tween the proposed modified ResNet-101 model
and various ResNet base models.
4. We further evaluate our proposed model alongside
state-of-the-art methods to validate its reliability
and effectiveness.
BIOSIGNALS 2024 - 17th International Conference on Bio-inspired Systems and Signal Processing
764
2 MATERIAL AND METHODS
Fig. 1 presents a schematic representation of the
methodology employed in this research. For a com-
prehensive understanding of each method and its
practical execution, please refer to the respective sec-
tions, namely Sections 2.1 to 2.3, where you can find
in-depth explanations and implementation specifics.
Figure 1: Block diagram of the proposed framework.
2.1 Data Collection
At the initial data collection phase, breast ultrasound
images were acquired from a cohort of female sub-
jects spanning an age range of 25 to 75 years. This
data acquisition process took place in the year 2018.
The total patient count stands at 600 individuals, all
of whom are female. The dataset encompasses a total
of 780 images, with each image possessing an aver-
age dimension of 500 pixels in both width and height
(500x500 pixels). The images are formatted in PNG
(Portable Network Graphics) format. Each original
image is accompanied by its corresponding ground
truth image. These images are systematically classi-
fied into three distinct classes, namely ’normal’, ’be-
nign’, and ’malignant’, facilitating the categorization
and analysis of breast conditions(Al-Dhabyani et al.,
2020). Exemplar samples from the breast ultrasound
dataset are presented in Fig. 2 for reference.
Figure 2: Few examples of dataset.
2.2 Data Preprocessing
The study incorporates a dataset of over 780 distinct
images, allocated for training, validation, and testing
in ratios of approximately 72: 13: 15, respectively.
Specifically, the training set consists of 563 images,
with 315 classified as benign, 152 as malignant, and
96 as normal. The validation set comprises 100 im-
ages, distributed as 56 benign, 27 malignant, and 17
normal cases. Additionally, the testing set encom-
passes 117 images, with 66 benign, 31 malignant, and
20 normal instances. To ensure precise evaluation,
a new folder was meticulously established for test-
ing due to the absence of a predefined testing folder.
In order to address the inherent imbalanced distribu-
tion within each group, data augmentation techniques
were systematically applied using an Image Gener-
ator. This approach was strategically employed to
enrich the dataset’s diversity and effectively mitigate
the challenge of class imbalance, thereby fostering a
more resilient and precise modeling for breast can-
cer prediction. Diverse data augmentation techniques
were tactically employed to accomplish this objective
2.3 Enhancing Breast Cancer
Prognostication Through Advanced
Predictive Modeling
In this section, we will elucidate our devised pre-
dictive modeling method for the classification of
breast cancer as shown in Fig. 3. In our proposed
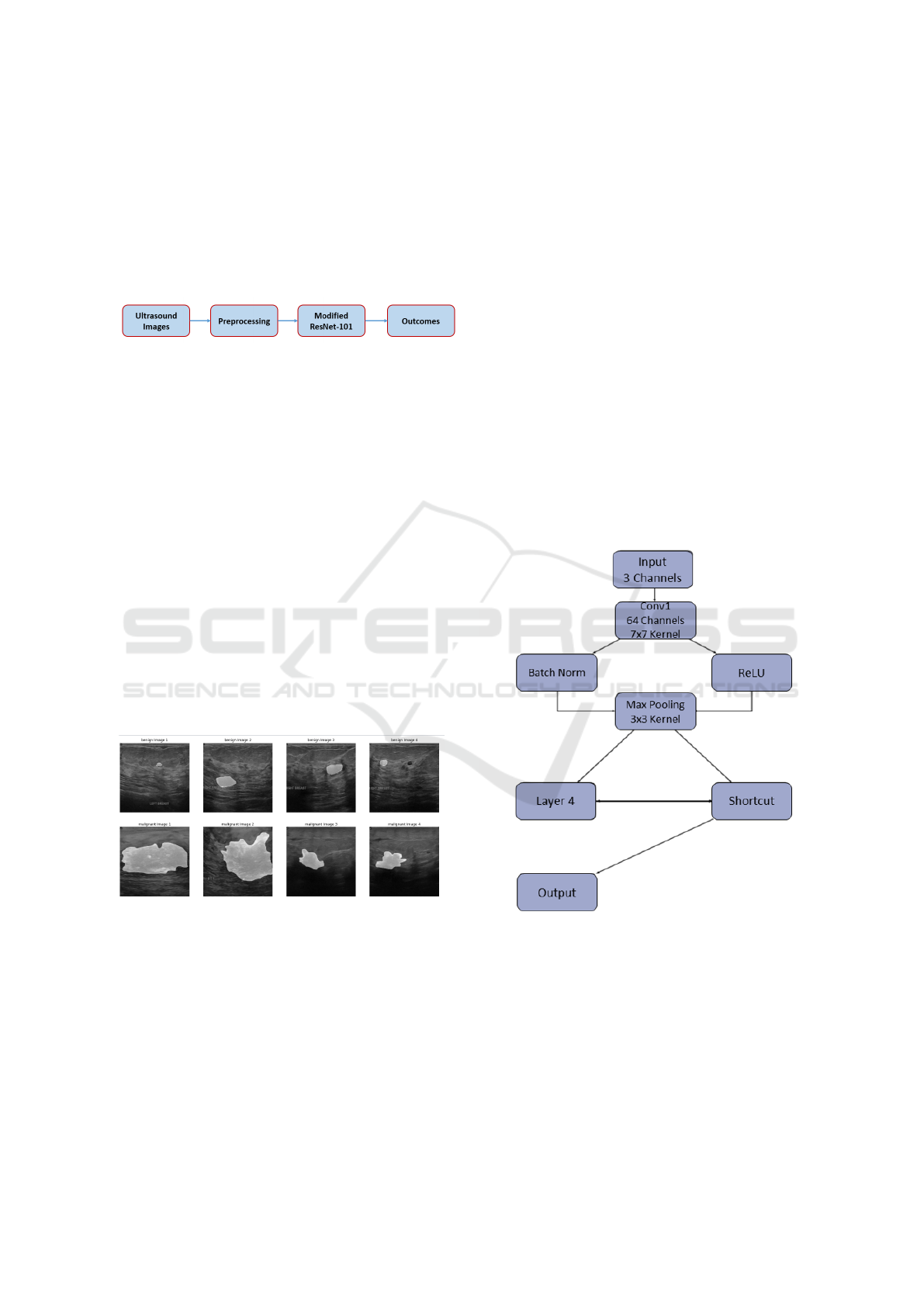
Figure 3: Our proposed modified ResNet-101 architecture.
model, we have made significant modifications to the
ResNet-101 architecture tailored specifically for clas-
sifying breast cancer ultrasound images. This adapted
ResNet-101 structure comprises multiple layers, each
carrying out a sequence of operations. To start, the
initial layer, denoted as ’conv1’, applies a 2D convo-
lution operation to the input with 3 input channels and
generates 64 output channels. It employs a kernel size
of 7x7 and a stride of 2. Following this, batch nor-
ResNet-101 Empowered Deep Learning for Breast Cancer Ultrasound Image Classification
765

malization (’bn1’) is applied, which standardizes the
activations, and a rectified linear unit (ReLU) activa-
tion function is used to introduce non-linearity. Sub-
sequently, the ’max pool’ layer conducts max pooling
with a kernel size of 3x3 and a stride of 2, reducing the
spatial dimensions of the data. The subsequent lay-
ers, namely ’layer1’, ’layer2’, ’layer3’, and ’layer4’,
are constructed as stacks of ’Bottleneck’ blocks. Each
of these blocks encompasses a series of convolutional
layers, combined with batch normalization and ReLU
activation functions. Additionally, they feature a dis-
tinctive ”shortcut” connection that allows information
to bypass certain layers. This is instrumental in miti-
gating the vanishing gradient problem, a common is-
sue in deep neural networks. The number of output
channels in each ’Bottleneck’ block varies, gradually
increasing, which enables the network to capture pro-
gressively complex features. Importantly, the archi-
tecture is purposefully designed to handle very deep
structures while remaining trainable, which is a cru-
cial factor in achieving effective learning and classi-
fication performance. The model employs the Cat-
egorical Cross Entropy loss function and undergoes
optimization via an Adam optimizer with a learning
rate set at 0.0001. The training procedure spans 20
epochs, employing a batch size of 8.
3 RESULTS AND DISCUSSIONS
To apply our model to the dataset by Dhabyani et
al.(Al-Dhabyani et al., 2020), we partitioned the data
into three categories: Benign tumor cell, Malignant
tumor cell, and Normal. The study encompasses more
than 563 unique images, allocating 100 for testing,
117 for validation, and 780 for training. The ratios
for training, validation, and testing were configured
at 72:13:15. The entire experiment was conducted
utilizing Google Colab and Jupyter Notebook. We
performed multiple model simulations to assess the
performance of our proposed system.
We evaluated several performance metrics to vali-
date our proposed modified ResNet-50 model, includ-
ing accuracy (Acc), precision (Pre), sensitivity (Sen)
or recall (Rec), and F1-score. These metrics were cal-
culated using the following formulas:
Acc =
T P + T N
T P + FP + FN + T N
(1)
Rec = Sen =
T P
T P + FN
(2)
Pre =
T P
FP + T P
(3)
F − measure =
2 ∗ Pre ∗ Rec
Pre + Rec
(4)
In our study, we conducted an evaluation of var-
ious parameters on each group of datasets, namely
Benign, Malignant, and Normal, as outlined in Ta-
ble 1 and this table provides precise performance met-
rics for our breast cancer classification model across
distinct groups. For ”Benign tumors,” the model
achieves a notable precision of 0.9565, indicating that
approximately 0.9565 of predicted benign cases were
accurately classified. Furthermore, the recall score of
1.0000 implies that all actual benign cases were suc-
cessfully identified. The F1-score, which balances
precision and recall, attains an impressive 0.9777.
Shifting to the ”Normal” category, both precision and
recall stand at a perfect 1.0000, signifying flawless
classification. In the realm of ”Malignant tumors,” the
model demonstrates impeccable precision at 1.0000,
affirming that all predicted malignant cases were in-
deed correct. However, the recall rate of 0.9032 sug-
gests that there were a few actual malignant cases
that the model missed. The F1-score in this category
amounts to 0.9491. The ”Overall average” metrics
amalgamate these assessments, yielding an average
precision of 0.9855, a recall of 0.9677, and an F1-
score of 0.9756. These aggregated metrics present a
comprehensive evaluation of the model’s proficiency
across all classification groups, portraying a robust
performance in the classification of breast cancer.
Table 1: Performance matrices.
Parameters/Group Precision Recall F1-score
Benign tumor 0.9565 1.0000 0.9777
Normal 1.0000 1.0000 1.0000
Malignant tumor 1.0000 0.9032 0.9491
Overall average 0.9855 0.9677 0.9756
In addition, we assessed testing and validation ac-
curacy using a dataset consisting of 100 images. The
results demonstrated a commendable accuracy level,
with both types of breast cancer groups achieving a
score of 0.9743. This indicates a robust performance
in accurately classifying cases within the dataset. Fig.
4 displays the predicted results obtained from our pro-
posed modified ResNet-101 model. Fig. 5 shown the
confusion matrix of our proposed model.
Table 2: Previous methods compared with our proposed
model.
Author Method Pre Rec F1-score Acc
Ramadan et al. (Ramadan et al., 2020) CNN - 0.9141 - 0.9210
Hakin et al. (Hakim et al., 2021) CNN - 0.8812 - 0.9034
Fanizzi et al. (Fanizzi et al., 2020) Random Forest - 0.8910 - 0.8855
Rehman et al. (Rehman et al., 2021) Fully connected CNN - 0.9700 - 0.8701
Zeiser et al. (Zeiser et al., 2020) CNN (U-NET) - 0.9230 - 0.8590
R. Rabiei et al. (Rabiei et al., 2022) Random Forest - 0.9304 - 0.8004
Proposed Method CNN (Resnet-101) 0.9855 0.9677 0.9756 0.9756
In Table 2, we present a comprehensive compar-
ative evaluation of various breast cancer classifica-
tion methodologies. Each approach was rigorously
BIOSIGNALS 2024 - 17th International Conference on Bio-inspired Systems and Signal Processing
766

Figure 4: Few predicted samples of our proposed model.
Figure 5: Confusion matrix of our proposed model.
examined for its capacity to accurately discern breast
cancer cases. The assessed methods, along with their
corresponding performance metrics, are meticulously
detailed. Ramadan et al. harnessed the power of a
CNN and attained a precision of 0.9141 and an ac-
curacy of 0.9210 (Ramadan et al., 2020). Hakin et
al. similarly employed a CNN, yielding a precision
of 0.8812 and an accuracy of 0.9034 (Hakim et al.,
2021). Fanizzi et al. opted for a Random Forest
technique, yielding a precision of 0.8910 and an ac-
curacy of 0.8855 (Fanizzi et al., 2020). Rehman et
al. implemented a Fully Connected CNN, achiev-
ing a remarkable precision of 0.9700, albeit with a
marginally lower accuracy of 0.8701 (Rehman et al.,
2021). Zeiser et al. employed a CNN with U-NET ar-
chitecture, securing a precision of 0.9230 and an ac-
curacy of 0.8590 (Zeiser et al., 2020). R. Rabiei et
al. utilized a Random Forest approach, resulting in a
precision of 0.9304 and an accuracy of 0.8004 (Rabiei
et al., 2022). However, our proposed method, leverag-
ing a CNN with ResNet-101 architecture, unequivo-
cally outperforms all alternative techniques. It attains
an exceptional precision of 0.9855, underscoring an
exceedingly low false positive rate. This precision
is of paramount importance in the medical domain,
where precise identification of positive cases holds ut-
most significance. Furthermore, the model exhibits
a commendable recall score of 0.9677, signifying its
adeptness in capturing all bonafide positive cases and
thereby minimizing false negatives. The F1-score, a
pivotal metric that strikes a balance between preci-
sion and recall, reaches an impressive 0.9756, further
corroborating the model’s robustness. The overall ac-
curacy, standing at 0.9756, unequivocally establishes
the proposed method’s superior performance. While
the other methods demonstrate commendable perfor-
mance, none surpasses the precision, recall, and F1-
score achieved by our proposed CNN with ResNet-
101 architecture.
Overall, our proposed breast cancer classification
model demonstrates superior performance compared
to the random forest-based approaches presented by
R. Rabiei et al. (Rabiei et al., 2022) and Fanizzi et
al. (Fanizzi et al., 2020) across key metrics includ-
ing accuracy, recall, precision, and F1 score. Fur-
thermore, our model achieves a competitive accuracy
when compared to the fully connected depth-wise
separable CNN model introduced by Rehman et al.
(Rehman et al., 2021), while also attaining higher re-
call, precision, and F1-score. These findings under-
score the efficacy of our model in the precise clas-
sification of breast cancer utilizing breast ultrasound
images.
3.1 Ablation Study
In this comprehensive ablation study, we systemat-
ically assessed the performance of various ResNet
architectures, as detailed in Table 3. The models
evaluated include ResNet-18, ResNet-34, ResNet-50,
ResNet-152, and ResNet-101, alongside a proposed
modified ResNet-101 specifically tailored for breast
cancer classification and segmentation tasks. The
baseline models demonstrated incremental improve-
ments in accuracy, precision, recall, and F1-score
with increasing depth, with ResNet-152 achieving
the highest accuracy. Notably, ResNet-101 exhibited
competitive accuracy and an impressive F1-score of
0.9760. However, the proposed modified ResNet-101
surpassed all models, showcasing its prowess with the
highest precision and recall at 0.9855, underscoring
its ability to capture nuanced features crucial for accu-
rate breast cancer classification, as evidenced in Table
3.
ResNet-101 Empowered Deep Learning for Breast Cancer Ultrasound Image Classification
767

Table 3: Performance metrics of diverse ResNet architec-
tures and the proposed modified ResNet-101.
Model/Parameters Pre Rec F1-score Acc
ResNet-18 0.8921 0.9123 0.9014 0.8898
ResNet-34 0.9040 0.9154 0.9102 0.9001
ResNet-50 0.9308 94.02 0.9405 0.9274
ResNet-152 0.9621 0.9614 0.9652 0.9847
ResNet-101 0.9542 0.9548 0.9760 0.9511
Proposed Mod-ResNet-101 0.9855 0.9855 0.9756 0.9756
Incorporating the segmentation task into the eval-
uation, the proposed modified ResNet-101 demon-
strated its versatility, as depicted in Figure 4. Seg-
mentation demands a nuanced understanding of im-
age features, and the model’s modifications evidently
contributed to its efficacy in delineating regions of in-
terest. The precision and recall metrics in the seg-
mentation task aligned with those of the classification
task, highlighting the model’s consistent performance
across both domains. This dual-task capability proves
instrumental in providing a holistic solution for breast
cancer analysis, where accurate localization and clas-
sification of abnormalities are paramount. While op-
portunities for fine-tuning and further improvement
exist, this study firmly establishes the proposed mod-
ified ResNet-101 as a robust and versatile architec-
ture for comprehensive breast cancer analysis, effec-
tively addressing both classification and segmenta-
tion tasks. By offering a comprehensive solution for
accurate localization and classification of abnormali-
ties, our meticulously designed architecture stands as
a beacon of innovation, paving the way for heightened
precision in medical image analysis and holding im-
mense promise for improving diagnostic outcomes in
breast cancer research and clinical practice.
4 CONCLUSION
Our proposed modified ResNet-101 architecture for
breast cancer classification via breast ultrasound im-
agery has demonstrated superior performance com-
pared to existing approaches. It exhibited height-
ened accuracy, recall, precision, and F1-score met-
rics, signifying its efficacy in precisely discerning
cases of breast cancer. These outcomes outperformed
random forest-based models and rivaled a fully con-
nected depth-wise separable CNN model. These re-
sults underscore the potential of deep learning archi-
tectures in breast cancer classification and provide a
solid groundwork for future investigations. Through
continued refinement and progression of these mod-
els, we can make significant strides in early and pre-
cise breast cancer identification, ultimately leading to
enhanced patient outcomes and reduced disease bur-
den.
Moving forward, we have outlined several areas
for potential improvement in breast cancer classifi-
cation models. A pivotal aspect of our agenda is to
leverage a distinct dataset sourced from our institution
(IGIMS Patna) to validate our model using authentic
clinical data. This endeavor will allow us to evalu-
ate the model’s performance and resilience in a real-
world healthcare setting, offering further substanti-
ation of its adeptness in accurately detecting breast
cancer.
REFERENCES
Aavula, R., Bhramaramba, R., and Ramula, U. S. (2019). A
comprehensive study on data mining techniques used
in bioinformatics for breast cancer prognosis. Journal
of Innovation in Computer Science and Engineering,
9(1):34–39.
Aboutalib, S. S., Mohamed, A. A., Berg, W. A., Zuley,
M. L., Sumkin, J. H., and Wu, S. (2018). Deep learn-
ing to distinguish recalled but benign mammography
images in breast cancer screening. Clinical Cancer
Research, 24(23):5902–5909.
Al-Dhabyani, W., Gomaa, M., Khaled, H., and Fahmy, A.
(2020). Dataset of breast ultrasound images. Data in
brief, 28:104863.
Althobaiti, M. M., Ashour, A. A., Alhindi, N. A., Al-
thobaiti, A., Mansour, R. F., Gupta, D., Khanna, A.,
et al. (2022). Deep transfer learning-based breast can-
cer detection and classification model using photoa-
coustic multimodal images. BioMed Research Inter-
national, 2022.
Chaurasia, V., Pal, S., and Tiwari, B. (2018). Prediction of
benign and malignant breast cancer using data mining
techniques. Journal of Algorithms & Computational
Technology, 12(2):119–126.
Fanizzi, A., Basile, T. M., Losurdo, L., Bellotti, R., Bottigli,
U., Dentamaro, R., Didonna, V., Fausto, A., Massafra,
R., Moschetta, M., et al. (2020). A machine learning
approach on multiscale texture analysis for breast mi-
crocalcification diagnosis. BMC bioinformatics, 21:1–
11.
Ferlay, J., Ervik, M., Lam, F., Colombet, M., Mery, L.,
Pi
˜
neros, M., Znaor, A., Soerjomataram, I., and Bray,
F. (2018). Global cancer observatory: cancer today.
Lyon, France: international agency for research on
cancer, 3(20):2019.
Hakim, A., Prajitno, P., and Soejoko, D. (2021). Micro-
calcification detection in mammography image using
computer-aided detection based on convolutional neu-
ral network. In AIP Conference Proceedings, volume
2346. AIP Publishing.
Jabeen, K., Khan, M. A., Alhaisoni, M., Tariq, U., Zhang,
Y.-D., Hamza, A., Mickus, A., and Dama
ˇ
sevi
ˇ
cius, R.
(2022). Breast cancer classification from ultrasound
images using probability-based optimal deep learning
feature fusion. Sensors, 22(3):807.
BIOSIGNALS 2024 - 17th International Conference on Bio-inspired Systems and Signal Processing
768

Kaushik, D. and Kaur, K. (2016). Application of data
mining for high accuracy prediction of breast tissue
biopsy results. In 2016 Third International Confer-
ence on Digital Information Processing, Data Mining,
and Wireless Communications (DIPDMWC), pages
40–45. IEEE.
Li, J.-w., Cao, Y.-c., Zhao, Z.-j., Shi, Z.-t., Duan, X.-
q., Chang, C., and Chen, J.-g. (2022). Prediction
for pathological and immunohistochemical character-
istics of triple-negative invasive breast carcinomas:
the performance comparison between quantitative and
qualitative sonographic feature analysis. European
Radiology, pages 1–11.
Loizidou, K., Elia, R., and Pitris, C. (2023). Computer-
aided breast cancer detection and classification in
mammography: A comprehensive review. Computers
in Biology and Medicine, page 106554.
Lotter, W., Diab, A. R., Haslam, B., Kim, J. G., Grisot, G.,
Wu, E., Wu, K., Onieva, J. O., Boyer, Y., Boxerman,
J. L., et al. (2021). Robust breast cancer detection in
mammography and digital breast tomosynthesis using
an annotation-efficient deep learning approach. Na-
ture Medicine, 27(2):244–249.
Misra, S., Jeon, S., Managuli, R., Lee, S., Kim, G., Lee, S.,
Barr, R. G., and Kim, C. (2021). Ensemble transfer
learning of elastography and b-mode breast ultrasound
images. arXiv preprint arXiv:2102.08567.
Rabiei, R., Ayyoubzadeh, S. M., Sohrabei, S., Esmaeili, M.,
and Atashi, A. (2022). Prediction of breast cancer us-
ing machine learning approaches. Journal of Biomed-
ical Physics & Engineering, 12(3):297.
Ragab, M., Albukhari, A., Alyami, J., and Mansour, R. F.
(2022). Ensemble deep-learning-enabled clinical de-
cision support system for breast cancer diagnosis
and classification on ultrasound images. Biology,
11(3):439.
Ramadan, S. Z. et al. (2020). Using convolutional neural
network with cheat sheet and data augmentation to de-
tect breast cancer in mammograms. Computational
and Mathematical Methods in Medicine, 2020.
Rehman, K. U., Li, J., Pei, Y., Yasin, A., Ali, S., and Mah-
mood, T. (2021). Computer vision-based microcalci-
fication detection in digital mammograms using fully
connected depthwise separable convolutional neural
network. Sensors, 21(14):4854.
Sharma, N., Kolekar, M. H., and Jha, K. (2020). Iterative
filtering decomposition based early dementia diagno-
sis using eeg with cognitive tests. IEEE Transactions
on Neural Systems and Rehabilitation Engineering,
28(9):1890–1898.
Ueda, D., Yamamoto, A., Onoda, N., Takashima, T., Noda,
S., Kashiwagi, S., Morisaki, T., Fukumoto, S., Shiba,
M., Morimura, M., et al. (2022). Development
and validation of a deep learning model for detec-
tion of breast cancers in mammography from multi-
institutional datasets. PLoS One, 17(3):e0265751.
Zeiser, F. A., da Costa, C. A., Zonta, T., Marques, N. M.,
Roehe, A. V., Moreno, M., and da Rosa Righi, R.
(2020). Segmentation of masses on mammograms us-
ing data augmentation and deep learning. Journal of
digital imaging, 33:858–868.
ResNet-101 Empowered Deep Learning for Breast Cancer Ultrasound Image Classification
769
