
Bone-Aware Generative Adversarial Network with Supervised Attention
Mechanism for MRI-Based Pseudo-CT Synthesis
Gurbandurdy Dovletov
1 a
, Utku Karadeniz
1 b
, Stefan L
¨
orcks
1 c
, Josef Pauli
1 d
,
Marcel Gratz
2,3 e
and Harald H. Quick
2,3
1
Intelligent Systems Group, Faculty of Computer Science, University of Duisburg-Essen, Duisburg, Germany
2
High-Field and Hybrid MR Imaging, University Hospital Essen, University of Duisburg-Essen, Essen, Germany
3
Erwin L. Hahn Institute for MR Imaging, University of Duisburg-Essen, Essen, Germany
Keywords:
Deep Learning, Image-to-Image Translation, Pseudo-CT Synthesis, Attention Mechanisms, Attention U-Net,
Generative Adversarial Network.
Abstract:
Deep learning techniques offer the potential to learn the mapping function from MRI to CT domains, allowing
the generation of synthetic CT images from MRI source data. However, these image-to-image translation
methods often introduce unwanted artifacts and struggle to accurately reproduce bone structures due to the
absence of bone-related information in the source data. This paper extends the recently introduced Atten-
tion U-Net with Extra Supervision (Att U-Net ES), which has shown promising improvements for the bone
regions. Our proposed approach, a conditional Wasserstein GAN with Attention U-Net as the generator, lever-
ages the network’s self-attention property while simultaneously including domain-specific knowledge (or bone
awareness) in its learning process. The adversarial learning aspect of the proposed approach ensures that the
attention gates capture both the overall shape and the fine-grained details of bone structures. We evaluate the
proposed approach using cranial MR and CT images from the publicly available RIRE data set. Since the
images are not aligned with each other, we also provide detailed information about the registration procedure.
The obtained results are compared to Att U-Net ES, baseline U-Net and Attention U-Net, and their GAN ex-
tensions.
1 INTRODUCTION
Computed Tomography (CT) and Magnetic Reso-
nance Imaging (MRI) are crucial medical imaging
techniques with significant roles in healthcare. While
both techniques are invaluable for diagnosing and
treating various medical conditions, they each offer
distinct advantages owing to their diverse underlying
physical principles.
A CT scan is acquired by employing a rotating
source tube, emitting X-rays from various angles dur-
ing its rotation. As these X-rays traverse the patient’s
body, they are attenuated and subsequently captured
by a rotating detector opposite the source. As a re-
sult, a CT image visually represents the attenuating
properties within the patient’s tissues. CT values are
expressed in Hounsfield Units (HU), a relative mea-
a
https://orcid.org/0000-0002-2401-8745
b
https://orcid.org/0009-0006-3456-1115
c
https://orcid.org/0000-0003-3641-4734
d
https://orcid.org/0000-0003-0363-6410
e
https://orcid.org/0000-0001-9723-5233
surement scale used to quantify the density of tissues
within the body.
On the other hand, MRI utilizes strong magnetic
fields and radiofrequency pulses to align the hydro-
gen nuclei present abundantly in the human body. Af-
ter the radiofrequency pulse is deactivated, the pro-
tons gradually realign themselves within the magnetic
field and simultaneously emit their radiofrequency
signal (or resonance), which detectors (or receiver
coils) capture.
A CT scan thus can offer superior visualization
of bone anatomy due to its high-density contrast,
whereas an MRI image excels in revealing soft tis-
sues and organs. Both modalities can complement
each other in some cases, ensuring a comprehensive
assessment of a patient’s condition.
In the context of Radiation Treatment (RT) plan-
ning, MRI offers a notable advantage over CT by de-
livering a highly detailed map of soft tissues and facil-
itating a precise delineation of both organs at risk and
treatment targets (e.g., tumors) (Schmidt and Payne,
2015). However, MRI cannot map electron densities,
which is essential for radiation dose calculations in
Dovletov, G., Karadeniz, U., Lörcks, S., Pauli, J., Gratz, M. and Quick, H.
Bone-Aware Generative Adversarial Network with Supervised Attention Mechanism for MRI-Based Pseudo-CT Synthesis.
DOI: 10.5220/0012370900003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 1, pages 223-235
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
223

RT planning. This necessitates an additional CT scan,
resulting in unwanted radiation exposure for patients,
which ideally should be reduced to zero, and, addi-
tionally, in increased healthcare costs.
One approach to mitigate these challenges is to
generate synthetic CT images directly from radiation-
free MRI data, often called pseudo-CT (pCT) images.
These synthetic CTs can also be used in Positron
Emission Tomography (PET) systems when com-
bined with MRI.
PET is a nuclear medicine imaging technique that
is used to reveal physiological and biochemical pro-
cesses within the body. It involves using a small
amount of a radioactive substance, known as a radio-
tracer, typically injected into the patient’s body. This
unstable radiotracer undergoes a radioactive decay
and emits positrons. When a positron collides with
an electron within the body, the annihilation process
produces two gamma rays, with 511 keV energy each,
that are emitted in opposite directions. While travers-
ing through some tissue or hardware parts (e.g., pa-
tient’s table) on their way to detectors, these photons
get attenuated. Thus, an Attenuation Correction (AC)
procedure is required for each PET image.
The absence of anatomical details in standalone
PET led to the development of integrated PET/CT
systems. In such hybrid modality imaging systems, a
complementary CT image is acquired within a single
gantry, allowing the generation of AC maps directly
from HUs by scaling the CT image’s energy level with
that of PET.
Superior soft tissue contrast and radiation-free
principles of MRI lead to PET/MRI systems (Quick,
2014; Paulus et al., 2015), where MRI-based pseudo-
CT is used for AC of PET.
Thus, accurate pseudo-CT synthesis, especially
for dense parts such as cortical bones, is crucial for
both AC of PET data and RT planning. At the same
time, it is a challenging task since standard T1- or T2-
weighted MRI cannot capture the signal from bone
regions (due to its relatively short relaxation time),
making it difficult to translate it into an accurate
pseudo-CT image.
2 RELATED WORK
In deep learning, synthesizing pseudo-CT images
from MRI scans is an image-to-image translation
problem. Several methods have been proposed to
tackle this challenging task.
(Nie et al., 2016) propose utilizing a Fully Con-
volutional Network (FCN) to preserve the neighbor-
hood information better while mapping from MR to
CT images. (Han, 2017) proposes adapting and using
the U-Net (Ronneberger et al., 2015) architecture for
MRI-based pseudo-CT synthesis. On the other hand,
(Wolterink et al., 2017) suggest employing Genera-
tive Adversarial Networks (GAN) (Goodfellow et al.,
2014) and their cyclic extension, CycleGAN (Zhu
et al., 2017), to achieve more realistic image synthe-
sis.
While synthesizing pseudo-CTs using FCNs,
U-Nets, or GANs is feasible, the resulting images of-
ten contain errors, particularly in the bone regions.
To address this challenge, a popular solution is
to incorporate different MRI sequences and contrasts
as additional sources of information to improve the
accuracy of bone representation in the synthesized
images. To this end, (Leynes et al., 2018) pro-
pose the utilization of Zero-Echo-Time (ZTE) images
in conjunction with Dixon MRI’s in-phase and out-
of-phase images to capture more information about
bone structures. In an alternative approach, (Torrado-
Carvajal et al., 2019) suggest using 2-echo Dixon im-
ages and explicitly emphasizing their fat- and water-
only derivatives. (Gong et al., 2018) propose to
efficiently make use of both Dixon and ZTE in-
puts using grouped convolutions (Xie et al., 2017)
in the deeper layers of U-Net. (Qi et al., 2020)
propose leveraging multiple imaging sequences, in-
cluding T1, T2, contrast-enhanced T1, and contrast-
enhanced Dixon T1 (water-only image), to enhance
the quality of the synthesized pseudo-CTs. Although
these methods can potentially improve the quality
of synthesized pseudo-CTs, it is important to note
that this improvement comes with the trade-off of in-
creased MR image acquisition costs and longer acqui-
sition times.
Another approach to improve the quality of syn-
thesized images involves utilizing attention mecha-
nisms during the training process of neural networks.
Generally speaking, attention mechanisms allow net-
works to focus on specific parts of the input data and,
thus, to capture important details and structures more
effectively.
Spatial attention extends this idea by refining the
focus to specific spatial regions within the input data.
Proposed by (Oktay et al., 2018) Attention U-Net
is a well-known semantic segmentation network that
incorporates a spatial attention mechanism in the form
of Attention Gates (AG) to self-focus on task-specific
features. One notable advantage of this approach is
that the model inherently possesses the ability to vi-
sualize learned attention maps, which enhances the
interpretability of models for human understanding.
Channel attention, such as e.g., Squeeze-and-
Excitation (SE) proposed by (Hu et al., 2018), gener-
BIOIMAGING 2024 - 11th International Conference on Bioimaging
224

ates attention masks along the channel dimension and
thus allows the feature recalibration for better use of
global abstract information for the classification.
Bottleneck Attention Module (BAM) (Park et al.,
2018), or its extension, Convolutional Block Atten-
tion Module (CBAM) (Woo et al., 2018), incorporates
both spatial and channel attention mechanisms. Thus,
AG, SE, BAM, or CBAM are attention mechanisms
capable of capturing the most valuable information on
their own without explicit guidance.
Although self-attention is generally preferable,
there are situations where attention mechanisms en-
hanced by domain-specific knowledge prove to be a
more effective choice.
To this end, (Xiang et al., 2018) adopt CycleGAN
and introduce structural dissimilarity loss to its learn-
ing process, which is calculated for both MRI and
CT domains based on the Structural Similarity Index
Measure (SSIM) (Wang et al., 2004). Alternatively,
(Ge et al., 2019) propose a modification that explic-
itly incorporates mutual information between MR and
synthesized CT images and enforces shape consis-
tency between these images using an additional seg-
mentation network. (Dovletov et al., 2022b) suggest
generating bone segmentations and utilizing them
for U-Net- and GAN-based models to penalize more
severely the errors in the bone regions. In (Dovle-
tov et al., 2022a), the same research group proposes
to use an additional classifier in combination with the
Grad-CAM (Selvaraju et al., 2017) technique to guide
their U-Net, forcing it to focus more on bone regions
without any auxiliary input.
In this contribution, we extend the Attention
U-Net with Extra Supervision (Dovletov et al.,
2023a), a technique that guides the model to learn at-
tention maps that closely resemble bone segmentation
maps. More specifically, we adopt this technique and
introduce it in the context of Generative Adversarial
Networks. Through experimentation, we demonstrate
that this extra supervision substantially reduces er-
rors in regions around bones compared to the baseline
GAN models.
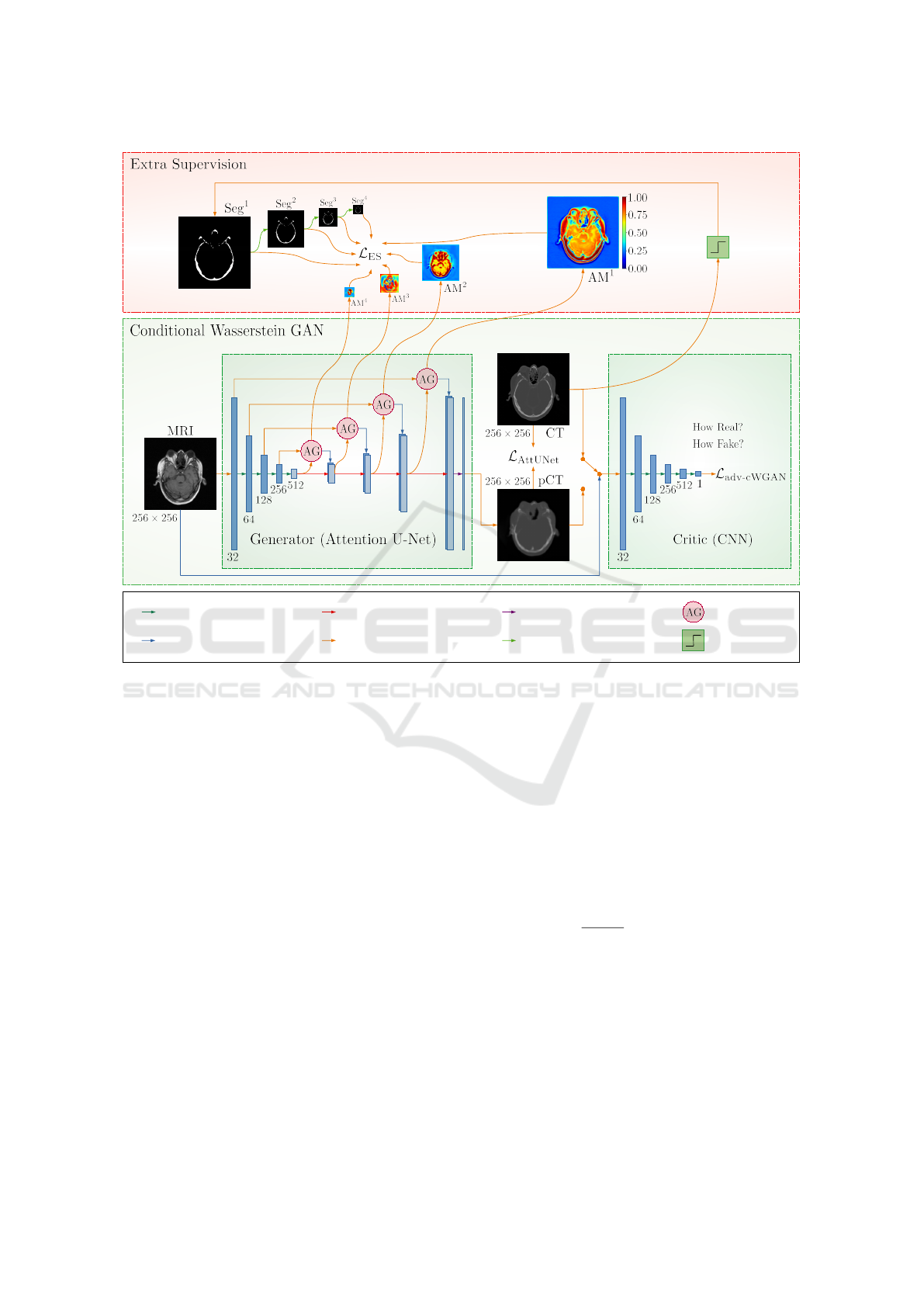
3 PROPOSED APPROACH
In this section, we first introduce and formulate our
baseline Attention U-Net model in the context of an
MRI-based pseudo-CT synthesis. Then, we extend it
and thus define our baseline conditional Wasserstein
Generative Adversarial Network. After that, the pro-
posed conditional Wasserstein GAN with Extra Su-
pervision (ES) is explained.
2xConvolution,
followed by Max Pooling
Skip Connection
(Concatenation)
Data Flow Arrow
(no operation)
Transposed Convolution,
followed by 2xConvolution
1x1 Convolution
Figure 1: Baseline U-Net for MRI-based pseudo-CT syn-
thesis.
3.1 U-Net and Attention U-Net
U-Net (Ronneberger et al., 2015) is a Fully Convolu-
tional Network (FCN) architecture that was initially
designed for biomedical image segmentation but has
found its place in a wide range of tasks, including
MRI-based pseudo-CT synthesis. As can be seen
from Figure 1, the network has a distinctive U-shaped
structure and consists of contracting (encoding) and
expansive (decoding) paths and skip connections be-
tween them. The encoding path of the network con-
sists of a series of convolutional layers followed by a
downsampling (or pooling) operation. It is responsi-
ble for capturing abstract features from the input MR
images. Hence, its task is similar to the feature ex-
traction part of traditional CNNs. The decoding path
takes these features (with lower spatial resolution) as
input and learns to produce the output pseudo-CT
image of the same size as the input image. It in-
volves a sequence of transposed convolutional layers
in combination with skip connections from the encod-
ing path. These skip connections are a crucial part
of U-Net since they allow the network to transfer de-
tailed information from the encoder’s layers and help
the model recover the fine-grained features in the de-
coder part.
Among its notable modifications, Attention
U-Net (Oktay et al., 2018) stands out as an exten-
sion that allows the network to self-focus on task-
specific regions. As shown in Figure 2 (middle block,
left side), the key difference compared to the original
U-Net is the incorporation of Attention Gates (AG).
These AGs are placed along the skip connections and
are responsible for selectively highlighting the most
task-relevant features and suppressing less relevant
details by learning suitable Attention Maps (AM).
Bone-Aware Generative Adversarial Network with Supervised Attention Mechanism for MRI-Based Pseudo-CT Synthesis
225

More specifically, this feature selection mechanism
is implemented using contextual information (repre-
sented as the gating signal) obtained at coarser scales.
The output of attention gates is the element-wise mul-
tiplication of input features and the attention coeffi-
cient of AMs. Thus, attention gates introduce a con-
cept of self-attention, where the network on its own
learns suitable AMs to solve the provided task better.
Both U-Net and Attention U-Net networks can be
used for MRI-based pseudo-CT synthesis. Thus, for
both baseline models, we choose Mean Absolute Er-
ror (MAE) to formulate their loss functions:
L
(Att)UNet
=
1
M · N
M
∑
i=1
N
∑
j=1
y
i j
− G(x)
i j
(1)
where G(x) represents the generated pseudo-CT im-
age with the size of M ×N pixels, x is the input MR
image, and y is its corresponding Ground Truth (GT)
CT image.
3.2 Conditional Wasserstein GAN
Generative Adversarial Network (GAN) (Goodfellow
et al., 2014) was initially proposed to create realistic
and high-quality data samples from noise. However,
its later adoptions, such as pix2pix (Isola et al., 2017),
can also be used for image-to-image translation tasks.
Thus, MRI-based pseudo-CT images can be synthe-
sized using generative models.
GANs consist of two networks: a generator and
a discriminator. The main goal of the generator G
is to synthesize data indistinguishable from the real
data (training data). The discriminator D, on the other
hand, has the task of distinguishing between real data
and synthetic data (created by the generator). Both G
and D networks are trained together by taking turns
and using an adversarial training approach, meaning
they compete against each other. While the generator
learns to synthesize data that can fool the discrimi-
nator, the discriminator strives to better differentiate
between real and fake data. The learning process for
G and D can be described using the adversarial objec-
tive function:
L
adv-GAN
=E
y
[log(D(y))]+E
x
[log(1−D(G(x)))] (2)
where x and y represent images from the source (MRI)
and target (CT) domain correspondingly, and E de-
notes the expected value. Thus, the generator is
trained to minimize the probability of the discrimina-
tor classifying its synthetic data as fake. In contrast,
the discriminator tries to maximize this probability by
correctly identifying synthesized data as a fake class.
In a well-trained GAN framework, the generator be-
comes so good at synthesizing data that the discrim-
inator cannot tell the difference between synthesized
fake data and real data.
Wasserstein GAN (WGAN) (Arjovsky et al.,
2017) is one of GAN’s modifications that heavily con-
tributes to the training stability and reduces the mode
collapse problem, where the generator only produces
a limited variety of synthetic data. These improve-
ments are achieved by using an alternative adversar-
ial loss function that approximates the Earth Mover’s
Distance and changing the discriminator’s role from a
binary classifier to a critic C, which assesses the de-
gree of realness by assigning continuous scores. An-
other extension of traditional GAN is a conditional
Generative Adversarial Network (cGAN) (Mirza and
Osindero, 2014), where both generator and discrim-
inator networks are provided with additional condi-
tioning information to better control the various as-
pects of the generative process.
Our baseline conditional WGANs (or cWGANs)
include image-based conditioning on the correspond-
ing critics to better preserve structural informa-
tion between the input MR image and synthesized
pseudo-CT image. More specifically, fake or real im-
ages are concatenated with the generator’s input im-
age before being propagated through the critic net-
work. Thus, our baseline adversarial objective is for-
malized as follows:
L
adv-cWGAN
= E
x,y
[C(x, y)] − E
x
[C(x, G(x ))] (3)
where x and y represent MR and CT images corre-
spondingly. While G tries to minimize L
adv-cWGAN
against adversarial critic C, the latter one attempts to
maximize the same objective. We use both U-Net and
Attention U-Net networks as generators, whereas a
CNN, depicted in Figure 2 (middle block, right side),
serves as the critic. For the sake of shortness, only
the architecture with Attention U-Net is depicted in
Figure 2 (middle block, left side). Thus, our final ob-
jectives for generator and critic networks can be sum-
marized as follows:
L
g
= L
(Att)UNet
+ λ
adv
L
adv-cWGAN
L
c
= L
adv-cWGAN
(4)
where λ
adv
denotes the weighting factor of the condi-
tional Wasserstein GAN’s objective. The generator’s
loss contains the previously introduced L
(Att)UNet
loss
term that penalizes the distance between the synthe-
sized outputs and ground truth data, further encour-
aging the generator to create plausible translation re-
sults.
BIOIMAGING 2024 - 11th International Conference on Bioimaging
226
2xConvolution,
followed by Max Pooling
Skip Connection
(Concatenation)
Data Flow Arrow
(no operation)
Transposed Convolution,
followed by 2xConvolution
1x1 Convolution
Attention Gate
2x2 Max Pooling
Thresholding
Figure 2: Proposed conditional Wasserstein Generative Adversarial Network with Extra Supervision for MRI-based
pseudo-CT synthesis. Compared to the baseline cWGAN (middle block), our approach has an additional Extra Super-
vision (ES) module (upper block), which forces the attention maps of the Generator (Attention U-Net) network (AM
l
,
l ∈ {1, 2, 3, 4}) to look as similar as possible to the (scaled) ground truth bone segmentation maps (Seg
l
, l ∈ {1, 2, 3, 4}).
3.3 Conditional Wasserstein GAN with
Extra Supervision
Our proposed network is based on the above-
mentioned conditional Wasserstein GAN architecture
utilizing the Attention U-Net network. We propose
imposing additional constraints on attention maps to
improve the generator’s ability to focus on crucial re-
gions, like the bone areas in MRI-based pseudo-CT
synthesis. Specifically, we adopt the Extra Supervi-
sion (ES) (Dovletov et al., 2023a) recently introduced
in the context of the pseudo-CT synthesis task and
utilize it for our generative model. The main idea of
ES is to force the Attention U-Net (or generator) to
pay more attention to the bone regions by using ad-
ditional supervision via coarse bone segmentations.
Our objective functions for generator G and critic C
networks can be summarized as follows:
L
g
= L
AttUNet
+ λ
ES
L
ES
+ λ
adv
L
adv-cWGAN
L
c
= L
adv-cWGAN
(5)
where L
ES
represents additional supervision for the
generator, and λ
ES
is a hyperparameter that can be
used to control its relative importance. Similarly, as
in the original ES paper, we propose calculating L
ES
as follows:
L
ES
=
L
∑
l=1
λ
l
· L
l
ES
=
=
L
∑
l=1
λ
l
·
1
M
l
· N
l
M
l
∑
i=1
N
l
∑
j=1
Seg
l
i j
− AM
l
i j
2
(6)
where AM
l
represents the attention map with size
M
l
× N
l
learned by the attention gate at the l-th im-
age resolution level (l ∈ {1, ..., L}), Seg
l
corresponds
to the ground truth segmentation scaled to match the
size of AM
l
, and L denotes the total number of reso-
lution levels in the network, excluding the bottleneck.
Since attention gates use the sigmoid function as the
final activation, the attention values learned during the
training fall within the range of zero to one. Hence,
Bone-Aware Generative Adversarial Network with Supervised Attention Mechanism for MRI-Based Pseudo-CT Synthesis
227
attention maps share the same value range as ground
truth segmentations, with zeros and ones represent-
ing non-bone and bone regions. We propose using
λ
l
to independently control the relative importance of
each extra supervision term L
l
ES
. These hyperparam-
eters, hence, have to be chosen manually. Thus, while
L
AttUNet
and L
adv-cWGAN
provide general supervision
and allow the image-to-image translation network to
learn the mapping from MRI to CT domains, L
ES
pro-
vides auxiliary guidance, enhancing the generator’s
ability to synthesize bone structures.
4 EXPERIMENTS
This section presents the data set utilized in the exper-
iments, followed by an overview of implementation
details and the metrics used for evaluation.
4.1 Data Set
The publicly available Retrospective Image Registra-
tion Experiment (RIRE) data set (West et al., 1997)
was initially introduced in the context of an image-
to-image registration task. It consists of cranial im-
age scans for sixteen patients, acquired with differ-
ent imaging techniques, such as MRI, CT, and PET.
Thus, the image volumes in this data set are not in-
herently aligned with each other, and the ground truth
registration data is also not included. Furthermore,
only a subset of CT scans within the data set contains
the patient’s table as part of the imagery. The images
are provided in the standard DICOM data format with
12-bit data representation.
We opted to use T1-weighted MRI scans with the
spatial size of 256 × 256 pixels, in conjunction with
CT images with a size of 512 × 512 pixels.
To register CT and MR volume pairs, we uti-
lized a mutual-information-based multi-resolution
algorithm (Mattes et al., 2003) using the Sim-
pleITK (Lowekamp et al., 2013; Yaniv et al., 2018;
Beare et al., 2018) framework. During the registration
process, due to its higher spatial resolution, the CT
volume was chosen as a fixed volume, while the cor-
responding MR volume was considered as a moving
one. Furthermore, linear interpolation was utilized to
resize the MR scans, and the Gradient Descent with a
learning rate of 0.01 was used to optimize the mutual
information between both scans.
After alignment, the registered volumes were first
brought to homogeneous voxel spacing. We then ad-
justed the field of view of each volume based on the
achieved spatial resolution. By cropping from the
center of the image or adding padding around its bor-
ders, we achieved an approximately equivalent field
of view. Next, we resized MR and CT image slices to
256 × 256 pixels, which is an input resolution for our
networks, and visually inspected them. Due to the dif-
fering initial fields of view between the unregistered
MR and CT volumes, specific MR/CT slices were left
without valid counterparts after registration. These
slices were typically located at the upper or lower ex-
tremities of the registered volumes (axial plane) and
were subsequently omitted. As a final validation step,
we examined the retained 553 MR-CT image pairs
(from all sixteen patients).
Table 1: Cross-validation details; Train / Valid / Test de-
note which patients were used during the training / valida-
tion / testing phase within each of four folds. The last col-
umn (Slice) represents the number of available paired slices
per patient. Patient IDs (Pat. ID) correspond to filenames in
the original data set.
Pat. ID Fold1 Fold2 Fold3 Fold4 Slice
001 Train Train Train Test 25
002 Train Test Train Train 24
003 Train Train Valid Test 19
004 Valid Test Train Train 18
005 Train Valid Test Train 26
006 Test Train Train Train 23
007 Test Train Train Valid 26
101 Train Valid Test Train 47
102 Train Train Train Test 48
103 Train Test Train Train 44
104 Train Train Valid Test 46
105 Valid Test Train Train 37
106 Train Train Test Train 44
107 Train Train Test Train 45
108 Test Train Train Train 40
109 Test Train Train Valid 41
Total number of slices in the data set:
∑
553
4.2 Experimental Details
All experiments were conducted in a four-fold cross-
validation manner, with four patients reserved for
each testing phase, while of the remaining twelve, ten
were used for training and two for validation. De-
tailed information regarding the utilized data split is
provided in Table 1.
To improve the model’s ability to generalize to un-
seen data, we enhanced image diversity by employing
data augmentation techniques in the form of random
rotations (within a range of ±7.5 degrees), scaling
(with a factor ranging from 1 to 1.15), and horizon-
tal flipping (with a 50% probability chance).
BIOIMAGING 2024 - 11th International Conference on Bioimaging
228
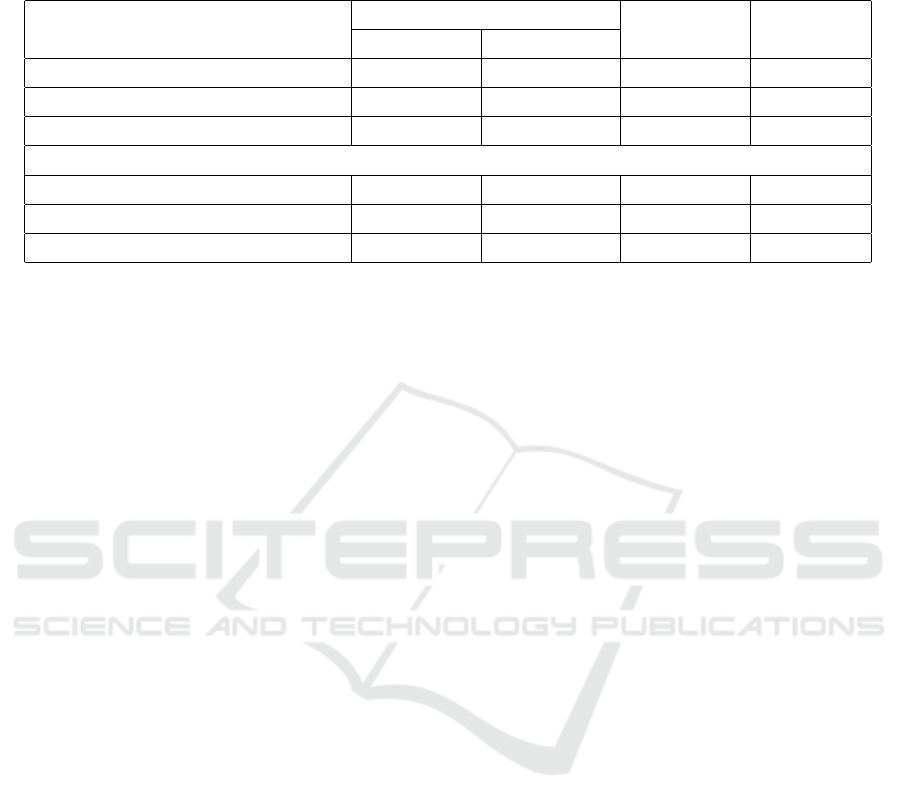
Table 2: Evaluation of pseudo-CT synthesis with respect to the images as a whole. Each evaluation metric is given with its
average value ± corresponding standard deviation. While MAE and MSE values are given in HU and HU
2
, SSIM and PSNR
values are reported in % and dB, respectively. The best results within U-Nets and cWGANs are highlighted bold.
Entire Image
Name ↓ MAE [HU] ↓ MSE [HU
2
] ↑ PSNR [dB] ↑ SSIM [%]
U-Net (Ronneberger et al., 2015) 101±35 69139±27664 24.3±1.9 79.6±6.8
Att U-Net (Oktay et al., 2018) 99±32 64919±22973 24.4±1.6 80.1±5.9
Att U-Net ES (Dovletov et al., 2023a) 99±35 61910±26966 24.8±2.0 80.2±6.3
cWGAN with U-Net 113±37 80507±31839 23.7±1.9 77.2±7.3
cWGAN with Att U-Net 114±39 75709±32252 23.9±2.0 76.5±7.6
cWGAN with Att U-Net ES 104±37 67125±29250 24.5±2.1 78.4±7.3
In our baseline U-Net implementation, we started
with 32 convolutional kernels of the size of 5× 5 pix-
els, and we doubled the number of learnable features
for each subsequent image resolution level. More-
over, we employed two consecutive convolutional
layers with zero-padding at each resolution level. In
the encoding path, max pooling with a window size of
2× 2 pixels and a stride of 2 pixels was utilized, while
in the decoding path, learnable transposed convolu-
tions were employed. At each upsampling step, the
number of output features was reduced by half com-
pared to the corresponding input channels. We ap-
plied the Rectified Linear Unit (ReLU) as a non-linear
activation function. We used 1 × 1 pixels convolution
as the final layer to generate a single-channel output
image.
The core architecture of our baseline Attention
U-Net remains the same, except for the embedding
of attention gates. Similarly, as in the original paper,
we used the sigmoid activation function to normalize
attention coefficients within the attention maps.
The previously described baseline U-Net and At-
tention U-Net architectures were used as the gener-
ator networks in our baseline Wasserstein GAN ap-
proaches. Furthermore, we included a hyperbolic tan-
gent (tanh) activation layer as the final activation layer
of the generators to enhance the effectiveness of the
training process.
In our critic architecture, we started with 32 con-
volutional kernels at the initial resolution level. As
suggested by (Radford et al., 2015), we utilized 4 × 4
pixels kernels with a stride of 2 pixels and 1-pixel
padding in both spatial dimensions instead of using
max pooling layers. Following each convolutional
layer, we applied the LeakyReLU non-linear activa-
tion function. The number of filters was doubled with
each subsequent image resolution. We utilized an ad-
ditional batch normalization layer at each resolution
level to enhance training stability, except for the first
one. Strided convolutional layers were iteratively em-
ployed until we obtained a single scalar value as the
output for each input image.
The exact same architecture as described previ-
ously (with Attention U-Net as a generator) was used
for the proposed conditional Wasserstein GAN ES.
We generated the required bone masks for ad-
ditional supervision by applying a global threshold-
based segmentation approach to the ground truth CT
images. We observed that the threshold value of
350 HU delivers reasonable results for the utilized
data set and is in the same range as suggested in the
literature (Buzug, 2009; Chougule et al., 2018; Wang
et al., 2019; Dovletov et al., 2023b; Yaakub et al.,
2023). However, since our GAN network expects nor-
malized images as input for its generator, this value
was mapped to the range between -1 and 1, which led
to the threshold value of -0.329.
When calculating the total loss function, we set
λ
adv
to 10, following (Isola et al., 2017), and we chose
300 for λ
ES
. We set all λ
l
(l ∈ {1, 2, 3, 4}) hyper-
parameters uniformly to 0.25, signifying the equal
importance of all attention gates of the Attention U-
Net network. We conducted additional experiments
with λ
l
values set to {0.012, 0.047, 0.118, 0.753} in
ascending and descending orders to analyze the im-
pact of hyperparameters on the quality of pseudo-CT
synthesis. These values were calculated by dividing
the pixel count of each attention map by the total num-
ber of pixels in all four attention maps, thus ensuring
a cumulative sum of one.
We implemented all our models in Python using
the PyTorch (Paszke et al., 2019) framework and exe-
cuted them on NVIDIA GTX 1080 TI GPUs equipped
with 11 GB VRAM.
The U-Net models were trained for 100 epochs us-
ing the Adam optimizer (β
1
= 0.9, β
2
= 0.999) with
a learning rate of 0.01. On the other hand, cWGAN
models were trained for 1000 epochs using the two
Bone-Aware Generative Adversarial Network with Supervised Attention Mechanism for MRI-Based Pseudo-CT Synthesis
229

time-scale update rule as suggested by (Heusel et al.,
2017). Thus, learning rates for generator and discrim-
inator networks were set to 0.0002 and 0.0004, re-
spectively. Moreover, in the case of cWGANs, we
incorporated an additional gradient penalty as pro-
posed by (Wu et al., 2018), and we used the RMSProp
optimizer during their training, as suggested by (Ar-
jovsky et al., 2017), to avoid instability issues. Addi-
tionally, one-sided label smoothing (Salimans et al.,
2016) was utilized. Both U-Net and cWGAN mod-
els were trained using mini-batches containing six-
teen images each.
4.3 Evaluation Metrics
Although the above-mentioned neural networks op-
erate in a 2D mode, it is crucial to evaluate using
3D volumes. Therefore, the produced 2D pseudo-CT
images of a patient were stacked to construct a 3D
volume before being compared to the desired ground
truth volume.
We chose Mean Squared Error (MSE) and Mean
Absolute Error (MAE) as pixel-wise quality assess-
ment metrics. These metrics were computed for both
entire volumes and specific regions of interest, the
head and bone regions.
To obtain the necessary head masks, we initially
generated them from MR images by applying Otsu’s
thresholding algorithm, followed by morphological
opening and closing operations. These masks were
subsequently validated and, if required, manually re-
fined. Initially, we employed a morphological open-
ing operation with a circular structuring element of 5
pixels in radius to eliminate minor artifacts from the
initial segmentations. Following that, we used a clos-
ing operation with a radius of 25 pixels to fill gaps in
the nasal areas. Lastly, a morphological dilation oper-
ation with a radius of 5 pixels was used to expand the
overall shape of the segments slightly.
For evaluating errors within the bone regions, we
utilized the same bone segmentation maps that had
been used during the training phase to guide the gen-
erator network (Attention U-Net).
Thus, bone masks allow quantifying errors in the
bone regions only, whereas head masks cover every-
thing except the background.
To facilitate a more comprehensive comparison of
the synthesized pseudo-CTs and ground truth CT im-
ages, we also computed the Peak Signal-to-Noise Ra-
tio (PSNR) (Hore and Ziou, 2010) as follows:
PSNR = 10 · log
10
I
2
MSE(CT, pCT)
(7)
where I represents the maximum intensity value for
the CT image. Thus, for the standard DICOM bit
depth of 12 bits, this value is set to 4095 (= 2
12
− 1).
Furthermore, we calculated the Structural Similarity
Index Measure (SSIM) (Wang et al., 2004) as follows:
SSIM =
(2µ
CT
µ
pCT
+C
1
)(2σ
CTpCT
+C
2
)
(µ
2
CT
+ µ
2
pCT
+C
1
)(σ
2
CT
+ σ
2
pCT
+C
2
)
(8)
where µ
pCT
and µ
CT
denote the mean HU values of
pseudo-CT and CT images, with σ
pCT
and σ
CT
repre-
senting their respective variances, while σ
CTpCT
sig-
nifies the covariance between two images. The pa-
rameters C
1
=(k
1
I)
2
and C
2
=(k
2
I)
2
are two variables
to stabilize division when dealing with weak denom-
inators (k
1
= 0.01, k
2
= 0.03). SSIM values vary be-
tween 0 and 1, and as the similarity between the gen-
erated pseudo-CT and the corresponding CT image
increases, the SSIM value approaches closer to 1.
To better assess the geometric accuracy of bone
structures, we also computed the Dice Similarity Co-
efficient (DSC) between binarized CT and pseudo-CT
images using the following equation:
DSC =
2 · |Seg
CT
∩ Seg
pCT
|
|Seg
CT
| + |Seg
pCT
|
(9)
where Seg
CT
and Seg
pCT
represent binarized bone
segmentations obtained from real CT and synthesized
pCT images, respectively. A higher DSC value repre-
sents a larger intersection between two segmentation
and thus indicates a greater similarity between the two
images.
5 RESULTS
We compare the performance of the six models
quantitatively using evaluation metrics from Subsec-
tion 4.3. The obtained results are outlined in Table 2
and Table 3, with the first table focusing on values re-
lated to the images as a whole and the latter on areas
of interest.
Although we conducted cWGAN experiments
with different λ
l
(l ∈ {1, 2, 3, 4}) settings (as de-
scribed in Subsection 4.2), we only report the results
for one experiment with lambdas set uniformly to
0.25 value. The main reason is that we did not notice
a substantial improvement when using other configu-
rations, which is consistent with findings in (Dovletov
et al., 2023a).
Our proposed approach of cWGAN with At-
tention U-Net ES outperforms all other conditional
Wasserstein GAN models in every evaluation met-
ric. When considering entire generated pseudo-CTs,
our approach introduces a gain of 8.8% in MAE and
11.3% in MSE compared to its counterpart, namely,
BIOIMAGING 2024 - 11th International Conference on Bioimaging
230
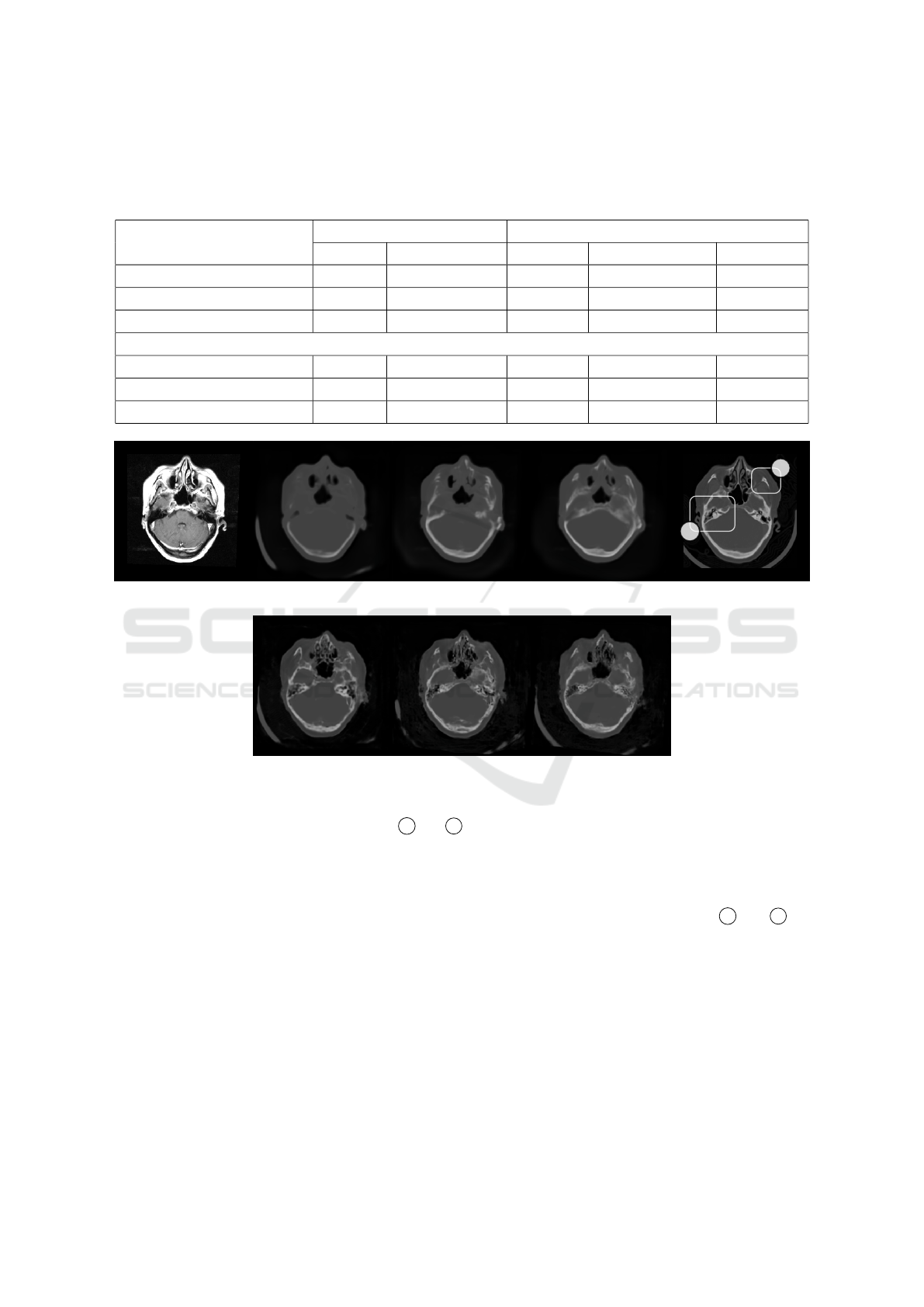
Table 3: Evaluation of pseudo-CT synthesis with respect to head and bone regions of interest. Each evaluation metric is given
with its average value ± corresponding standard deviation. While MAE and MSE values are given in HU and HU
2
, SSIM
and PSNR values are reported in % and dB, respectively. DSC values are also reported in %. The best results within U-Nets
and cWGANs are highlighted bold.
Head Area Bone Area
Name ↓ MAE ↓ MSE ↓ MAE ↓ MSE ↑ DSC [%]
U-Net 180±30 131393±38343 595±120 532695±198331 60.1±9.4
Att U-Net 183±29 131679±32502 548±90 447014±122953 62.4±9.0
Att U-Net ES 173±30 117800±33330 432±88 310223±111930 67.3±7.7
cWGAN with U-Net 202±34 154101±42147 493±90 408417±131774 61.5±9.4
cWGAN with Att U-Net 193±32 138151±36758 463±91 357665±119125 63.8±8.6
cWGAN with Att U-Net ES 178±34 124601±37336 438±88 325305±115481 66.7±8.7
(a) MR. (b) U-Net. (c) Att U-Net. (d) Att U-Net ES.
A
B
(e) CT.
(f) cWGAN with
U-Net.
(g) cWGAN with
Att U-Net.
(h) cWGAN with
Att U-Net ES.
Figure 3: Synthetic pseudo-CT images. (a) Input MR image; Pseudo-CTs from (b-d) U-Nets and (f-h) cWGANs; (e) Corre-
sponding ground truth CT image. Bounding boxes A and B annotate the temporal and zygomatic bone, correspond-
ingly. Note the improved synthesis of bone structures from the proposed cWGAN approach with Attention U-Net
and ES in (h) compared to the results from baseline models in (f and g).
cWGAN with Attention U-Net, without additional su-
pervision. With 0.6 dB and 1.9% gain, the corre-
sponding PSNR and SSIM values are also slightly im-
proved.
More importantly, we achieved significant im-
provements in bone areas. Specifically, DSC is 5.2%
and 2.9% higher when compared to cWGAN with U-
Net and cWGAN with Attention U-Net, respectively.
Additionally, the fact that results for the head area are
also better in our approach implies that the improve-
ment around the bone area is not coming at the cost
of error in other regions.
These findings can be further supported by the vi-
sual comparison of pseudo-CTs in Figure 3 (bottom
row) and by looking more closely at
A and B re-
gions in these images. It can be noted that both base-
line cWGAN models are capable of producing air-
filled cavities within the temporal bone of the cranium
at the correct positions. However, the bone structures
are not always correctly synthesized, such as in the
right half of the images. Moreover, the baseline mod-
els produce some bone artifacts around the zygomatic
bones (or cheeks). In contrast, our approach can more
accurately synthesize the previously mentioned bone
structures.
To investigate the impact of additional guid-
Bone-Aware Generative Adversarial Network with Supervised Attention Mechanism for MRI-Based Pseudo-CT Synthesis
231
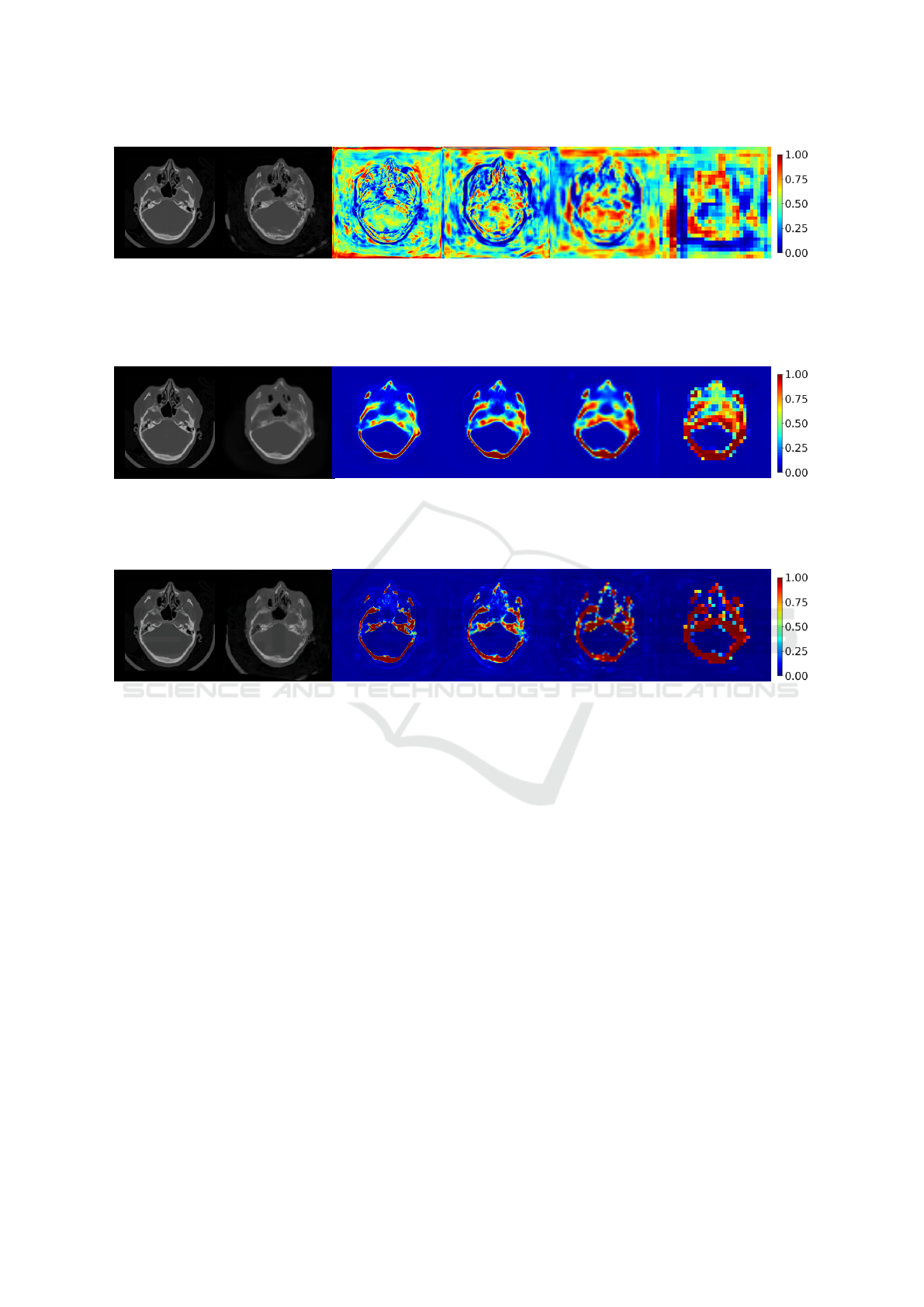
(a) CT. (b) Pseudo-CT. (c) AM
1
. (d) AM
2
. (e) AM
3
. (f) AM
4
.
Figure 4: Learned Attention Maps (AMs) from cWGAN with Attention U-Net. (a) CT image; (b) Pseudo-CT; (c-f) AMs from
the corresponding generator (Attention U-Net). The superscript l in AM
l
indicates the attention map learned by the attention
gate at the l-th image resolution level as described in Subsection 3.3. The focus of attention maps is distributed along the
entire image space. Attention at lowest resolution levels (corresponds to abstract features) partially covers the bone regions.
(a) CT. (b) Pseudo-CT. (c) AM
1
. (d) AM
2
. (e) AM
3
. (f) AM
4
.
Figure 5: Learned Attention Maps (AMs) from Attention U-Net ES (Dovletov et al., 2023a). (a) CT image; (b) Pseudo-CT;
(c-f) AMs from Attention U-Net ES. Attention maps at all resolutions levels focus on the overall shape of bones without
capturing fine details. As a result, this limitation is inherited by the synthesized pseudo-CT images.
(a) CT. (b) Pseudo-CT. (c) AM
1
. (d) AM
2
. (e) AM
3
. (f) AM
4
.
Figure 6: Learned Attention Maps (AMs) from cWGAN with Attention U-Net with Extra Supervision. (a) CT image;
(b) Pseudo-CT; (c-f) AMs from the corresponding generator (Attention U-Net ES). Attention maps at all resolution level
more accurately cover the bone regions, while at the same time almost completely ignore (zero values) irrelevant background
information.
ance, we visualize attention maps from the base-
line cWGAN with Attention U-Net as the generator
and from the proposed cWGAN with ES. Attention
maps from all four resolution levels from the baseline
model are depicted in Figure 4. As can be seen, the
generator’s focus is distributed along the whole image
space, and only the attention map at the lowest resolu-
tion level (see Figure 4f) partially focuses on the bone
structures. Such behavior implies that the network
relies more on high-level features when learning the
mapping from MRI to CT domain. In comparison, at-
tention maps from cWGAN with Attention U-Net ES
in Figure 6 clearly emphasize bone regions at all res-
olution levels. We also note that the extra supervision
allows the generator network to learn suitable AMs
that ignore other details, such as background noise or
the patient’s table.
Another important finding is seen when compar-
ing Attention U-Net ES (Dovletov et al., 2023a) with
the proposed approach, with the latter using an identi-
cal network as the generator in a conditional Wasser-
stein GAN setting. Both approaches clearly outper-
form the rest of the models when it comes to head
and bone areas. However, Attention U-Net ES is al-
ways quantitatively better than its GAN extension,
with the most significant difference (percentage-wise)
being the MSE value around the head area at 5.4%.
The main reason for this discrepancy can be seen
in Figure 3, specifically when comparing images in
Figures 3d and 3h and their corresponding attention
maps. As seen from Figure 5, Attention U-Net ES
focuses well on areas with bones. However, it pays
little attention to details but instead captures the over-
all shape of the bones. This results in the network
distributing its values in every position where there
might be a bone, allowing it to gain the stat-wise ad-
BIOIMAGING 2024 - 11th International Conference on Bioimaging
232

vantage over the proposed approach. As a result, the
corresponding pseudo-CT in Figure 3d also lacks de-
tails. Moreover, bone structures appear slightly in-
creased in form, with a smoothly curved outer shape
and blurry inner parts. In comparison, the proposed
approach not only learns to focus on bone regions but
also learns to pay particular attention to details. This
statement can be supported by visually inspecting the
corresponding attention maps in Figure 6 that delin-
eate the bone structures in more detail. Furthermore,
attention values are more clearly distributed in two
high-density regions (two distinct peaks with values
close to zero or one), indicating that the proposed net-
work focuses more reliably on bone regions and thus
produces more realistic fine-grained pseudo-CTs.
6 CONCLUSION
This paper presents a conditional Wasserstein GAN
approach that utilizes an Attention U-Net network as
the generator and includes a domain-specific atten-
tion mechanism for more accurate synthesis of bone
structures when generating pseudo-CT images from
the given MR images. The adopted attention mecha-
nism has been recently published in (Dovletov et al.,
2023a) and leverages the bone segmentation masks
obtained by thresholding from ground truth CTs to
guide the image-to-image translation network to learn
a better mapping function. Although this attention
mechanism improves the quantitative results within
the bone regions, the synthesized bone structures ap-
pear blurry and lack details. The proposed generative
approach allows for overcoming this limitation. The
presented qualitative and quantitative results confirm
that incorporating additional domain knowledge can
significantly reduce errors in bone regions and, thus,
provide more accurate pseudo-CT compared to two
baseline conditional Wasserstein GAN models.
REFERENCES
Arjovsky, M., Chintala, S., and Bottou, L. (2017). Wasser-
stein Generative Adversarial Networks. In Interna-
tional conference on machine learning, pages 214–
223. PMLR.
Beare, R., Lowekamp, B., and Yaniv, Z. (2018). Image
Segmentation, Registration and Characterization in R
with SimpleITK. Journal of statistical software, 86.
Buzug, T. M. (2009). Computed Tomography: From Photon
Statistics to Modern Cone-Beam CT.
Chougule, V., Mulay, A., and Ahuja, B. (2018). Clini-
cal Case Study: Spine Modeling for Minimum Inva-
sive Spine Surgeries (MISS) using Rapid Prototyping.
Bone (CT), 226:3071.
Dovletov, G., Karadeniz, U., L
¨
orcks, S., Pauli, J., Gratz,
M., and Quick, H. H. (2023a). Learning to Pay Atten-
tion by Paying Attention: Attention U-Net with Ex-
tra Supervision for MRI-based Pseudo-CT Synthesis.
In Scandinavian Conference on Image Analysis, pages
229–242. Springer.
Dovletov, G., L
¨
orcks, S., Pauli, J., Gratz, M., and Quick,
H. H. (2023b). Double Grad-CAM Guidance for Im-
proved MRI-based Pseudo-CT Synthesis. In BVM
Workshop, pages 45–50. Springer.
Dovletov, G., Pham, D. D., L
¨
orcks, S., Pauli, J., Gratz,
M., and Quick, H. H. (2022a). Grad-CAM Guided
U-Net for MRI-based Pseudo-CT Synthesis. In 2022
44th Annual International Conference of the IEEE En-
gineering in Medicine & Biology Society (EMBC),
pages 2071–2075. IEEE.
Dovletov, G., Pham, D. D., Pauli, J., Gratz, M., and Quick,
H. H. (2022b). Improved MRI-based Pseudo-CT Syn-
thesis via Segmentation Guided Attention Networks.
In Proceedings of the 15th International Joint Confer-
ence on Biomedical Engineering Systems and Tech-
nologies - BIOIMAGING, pages 131–140. INSTICC,
SciTePress.
Ge, Y., Wei, D., Xue, Z., Wang, Q., Zhou, X., Zhan,
Y., and Liao, S. (2019). Unpaired MR to CT Syn-
thesis with Explicit Structural Constrained Adversar-
ial Learning. In 2019 IEEE 16th International Sym-
posium on Biomedical Imaging (ISBI 2019), pages
1096–1099. IEEE.
Gong, K., Yang, J., Kim, K., El Fakhri, G., Seo, Y., and
Li, Q. (2018). Attenuation Correction for Brain PET
Imaging Using Deep Neural Network Based on Dixon
and ZTE MR Images. Physics in Medicine & Biology,
63(12):125011.
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B.,
Warde-Farley, D., Ozair, S., Courville, A., and Ben-
gio, Y. (2014). Generative Adversarial Nets. Advances
in neural information processing systems, 27.
Han, X. (2017). MR-based Synthetic CT Generation Using
a Deep Convolutional Neural Network Method. Med-
ical physics, 44(4):1408–1419.
Heusel, M., Ramsauer, H., Unterthiner, T., Nessler, B., and
Hochreiter, S. (2017). GANs Trained by a Two Time-
Scale Update Rule Converge to a Local Nash Equi-
librium. Advances in neural information processing
systems, 30.
Hore, A. and Ziou, D. (2010). mage Quality Metrics: PSNR
vs. SSIM. In 2010 20th international conference on
pattern recognition, pages 2366–2369. IEEE.
Hu, J., Shen, L., and Sun, G. (2018). Squeeze-and-
Excitation Networks. In Proceedings of the IEEE con-
ference on computer vision and pattern recognition,
pages 7132–7141.
Isola, P., Zhu, J.-Y., Zhou, T., and Efros, A. A. (2017).
Image-to-Image Translation with Conditional Adver-
sarial Networks. In Proceedings of the IEEE con-
ference on computer vision and pattern recognition,
pages 1125–1134.
Bone-Aware Generative Adversarial Network with Supervised Attention Mechanism for MRI-Based Pseudo-CT Synthesis
233

Leynes, A. P., Yang, J., Wiesinger, F., Kaushik, S. S.,
Shanbhag, D. D., Seo, Y., Hope, T. A., and Lar-
son, P. E. (2018). Zero-echo-time and Dixon Deep
Pseudo-CT (ZeDD CT): Direct Generation of Pseudo-
CT Images for Pelvic PET/MRI Attenuation Correc-
tion Using Deep Convolutional Neural Networks with
Multiparametric MRI. Journal of Nuclear Medicine,
59(5):852–858.
Lowekamp, B. C., Chen, D. T., Ib
´
a
˜
nez, L., and Blezek, D.
(2013). The Design of SimpleITK. Frontiers in neu-
roinformatics, 7:45.
Mattes, D., Haynor, D. R., Vesselle, H., Lewellen, T. K., and
Eubank, W. (2003). PET-CT Image Registration in the
Chest Using Free-form Deformations. IEEE transac-
tions on medical imaging, 22(1):120–128.
Mirza, M. and Osindero, S. (2014). Conditional Generative
Adversarial Nets. arXiv preprint arXiv:1411.1784.
Nie, D., Cao, X., Gao, Y., Wang, L., and Shen, D. (2016).
Estimating CT Image from MRI Data Using 3D Fully
Convolutional Networks. In Deep Learning and Data
Labeling for Medical Applications: First Interna-
tional Workshop, LABELS 2016, and Second Inter-
national Workshop, DLMIA 2016, Held in Conjunc-
tion with MICCAI 2016, Athens, Greece, October 21,
2016, Proceedings 1, pages 170–178. Springer.
Oktay, O., Schlemper, J., Folgoc, L. L., Lee, M., Hein-
rich, M., Misawa, K., Mori, K., McDonagh, S., Ham-
merla, N. Y., Kainz, B., et al. (2018). Attention U-
Net: Learning Where to Look for the Pancreas. arXiv
preprint arXiv:1804.03999.
Park, J., Woo, S., Lee, J.-Y., and Kweon, I. S. (2018).
BAM: Bottleneck Attention Module. arXiv preprint
arXiv:1807.06514.
Paszke, A., Gross, S., Massa, F., Lerer, A., Bradbury, J.,
Chanan, G., Killeen, T., Lin, Z., Gimelshein, N.,
Antiga, L., et al. (2019). Pytorch: An imperative
style, high-performance deep learning library. In
Advances in neural information processing systems,
pages 8026–8037.
Paulus, D. H., Quick, H. H., Geppert, C., Fenchel, M.,
Zhan, Y., Hermosillo, G., Faul, D., Boada, F.,
Friedman, K. P., and Koesters, T. (2015). Whole-
body PET/MR Imaging: Quantitative Evaluation
of a Novel Model-based MR Attenuation Correc-
tion Method Including Bone. Journal of Nuclear
Medicine, 56(7):1061–1066.
Qi, M., Li, Y., Wu, A., Jia, Q., Li, B., Sun, W., Dai, Z.,
Lu, X., Zhou, L., Deng, X., et al. (2020). Multi-
sequence MR Image-based Synthetic CT Generation
Using a Generative Adversarial Network for Head
and Neck MRI-only Radiotherapy. Medical physics,
47(4):1880–1894.
Quick, H. H. (2014). Integrated PET/MR. Journal of mag-
netic resonance imaging, 39(2):243–258.
Radford, A., Metz, L., and Chintala, S. (2015). Un-
supervised Representation Learning with Deep Con-
volutional Generative Adversarial Networks. arXiv
preprint arXiv:1511.06434.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-Net:
Convolutional Networks for Biomedical Image Seg-
mentation. In International Conference on Medical
image computing and computer-assisted intervention,
pages 234–241. Springer.
Salimans, T., Goodfellow, I., Zaremba, W., Cheung, V.,
Radford, A., and Chen, X. (2016). Improved Tech-
niques for Training GANs. Advances in neural infor-
mation processing systems, 29.
Schmidt, M. A. and Payne, G. S. (2015). Radiotherapy
Planning Using MRI. Physics in Medicine & Biology,
60(22):R323.
Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R.,
Parikh, D., and Batra, D. (2017). Grad-CAM: Visual
Explanations from Deep Networks via Gradient-based
Localization. In Proceedings of the IEEE interna-
tional conference on computer vision, pages 618–626.
Torrado-Carvajal, A., Vera-Olmos, J., Izquierdo-Garcia, D.,
Catalano, O. A., Morales, M. A., Margolin, J., Sori-
celli, A., Salvatore, M., Malpica, N., and Catana,
C. (2019). Dixon-VIBE Deep Learning (DIVIDE)
Pseudo-CT Synthesis for Pelvis PET/MR Attenuation
Correction. Journal of nuclear medicine, 60(3):429–
435.
Wang, Y., Liu, C., Zhang, X., and Deng, W. (2019). Syn-
thetic CT Generation Based on T2 Weighted MRI
of Nasopharyngeal Carcinoma (NPC) Using a Deep
Convolutional Neural Network (DCNN). Frontiers in
oncology, 9:1333.
Wang, Z., Bovik, A. C., Sheikh, H. R., and Simoncelli, E. P.
(2004). Image Quality Assessment: From Error Vis-
ibility to Structural Similarity. IEEE transactions on
image processing, 13(4):600–612.
West, J., Fitzpatrick, J. M., Wang, M. Y., Dawant, B. M.,
Maurer Jr, C. R., Kessler, R. M., Maciunas, R. J., Bar-
illot, C., Lemoine, D., Collignon, A., et al. (1997).
Comparison and Evaluation of Retrospective Inter-
modality Brain Image Registration Techniques. Jour-
nal of computer assisted tomography, 21(4):554–568.
Wolterink, J. M., Dinkla, A. M., Savenije, M. H., Seevinck,
P. R., van den Berg, C. A., and I
ˇ
sgum, I. (2017). Deep
MR to CT Synthesis Using Unpaired Data. In Interna-
tional Workshop on Simulation and Synthesis in Med-
ical Imaging, pages 14–23. Springer.
Woo, S., Park, J., Lee, J.-Y., and Kweon, I. S. (2018).
CBAM: Convolutional Block Attention Module. In
Proceedings of the European conference on computer
vision (ECCV), pages 3–19.
Wu, J., Huang, Z., Thoma, J., Acharya, D., and Van Gool,
L. (2018). Wasserstein Divergence for GANs. In Pro-
ceedings of the European conference on computer vi-
sion (ECCV), pages 653–668.
Xiang, L., Li, Y., Lin, W., Wang, Q., and Shen, D. (2018).
Unpaired Deep Cross-Modality Synthesis with Fast
Training. In Deep Learning in Medical Image Anal-
ysis and Multimodal Learning for Clinical Decision
Support: 4th International Workshop, DLMIA 2018,
and 8th International Workshop, ML-CDS 2018, Held
in Conjunction with MICCAI 2018, Granada, Spain,
September 20, 2018, Proceedings 4, pages 155–164.
Springer.
Xie, S., Girshick, R., Doll
´
ar, P., Tu, Z., and He, K. (2017).
Aggregated Residual Transformations for Deep Neu-
ral Networks. In Proceedings of the IEEE conference
BIOIMAGING 2024 - 11th International Conference on Bioimaging
234

on computer vision and pattern recognition, pages
1492–1500.
Yaakub, S. N., White, T. A., Roberts, J., Martin, E., Verha-
gen, L., Stagg, C. J., Hall, S., and Fouragnan, E. F.
(2023). Transcranial Focused Ultrasound-mediated
Neurochemical and Functional Connectivity Changes
in Deep Cortical Regions in Humans. Nature Commu-
nications, 14(1):5318.
Yaniv, Z., Lowekamp, B. C., Johnson, H. J., and Beare,
R. (2018). SimpleITK Image-analysis Notebooks:
a Collaborative Environment for Education and Re-
producible Research. Journal of digital imaging,
31(3):290–303.
Zhu, J.-Y., Park, T., Isola, P., and Efros, A. A. (2017).
Unpaired Image-to-Image Translation Using Cycle-
Consistent Adversarial Networks. In Proceedings of
the IEEE international conference on computer vi-
sion, pages 2223–2232.
Bone-Aware Generative Adversarial Network with Supervised Attention Mechanism for MRI-Based Pseudo-CT Synthesis
235
