
Predicting the Level of Co-Activation of One Muscle Head from the
Other Muscle Head of the Biceps Brachii Muscle by Linear Regression
and Shallow Feedforward Neural Networks
Nils Grimmelsmann
1,2,∗ a
, Malte Mechtenberg
1,2 b
, Markus Vieth
3 c
, Alexander Schulz
3 d
,
Barbara Hammer
3 e
and Axel Schneider
1,2 f
1
Biomechatronics and Embedded Systems Group, University of Applied Sciences and Arts, Bielefeld, Germany
2
Institute of System Dynamics and Mechatronics, University of Applied Sciences and Arts, Bielefeld, Germany
3
Machine Learning Group, Bielefeld University, Bielefeld, Germany
Keywords:
sEMG, Muscle Model, Limb Movement Prediction, Virtual Sensor, Linear Regression, Regression.
Abstract:
One of the challenges in close-to-body robotics is the intuitive control of exoskeletal devices which requires
lag-free responses of its actuated joints. A frequently used signal domain to satisfy the required control
properties is surface electromyography (sEMG). By using a Hill-type model of the muscle mainly responsible
for the movement of a biological joint, which is excited by the corresponding sEMG of this muscle, the joint
movement can be pre-calculated. If the muscle internal delays are used, this information can be used for an
intuitive and lag-free control. So far, biomechanical limb and joint models including Hill-type muscle sub-
model were used. In current studies, state-of-the-art machine learning models are evaluated for this problem.
Both types, classical and machine learning models, depend on the measured sEMG signals of all muscle heads
of a relevant muscle and on their respective signal quality.
This work introduces a method to train a virtual sEMG-sensor as a replacement for the real sEMG signal of
a muscle head, thus reducing the number of real sensor electrodes on a given muscle. The virtual sensor is
trained based on data from the remaining sensor. This method allows to compare the measured sEMG signal
with the virtual sensor output to assess the measured signal. Furthermore, this study explains the training
process and evaluates the use of the virtual sensor in a biomechanical limb model.
.
1 INTRODUCTION
The use of active exoskeletons and wearables
provides a way to support the wearer during force-
intensive movements. This requires time critical
motion prediction so that the active exoskeleton
can be controlled in a way to follow the postures
of the wearer without delay. Electromyography
(EMG) signals provide a source of information for
the required movement prediction which can also
be measured before the muscle contracts and an
actual limb movement or force interaction with the
a
https://orcid.org/0000-0002-4864-4978
b
https://orcid.org/0000-0002-8958-0931
c
https://orcid.org/0000-0003-1707-6231
d
https://orcid.org/0000-0002-0739-612X
e
https://orcid.org/0000-0002-0935-5591
f
https://orcid.org/0000-0002-6632-3473
exoskeleton occurs. EMG signals can be measured
invasively with needles inserted into the muscle
(Merletti and Farina, 2008). This method requires
special medical knowledge on the correct insertion of
the needle into the muscle. It provides measurements
that are less affected by crosstalk, so the signal
amplitude from the selected muscle head is higher
than that from the surrounding muscle heads. The
surface electromyography (sEMG), on the other hand,
requires no special medical knowledge and is non-
invasive. Measuring EMG on the surface of the
skin means that only superficial muscles or muscle
heads can be measured. Muscles or muscle heads
that lie deep below other muscle tissue are less
accessible with this measurement method. Therefore,
not all muscles or muscle heads involved in a limb
movement can be used in a model to predict the limb
movement when sEMG is the source of information.
Grimmelsmann, N., Mechtenberg, M., Vieth, M., Schulz, A., Hammer, B. and Schneider, A.
Predicting the Level of Co-Activation of One Muscle Head from the Other Muscle Head of the Biceps Brachii Muscle by Linear Regression and Shallow Feedforward Neural Networks.
DOI: 10.5220/0012368700003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 1, pages 611-621
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
611

For example, if the movement of the forearm
is considered via the elbow joint, which has one
rotatory degree of freedom, mainly three muscles are
used for flexing: the two-headed biceps brachii, the
single-headed brachioradialis and the single-headed
brachialis. The extension of the elbow is mainly
performed by two muscles: the three-headed triceps
brachii and the single-headed anconeus.
The two biceps heads (biceps short head and
biceps long head) are located below the skin surface.
Both muscle heads end distally in a common tendon.
Proximally, each muscle head ends in its own tendon.
The tendons terminate at different points on the
shoulder bone, called scapular. Both heads can
flex the elbow but also have secondary functions.
The proximal tendon of the biceps long head wraps
around the shoulder joint and stabilises the shoulder
(Sch
¨
unke et al., 2010).
As shown in previous work, limb movement can
be predicted with only the two surface muscles biceps
and triceps. Firstly, with a biomechanical limb model
based on a Hill-type (Hill, 1964; Zajac, 1989) muscle
model (Grimmelsmann et al., 2023). Secondly, with
a purely data-driven (black box) approach (Leserri
et al., 2022). Depending on the experiment and data
set, a different set of the five main muscles involved
in the flexion and extension of the elbow are used
for elbow movement prediction (e.g. (Koo and Mak,
2005) uses biceps, brachioradialis and the tricpes).
However, this work also shows that the signal
quality and signal integrity can be different for two
muscle heads of the same muscle, e.g. due to
changing quality of the respective electrode skin
contact. This can potentially lead to a failed
prediction depending on the overall model structure.
Due to the common distal tendon, both biceps
heads show similarities in the time courses of their
respective neuronal activation.
For this reason, a method is proposed that exploits
the close similarity between the two biceps heads
to create a virtual sensor for one head based only
on the meassurement of the sEMG of the other.
This allows the use of only one sensor, namely the
one with the better signal quality. In principle,
the concept of a virtual sensor is also suited to
derive unavailable sEMG measurements, e.g. of
deep lying muscle heads, from easy to measure more
superficially located muscle heads, if their common
function suggests a similar activation in terms of time.
To test the concept of a virtual sensor, this work
follows the former of the above two applications and
replaces one of the two biceps heads, although sEMG
measurements of both superficially located heads are
available. Here, the unknown signal quality of the
two measured heads is an additional challenge (see
above). The signal of the virtual sensor for one head is
derived by linear regression and shallow feedforward
neural network (FFN) using the measurement of the
other head. To reflect the secondary functions of the
two heads, additional features are added as input to
the regression. These additional features are related
to the dynamics of the elbow and are the elbow angle,
the upper arm angle (w.r.t. the gravitational vector)
and the overall weight of lower arm plus hand and an
additional weight (dumb bell). After the regression
step, these virtual sensors are also used as input to
the biomechanical limb model (domain-knowledge
based model) to prove the suitability for the limb
movement prediction. In previous works, different
strategies were used to train virtual sensors. One
approach is to use recurrent neural network (RNN)
such as long short-term memory (LSTM) to estimate
the virtual sEMG channel (Machado et al., 2019).
This approach focuses mainly on the performance
w.r.t. the classification of a hand movement and
not on the interpretation of the underlying sEMG
data. The method proposed in this work uses linear
regression and shallow FFN.
Based on an extensive data set (Mechtenberg
et al., 2023), the outputs of the virtual sensor can
be compared with the neuronal activation calculated
from the real sEMG measurements of the replaced
muscle head on the one hand and evaluated w.r.t
their contribution to the predicted joint movement
on the other hand, as the virtual sensor output is
fed into the model instead of the replaced head. To
maintain interpretability the architecture/ complexity
is gradually extended in this work. Besides
interpreting the results of the virtual sensor, the sensor
is also validated using a domain-knowledge based
model.
Other methods such as (Kim et al., 2019) use
virtual sensors for a signal-assisted classification.
The general idea of using a trained regression for
enhancing the signal integrity matches parts of our
approach (sEMG assessment).
The methods section starts with an overview of
the sEMG data set and the biomechanical model of
the upper arm and the elbow joint. The internal
signal neuronal activation from this model serves as
a foundation for the domain knowledge-based feature
used in the regression (section 2.2). A description of
the different regression setups follows in section 2.3
and section 2.4. The virtual sensor will be used in a
domain knowledge-based model. The explanation of
the validation process for this purpose can be found
in section 2.5. The methods section ends with an
exemplary use case of the virtual sensor, the sEMG
BIOSIGNALS 2024 - 17th International Conference on Bio-inspired Systems and Signal Processing
612

post measurement assessment. The results of the two
setups are discussed in section 3.1 and section 3.2.
The results section also shows the validation results
and the assessment for two different subjects.
2 METHODS
As a basis for understanding the nature of the data
on which this study is based on (Mechtenberg et al.,
2023), a brief summary of the experiments used to
collect the data is given first. In these experiments,
the sEMG of both biceps and the two triceps heads
were recorded. The sEMG signals contain frequency
components from about 10 Hz to 400 Hz (Merletti
et al., 2018).
The periodic movement of the elbow (dumbbell
curls), however, is at about 0.5 Hz. Because of
this large difference in dynamics, the sEMG was
not directly used as the target value of the virtual
sensor regression. It is further converted into the
activation of the muscle. In the domain model, this is
achieved by a nonlinear low-pass filter that represents
the activation dynamics of the muscle head (Zajac,
1989). The resulting signal thus has lower frequency
components than the sEMG. The calculation of this
activation dynamics is briefly described in section 2.2.
In sections 2.3 and 2.4 a description of the two setups
for learning the virtual sensor is presented. For
validation of the virtual sensor, the sensor is used
as a replacement input in a domain-knowledge based
model (described in section 2.5). At the end of the
methods section a potential use case (the sEMG post-
measurement assessment) of the trained virtual sensor
is described (see section 2.6).
2.1 sEMG Data Set and Biomechanical
Overview
The used data set (Mechtenberg et al., 2023) involved
31 healthy subjects performing different motion
sequences, with 29 choosing their right arm as their
dominant arm and 2 choosing the left arm. They
were randomly labeled with an identification number
(subject id) starting from id=20. The subject ids
[23, 27, 35] were not assigned. The positions of the
acromion at the shoulder and the lateral epicondyle at
the elbow were used as reference points to calculate
the length of the upper arm. The positions of the
medial epicondyle and the processus styloideus ulnae
at the wrist were used to calculate the length of the
forearm.
Two wireless sEMG sensors (Delsys Trigno,
Delsys, Inc., Boston, MA, USA) were attached to
the skin surface above the biceps brachii and triceps
brachii. The sensors used a sampling period of
900µs and 16-bit resolution. The sensors were
applied such that their electrodes were placed on
the muscle belly proximal to the innervation zone.
The individual muscle heads of biceps and triceps
were palpated by an experienced experimenter. A
schematic representation of the two biceps heads is
shown in fig. 1 (D). After the skin preparation and
alignment, the sensors were attached to the subjects’
skin with double-sided adhesive tape. The electrical
quality of the interface between the skin and the
electrodes was evaluated in a preliminary experiment
by instructing the subjects to contract the flexors and
extensors of the upper arm while the experimenter
visually checked the signal quality.
A passive measurement orthosis was used to
measure the elbow angle θ synchronously with sEMG
recordings. The orthosis was custom-designed and
3D-printed in-house from PLA plastic so that it
could be adapted to different arm sizes. The elbow
angle was determined using a 10-bit magnetic rotary
position encoder (AS5043, ams AG, Premstaetten,
Austria) integrated into the joint and aligned to the
subject’s rotary axis of the elbow joint. The analog
output was fed into a Trigno Analog Adapter for
synchronous recording. Calibration was performed
by the supervisor during the initial experiment.
Figure 1(A) shows the measurement orthesis as well
as the sEMG sensors.
All subjects performed the same movement
sequences, which involved periodic movements of
the dominant forearm. Two different postures of the
upper arm were adopted and two different weighted
dumbbells were used, which were held by the subject.
The subjects were instructed to align the longitudinal
axis of the forearm orthogonally to the longitudinal
axis of the upper arm, resulting in an initial elbow
angle of θ
0
= 90
◦
. In the lower posture (see fig. 1
(B), the upper arm was held vertically pointing
downwards. In the upper posture, the upper arm was
held vertically pointing upwards. For this study, only
the lower posture (flexor muscles dominant) is used.
In the experiments, four different weights were
held by the subject (w = [2 kg, 4kg]). After the initial
static phase with an elbow angle of θ
0
= 90
◦
, subjects
were instructed to move the forearm up and down
rhythmically about the axis of the elbow joint at
a constant angular velocity, resulting in a sine-like
modulation of the elbow joint angle. After ∆t = 30 s
of dynamic movement, subjects were instructed to
stop and rest for at least one minute before repeating
the trials with a different weight. For each weight,
experiments were performed at [0.25 Hz (slow) and
Predicting the Level of Co-Activation of One Muscle Head from the Other Muscle Head of the Biceps Brachii Muscle by Linear Regression
and Shallow Feedforward Neural Networks
613

0.5Hz (fast)]. One experiment contained an unusual
arm movement and was rejected (2 kg, slow speed,
id=50).
2.2 Domain Knowledge-Based Model
for Feature Extraction and Virtual
Sensor Validation
In perspective, the virtual sensor is intended to serve
as an input for a biomechanical model for limb motion
prediction. In the following method section different
virtual sensors are trained. All virtual sensors predict
a neuronal activation of one biceps head but have
different training setups and input configurations. The
biomechanical model used in section 3.3 for the
validation is described in the following overview. A
detailed description of all subsystems of the model
can be found in (Grimmelsmann et al., 2023). Parts
of this model were also used for the calculation of the
neuronal activation from the sEMG signals.
The signal flow in the model shown in fig. 2
is from left to right. The model contains different
submodels which represent the biological parts of
muscle, joint, and biomechanics. From the left, four
sEMG channels are shown, which are divided into
subsystems. The upper path (two shades of red)
represents the two biceps heads. The lower path (two
shades of blue) represents the triceps heads. The
original model also uses all 4 sEMG channels for
validation in this work. However, the virtual sensors
are only trained for biceps activation. Both paths
converge on the right side of the figure to a torque that
moves the elbow joint. The angle of the elbow joint is
then calculated from the superimposed torque of the
two muscles using the respective dynamics equations.
This is usually used as a prediction signal.
Within the model, the neural activation is available
at the output of the activation dynamics. Up to this
point, the EMG signal has been processed in several
steps. First, it was amplified by a factor of k. The
amplified signal was then filtered with a 4th order
Butterworth bandpass (cut-off frequencies of f
low
=
4Hz and f
high
= 400 Hz) (del Toro et al., 2019) to
reduce noise. Rectification of the signal as introduced
in (Zajac, 1989) resulted in the neuronal excitation e,
which is mostly between zero and one.
The excitation e leads to a neuronal activation u.
Neural activation and neuronal excitation are related
as formulated by (Zajac, 1989) and as shown in
eq. (1).
du(t)
dt
+
β + [1 − β]e(t)
τ
act
·
· u(t) =
e(t)
τ
act
0 < β = const. < 1 (1)
The time constant τ
act
defines the rise time
response of the activation. β is a dimensionless
parameter that defines the fall time response. τ
act
(17 ms) and β (0.35) were set, as described in
(Grimmelsmann et al., 2023). As a result, two
neuronal activations are available, one from the long
biceps head, one from the short biceps head.
2.3 Setup 1: Linear Regression
Individually for Each Experiment
For the regression, the neuronal activation of one head
of the biceps is used as the input. The neuronal
activation of the other head of the biceps is used as the
target. Therefore, there are two possible directions for
the regression. From the short head of the biceps to
the long head of the biceps and vice versa.
In the first setup, the regression is done with a
linear regression with only the neuronal activation as
input. In this setup, the training is done individually
for each experiment. This setup should require
the least amount of generalisation. For example,
the experimental condition slow, 2kg, subject id=24
is split into test and training set. The first setup
aims to get an impression of the structure and basic
dependencies, i.e., how good is the performance of
the regression result for the two directions [long to
short, short to long] (see fig. 3), by the experiments
[[2kg, 4k g], [slow movement, fast movement]] or by
the subject numbers [21,...,53]. In total, 2 · 2 · 2 · 31 =
248 experiment permutations are possible. However,
one experiment failed and is rejected from the used
data set.
From the remaining 246 experiment permutations
the mean is subtracted and they are scaled to unit
variance (standard scaler). After the scaling, the
individual experiments are split into 70% training
data and 30% test data. The experiments vary in
length, but usually last about 30 s @1.1 kHz. The split
is based on the sample so the training data contains
about 23.000 samples.
In the training process, a linear model is fitted
to minimize the residual sum of squares between
the targets and the predicted data by the model
(Pedregosa et al., 2011). After training, the mean
absolute error (MAE) of the test prediction is
calculated (in eq. (2)) and presented in section 3.1.
The test prediction is the predicted neuronal activation
for the test data set.
MAE =
∑
n
i=1
y
i
− x
i
n
(2)
With n is the number of samples, y
i
the sample of the
predicted value and x
i
is the predicted sample.
BIOSIGNALS 2024 - 17th International Conference on Bio-inspired Systems and Signal Processing
614
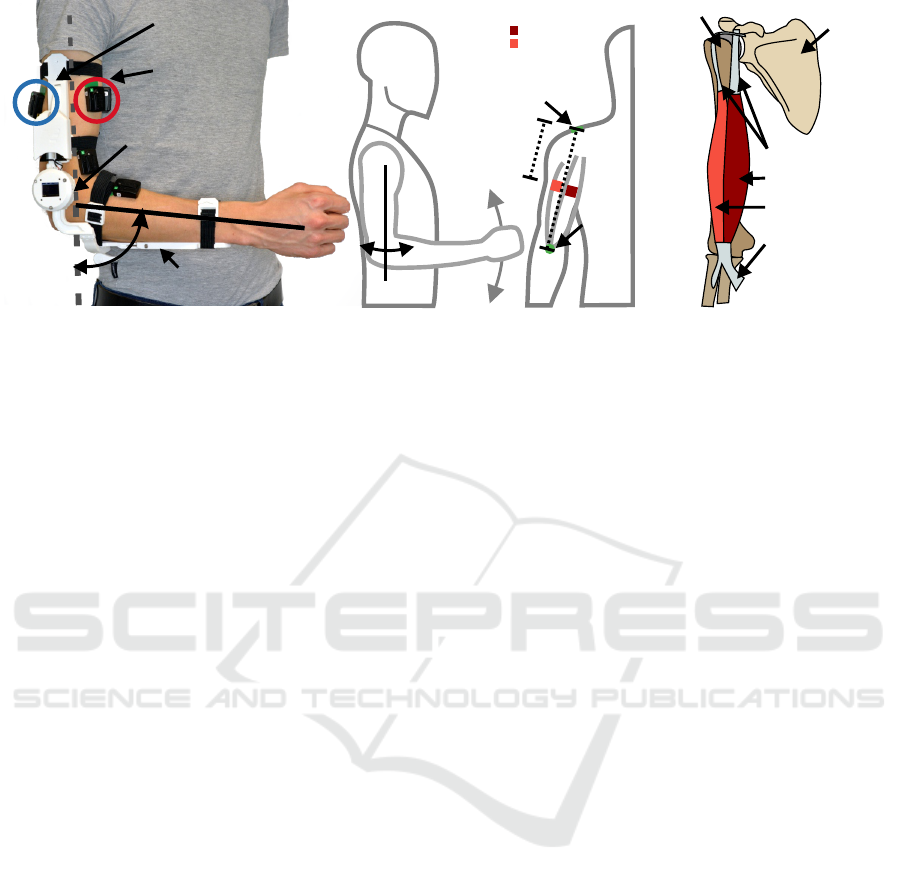
(A) (B) (C)
q
α = 0
orthesis
(D)
anterior
rotary encoder
sEMG sensor
biceps
sEMG sensor
triceps
bic short h.
bic long h.
di_bic
ac
L
iz
bic
L
humerus
distal tendon
proximal tendon
scapular
bic short h.
bic long h.
Figure 1: (a) shows the measurement orthosis on the elbow using flexible straps, allowing for adaptation to different subjects.
The sEMG sensors were placed on the short and long head of the biceps and long and lateral head of the triceps, with the
wrist rotation in a neutral position. The lower experimental posture is shown in (A) and (B), with the angle of the upper arm
(α) being zero (long axis of the upper arm is pointing towards the ground). In (C) the right arm is shown in the coronal plane
from an anterior perspective, with the color code of red indicating the biceps sensors. The distance from the acromion to the
innervation zone is also noted. (D) shows a schematic depiction of the long and the short head of the biceps brachii. The right
shoulder is shown from the anterior perspective. At the top the scapular is shown where the two tendons for the individual
biceps heads are attached. The long head tendon wraps around the humerus to add stability to the joint. Whereas the short
head is connected to the anterior structure of the scapular and links directly down to the muscle belly. At the distal end of the
muscle belly, both heads merge into a common tendon. (A), (B), and (C) were adopted and modified from (Grimmelsmann
et al., 2023). (D) was modified based on (Sch
¨
unke et al., 2010).
To be able to evaluate the size of the error, the
test prediction is compared to a baseline. To obtain a
baseline, the regression output is set to the regression
input.
2.4 Setup 2: Leave-One-out, Subject as
Variable
In the second setup, the regression is done with (I)
a linear regression with only the activation as input,
(II) a linear regression with activation, elbow angle,
elbow angular velocity, the weight of the dumbbell,
and the angle of the upper arm, and (III) a FFN
with the rectified linear unit as activation function
and the same five inputs as (II). The motivation for
these three different configurations was a granular
extension of the architecture. The elbow angle is
chosen as an input caused by the different utilisation
of the muscles over the elbow angle (Chang et al.,
1999). One property of the muscle fibre itself is
the velocity-dependent force generation. This is why
the elbow velocity is taken as an input. The two
biceps heads attach via their respective tendons to
different locations on the shoulder (Sch
¨
unke et al.,
2010). Therefore, the angle of the upper arm has
potentially an impact on the activation for the short
head of the biceps. The long head of the biceps needs
to stabilise the shoulder more with additional weight
in the hand.
The train/ test split strategy is different from setup
1. Here, the training is performed on all experiments
from all subjects excluding all experiments of one
subject. This excluded subject is used as the test
data set. This training/ test split is done ones for the
direction long head to short head and ones for the
other direction. As a result, 31 · 2 training and test
sets are created.
The input activation is scaled for each subject with
a standard scaler. The elbow angle (θ), the elbow
angular velocity (ω), the weight of the dumbbell, and
the angle of the upper arm (α) are scaled combined
over all subjects again with a standard scaler.
Before training, the activation is filtered with a
symmetrical rolling mean. The whole training of
setup 2 was optimised using the adam algorithm
(Kingma and Ba, 2015) with a learning rate of 0.001
and the MAE as loss function.
2.5 Validation of the Virtual Sensor via
a Domain Knowledge Based Model
The model shown in fig. 2 was also used for validation
of the virtual sensor but with one input channel
(biceps) disabled. The remaining biceps channel was
fed into the model at the point where the neural
activation was computed as described in fig. 3(B,C).
Predicting the Level of Co-Activation of One Muscle Head from the Other Muscle Head of the Biceps Brachii Muscle by Linear Regression
and Shallow Feedforward Neural Networks
615
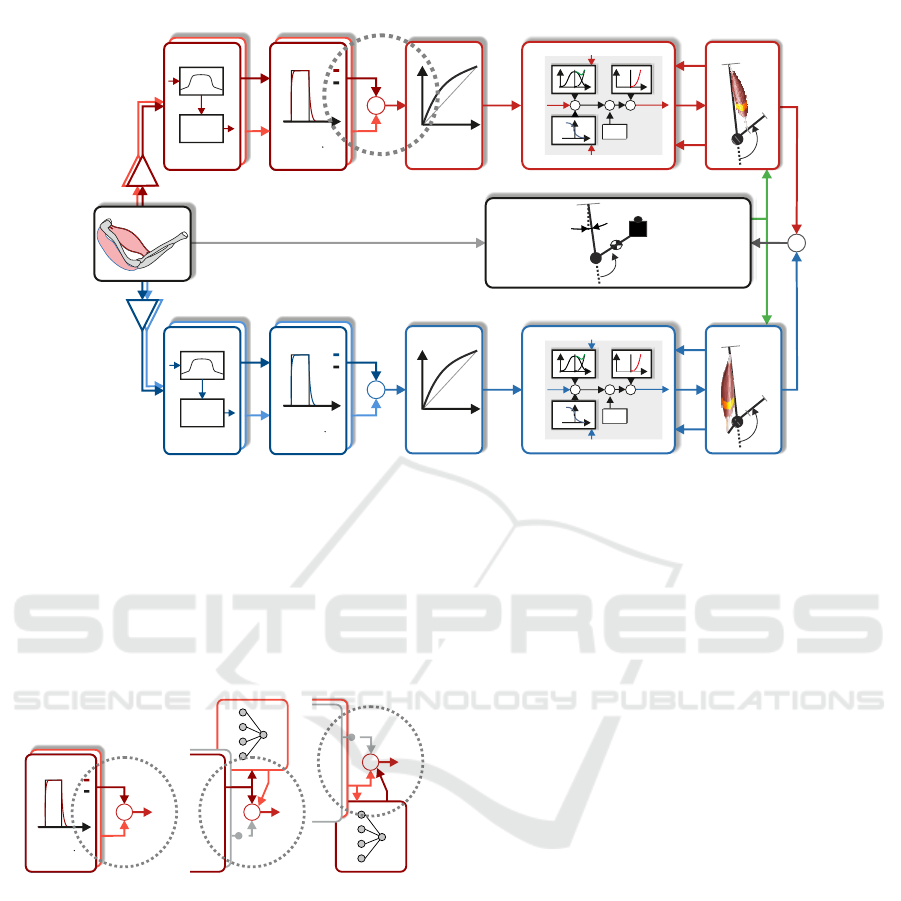
F
M
contraction dynamics
muscle
force
passive elast.force-l.
force-v.
P
F
max
muscle
activation
muscle length
muscle velocity
S
P
contraction dynamics
muscle
force
passive elast.force-l.
force-v.
P
F
max
muscle
activation
muscle length
muscle velocity
S
P
F
M
a
muscle
activation
a
muscle
activation
neural act.
A-model
neural
activation
neural act.
A-model
muscle act.
neural
activation
muscle act.
T
u
S
T
T
u
upper arm angle α
activation dyn.
activation level
time
u = f(e,u,u)
u(t)
e(t)
activation dyn.
joint geometry
q
L
M
elbow
L
T
eqns. of motion
CoG
q
kg
elbow
neural sig.
neural sig.
sEMG & angle
muscle 1
muscle 2
activation level
time
u = f(e,u,u)
u(t)
e(t)
preprocessing
activation level
preprocessing
|abs|
Butterworth
|abs|
Butterworth
S
activation level
time
u = f(e,u,u)
u(t)
e(t)
S
joint geometry
q
L
M
elbow
L
T
EMG
k
EMG
k
α
q
L
M
v
M
L
M
v
M
e
e
e
e
Figure 2: Depiction of the signal flow of the biomechanical model. The biomechanical model which is used to calculate the
neuronal activation and to validate the virtual sensor is shown. The signal chart starts on the left with the four sEMG channels.
The two biceps channels are shown in two shades of red at the top of the arm box. The triceps is shown in two shades of
blue at the bottom. The sEMG signals are amplified and fed into a preprocessing submodel. The next submodel calculates
the neuronal activation for each of the muscle heads. One of these activations is used as an input for the regression. The other
activation is used as the target for the regression. In the biomechanical model, the activation is fed into the next submodels
until the torque of the biceps and the triceps sum up to a superimposed torque. This torque is used with dynamics equations to
calculate the angle of the elbow. This angle is in the validation step compared to the measured angle. The dotted, grey circle
marks the spot where in fig. 3 different structures replace the original structure (Grimmelsmann et al., 2023).
u
S
u
S
u
activation level
time
u = f(e,u,u)
u(t)
e(t)
activation level
time
u = f(e,u,u)
u(t)
e(t)
S
(A) (B) (C)
virtual sensor
virtual sensor
Figure 3: (a) original structure of how neural activation of
two heads was added as shown in fig. 2 (cmp. dotted, grey
circle). (B) and (C) are the possible replacements for figure
(A). (A) uses both neuronal activations from the activation
dynamics. (B) shows the structure when using a virtual
sensor for the long head of the biceps. In (C) the structure is
shown for a virtual sensor for the short head of the biceps.
This activation was fed into the pre-trained virtual
sensor from section 2.4 to get the replacement channel
for the disabled one. The performance of the
virtual sensor was then measured by comparing the
simulated elbow angle θ with the measured elbow
angle θ
meas
. As introduced in (Grimmelsmann
et al., 2023), the quality score (QS) was used as
a performance indicator. The indicator QS allows
for comparison between different lengths, ranges and
shapes of a limb movement. A quality score QS of
zero means that no voluntary movement is visible,
whereas a QS=1 represents an optimal movement
prediction.
Two different configurations can be compared
with the baseline. The baseline is the simulation
where none of the two biceps channels are disabled.
Here, all available measured information is used.
In the first configuration, the sEMG of the long
head was disabled and the activation of this head
was provided via the virtual sensor (trained on the
direction from short head to long head). In the second
configuration, the short head was disabled and the
activation was provided via the virtual sensor (trained
on the direction long head to the short head).
2.6 sEMG Post Measurement
Assessment
The sEMG post measurement assessment was done
via the virtual sensor from setup 2 (section 2.4).
The goal was to use the general training (on all
subject but one) in comparison to the specific test
BIOSIGNALS 2024 - 17th International Conference on Bio-inspired Systems and Signal Processing
616
(only one subject). This is based on the assumption
that both biceps heads co-contract as a result of the
shared distal tendon. The test prediction from setup
2 was used and plotted to the measured activation.
The results are shown in section 3.4 and evaluated
manually. This method can be used to provide an idea
of the signal quality to an experimenter after or during
the sEMG measurement.
3 RESULTS
The result section mirrors the structure of the methods
section. First, the results for the regression on
individual experiments are shown. After that, the
results from the leave-one-out strategy are described.
The next part is the validation of the trained sensors
by using it in a biomechanical model of the human
elbow joint. In the end of the result section
the possibility of the sEMG assessment using the
introduced method is shown.
3.1 Regression on Individual
Experiment Varies Between
Subjects
The 246 training MAE and the 246 test MAE of setup
1 show a maximal absolute difference of 0.012. The
test errors are sorted in two different ways.
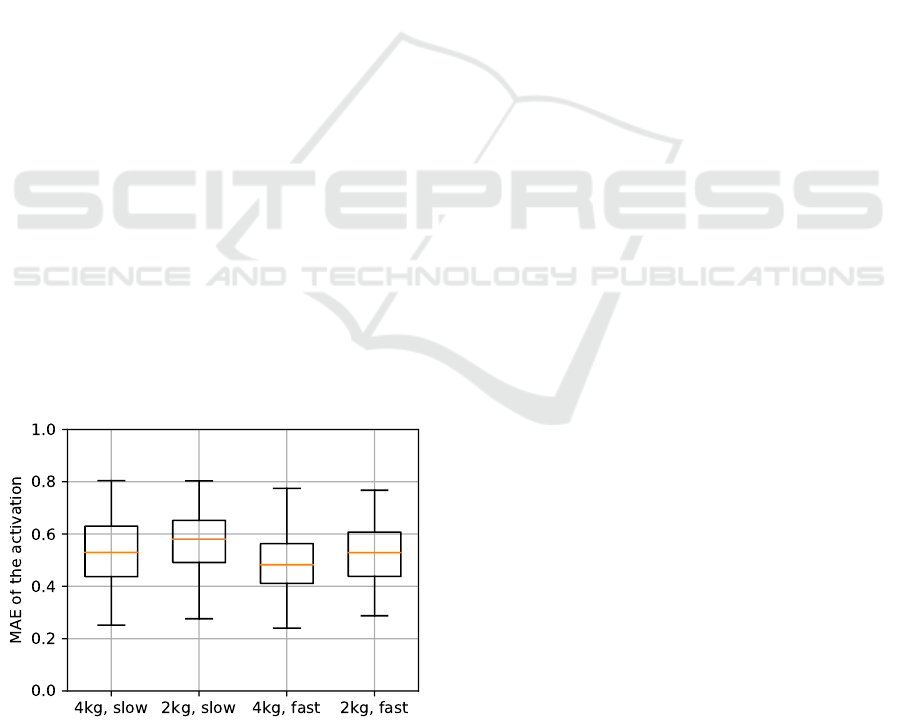
First, they are sorted by the four experiments as
shown in fig. 4. The variation between experimental
conditions is not significant. Second, the training and
test errors are sorted by the subjects. This results in
31 distributions as shown in fig. 5. The individual
distributions show greater variations in MAE, in the
Figure 4: The figure shows the distribution of the MAE
over the experiment type in a box and whisker plot. The
box marks the interquartile range (IQR), and the whiskers
represent data of 1.5· IQR. The MAE for the test set is
shown. All four experiments results in similar distributions.
mean as well as also in the range of the distribution.
One reason for this is the lower number of data points
within each distribution. Compared to the baseline,
the performance is only better for some subjects. The
subject with the id 28 has a lower MAE compared
to the baseline. In contrast to the subject with the id
26 which shows similar MAE values. Some of the
subjects (e.g. subject id 26) have significantly lower
MAE compared to others. The resulting mean of the
MAE for the baseline is equal to mean of the MAE
for the virtual sensor (Baseline MAE=0.601, setup 1:
virt. short MAE=0.538, virt. long MAE=0.546)
3.2 Small Extension in the Architecture
Makes Interpretation Possible
The results for setup 2 are more diverse. The training
was performed with all but one subject to obtain a
general model, which could then be tested with the
left-out subject. The setup contains three different
configurations. Configuration (I) is a linear regression
with only the activation as input. Configuration (II)
is a linear regression with activation, elbow angle,
elbow angular velocity, weight of the dumbbell, and
the angle of the upper arm. Finally, configuration (III)
is a feed-forward net with rectified linear unit as an
activation function and the same five inputs as for (II).
The first configuration is a model comparable
to the model from setup 1. However, the trained
virtual sensor should generalise over more subjects
and experiments than in setup 1. The results are
shown in fig. 6.
Comparing setup 2 configuration (I) with the
baseline shows some performance improvement
(baseline MAE=0.601, setup 2 (I): test virt. short
MAE=0.476, test virt. long: MAE=0.480, training
virt. short: 0.479, training virt. MAE=long: 0.484).
In configuration (II), the input dimension is
expanded to five. The additional information leads
to larger differences between the two directions for
some subjects (e.g. subject ids [32,40]).
The expansion of the input dimension leads to
similar MAEs (baseline MAE=0.601, setup 2 (I): test
virt. short MAE=0.468, test virt. long: MAE=0.482,
training virt. short: 0.465, training virt. MAE=long:
0.481).
The last expansion step in the configuration is to
add non-linearity via an activation function (rectified
linear unit). The results of this configuration (III)
match the trend shown for configurations (I) and
(II). The distinction between the two directions is
clearer with this extension. The overall training
MAE and test MAE are lower than in the previous
configurations (baseline MAE=0.601, setup 2 (I) test
Predicting the Level of Co-Activation of One Muscle Head from the Other Muscle Head of the Biceps Brachii Muscle by Linear Regression
and Shallow Feedforward Neural Networks
617
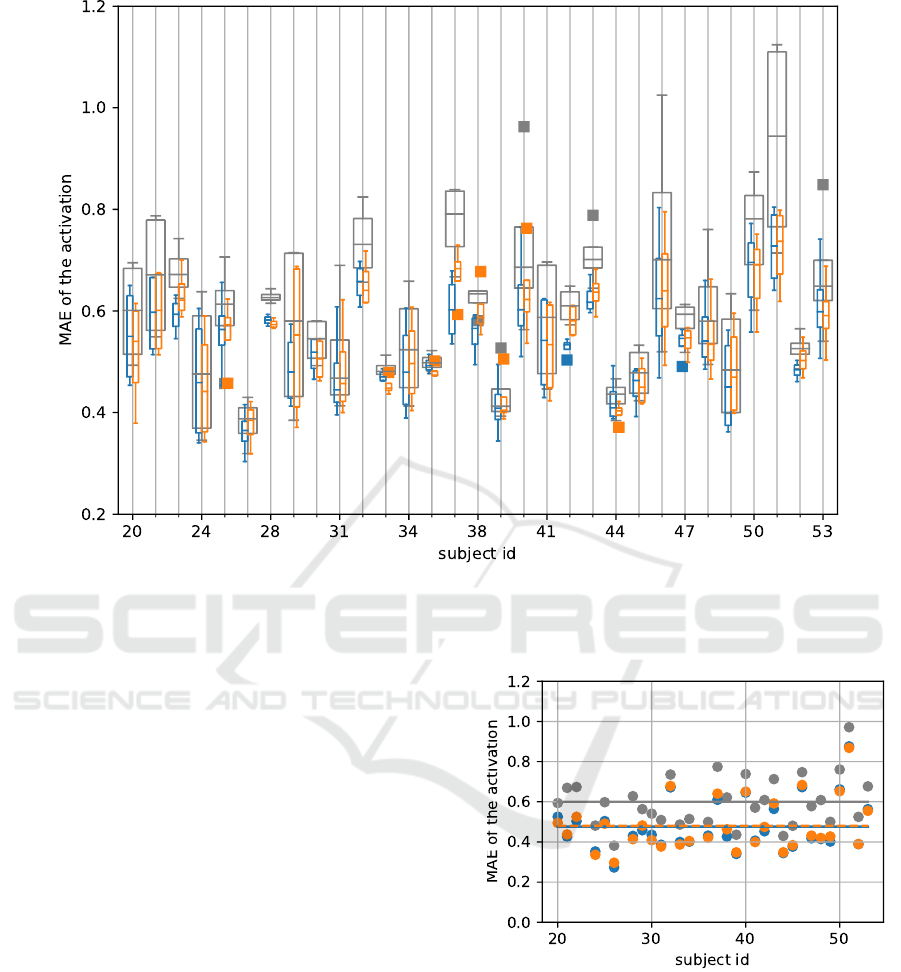
Figure 5: Box and whisker plot of the MAE over the subject id. The general structure is the same as in fig. 4. There are outliers
in these distributions marked by boxes. The color represents the direction of the regression. Blue shows the distribution from
the virtual sensor for the short head. Orange shows the virtual sensor for the long head. Overall more variation of the median
and the IQR is present in this plot. The corresponding data for the training is shown in grey.
virt. short MAE=0.405, test virt. long: MAE=0.367,
training virt. short: 0.372, training virt. MAE=long:
0.347).
3.3 The Virtual Sensor Is Viable for
Using It in a Biomechanical Model
for Movement Prediction
To validate the previously trained virtual sensor, the
sensor is used as a replacement input channel in the
biomechanical model. For that, the virtual sensor
trained with the leave-one-subject-out-strategy and
configuration (III) is used. In contrast to the results
before, the loss function is replaced by the quality
score (QS) between the measured elbow angle and
the predicted elbow angle. A higher value means
a better result. The data shown in fig. 9 uses
the two directions (from long head to short head
and vice versa) and the baseline prediction of the
biomechanical model. Some variations between these
three results are visible for individual subjects. In
general, two main trends can be identified. The
direction long head to short head tend to increase the
prediction performance of the biomechanical model
Figure 6: MAE plotted over subject id. The different colors
represent the training MAE (grey) and the test MAE for the
different directions (from short to long head = orange and
vice versa = blue). The lines indicate the mean of the error
over all subjects. The subject dependence is similar to the
dependence shown in fig. 5. Minor differences between the
two directions can be seen for some subjects.
slightly. Using the other direction the biomechanical
model shows similar results as with the baseline (both
sEMG channels used). The respective values are:
baseline: QS=0.324, virt. short: QS=0.346, virt.
long: QS=0.324.
BIOSIGNALS 2024 - 17th International Conference on Bio-inspired Systems and Signal Processing
618
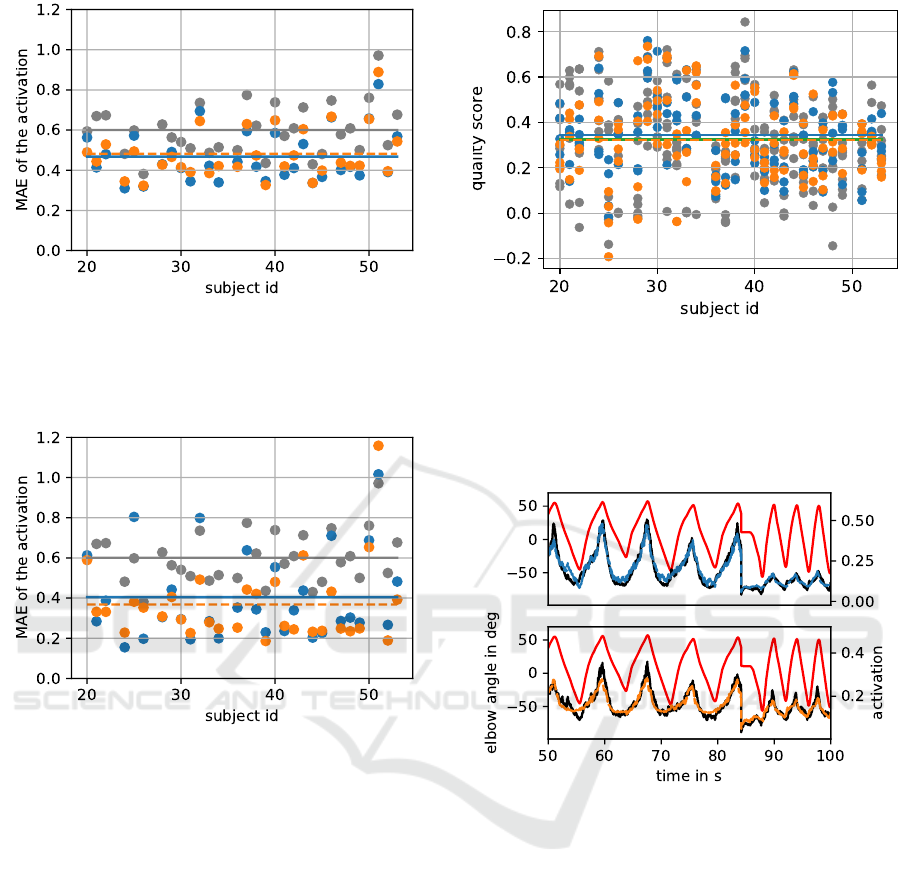
Figure 7: The structure of the figure is the same as fig. 6.
The only difference is the underlying data. The overall
training MAE and test MAE are similar to configuration (I).
The training MAE is slightly lower than the test MAE which
indicates a small overfitting.
Figure 8: Same structure as figure fig. 6. Configuration
(III) is used for the underlying data. This increase in
model complexity leads to a better distinction between
the directions (from short to long head = orange and vice
versa = blue). The training MAE is shown in grey. The
lines indicate the mean of the error over all subjects.
The loss is generally lower as compared to the other two
configurations.
3.4 Virtual Sensor Compared to
Measured Signal Can Serve as
sEMG Assessment
A deeper analysis of the results from the virtual sensor
with configuration (III) allows for an assessment
of the sEMG. Therefore, the outputs of the two
virtual sensors for the two respective directions were
compared. Two exemplary results are chosen from
different subjects. Figure 10 shows one of the subjects
for which the virtual sensor performed well. The
two virtually created activations of this subject are in
phase with each other and with the elbow angle.
In contrast to that, the course of the activation
Figure 9: Prediction performance of the biomechanical
model utilising: both measured sEMG channel (grey),
measured sEMG from the long head plus the virtual sensor
for the short head (blue), measured sEMG from the short
head plus the virtual sensor for the long head (orange). The
subject id is shown vs. the quality score. The mean score is
shown as a line over all subject ids.
Figure 10: Time series of the two virtual sensors for subject
id 24 and the two activations from the measured sEMG.
In the upper panel both signals for the short head are
shown. The lower panel shows the signals for the long
head. The blue line represents the virtual sensor for the
virtual long head. In orange the virtual sensor for the short
head is shown. As a reference, the angle of the elbow is
plotted in red and the activation derived from the measured
sEMG plotted in black. The different experiments were
concatenated. The discontinuity at time 83 s is the change
of the experiment.
from the virutal sensor for the two directions can
be different for a subject. One of these subjects is
shown on fig. 11. Comparing the two virtually created
activations for these subjects shows a discrepancy in
the phases.
The discrepancy is also present across all
experiments. With this plot, the used sEMG can be
manually assessed.
Predicting the Level of Co-Activation of One Muscle Head from the Other Muscle Head of the Biceps Brachii Muscle by Linear Regression
and Shallow Feedforward Neural Networks
619
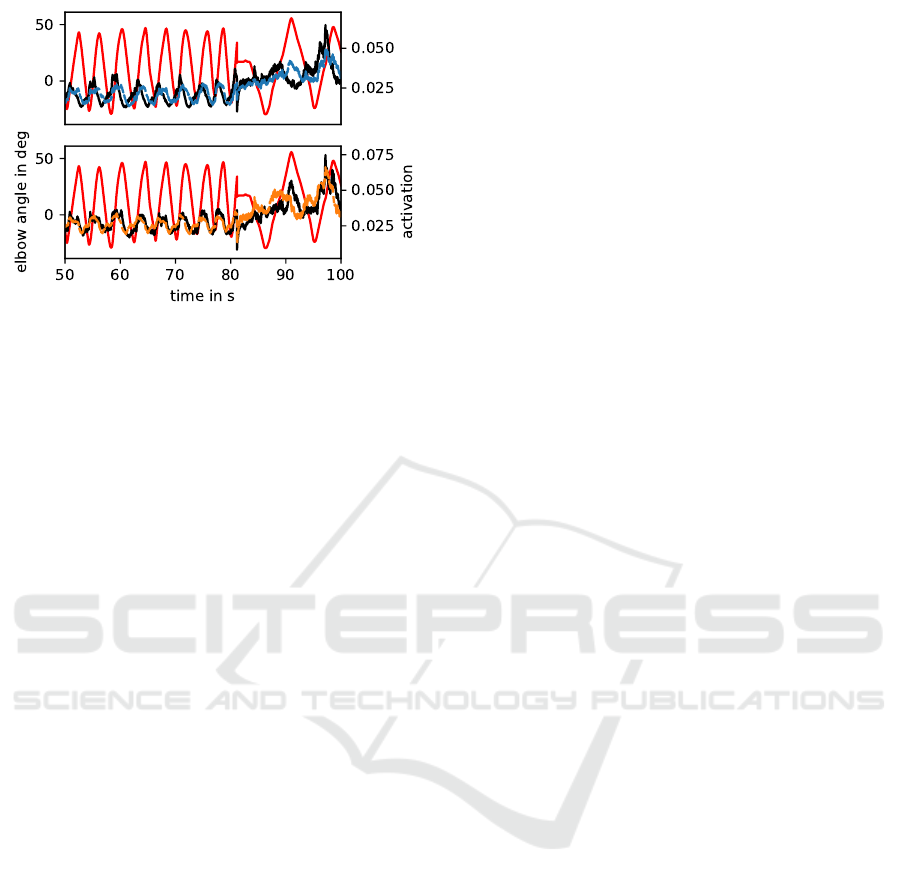
Figure 11: (Cmp. to fig. 10) Time series of the two virtual
sensors and the two activation derived from the measured
sEMG of subject id 32. The color code is consistent
with fig. 10. Here, the experiments changes at time 81s.
The activation from the measured short head (black, lower
panel) is slightly out of phase. Whereas the activation from
the measured long head (black, upper panel) has a higher
phase angle. The corresponding virtual sensor shows the
same phase. By comparing the measure (black) with the
virtual course (blue), the discrepancy is visible.
4 DISCUSSION
In this work, different architectures and training
strategies for the regression of two muscle heads
are presented. The architecture is expanded from
a linear regression with one input to a shallow ffn
with five inputs. The presented strategies started with
the training based on the individual experiments and
ended with the training on all but one subject.
As a result of the regression, virtual sensors are
obtained that are suitable as a replacement input to a
biomechanical model for limb movement prediction.
The underlying architecture of the virtual sensor
contains linear regression and shallow FFN. The
results of the linear regression fit the general
hypothesis that the two biceps heads are usually co-
activated in the described experimental paradigm.
Using an architecture with more degrees of
freedom, it is possible to differentiate between the
two muscle heads. This will be crucial when
creating a virtual sensor for a different muscle (like
the brachialis). Additionally, the virtual sensors
generated via leave-one-out learning allow for basic
sEMG assessment.
A slight overfitting in the leave-one-out strategy
with the configuration (III) is visible. The manual
inspection of each test set confirms comparable
performance between the training and the test set.
One reason for the overfitting could be the relatively
high sample frequency of 1.1kHz.
The results of this study can be used as a simple
sEMG assessment tool for data centric AI approaches.
The shallow structure of the used FFN allows other
researchers to train a similar model for their own data.
Furthermore, the results lay the foundation to
push the boundaries of sEMG measurement as a next
step. The general problem of measuring deep muscles
with sEMG (such as the brachialis) may be partially
solved by extending the presented method via other
architectures beside FFN. In this use case, the virtual
sensor will be learned from muscle to muscle to
provide a signal of an unknown muscle head for a
biomechanical model. A potential time dependency
between different muscles activations could require
model with a RNN or LSTM architecture.
ACKNOWLEDGEMENTS
This work has been supported by the research training
group “Dataninja” (Trustworthy AI for Seamless
Problem Solving: Next Generation Intelligence Joins
Robust Data Analysis) and by the “TransCareTech”
project (Transformation in Care & Technology), both
funded by the German federal state of North Rhine-
Westfalia.
We would like to thank Philipp J
¨
unemann and
Irina L
¨
owen for their valuable contributions during
proofreading.
REFERENCES
Chang, Y.-W., Su, F.-C., Wu, H.-W., and An, K.-N. (1999).
Optimum length of muscle contraction. Clinical
Biomechanics, 14(8):537–542.
del Toro, S. F., Wei, Y., Olmeda, E., Ren, L., Guowu,
W., and D
´
ıaz, V. (2019). Validation of a low-cost
electromyography (EMG) system via a commercial
and accurate EMG device: Pilot study. Sensors,
19(23):5214.
Grimmelsmann, N., Mechtenberg, M., Schenck, W.,
Meyer, H. G., and Schneider, A. (2023). sEMG-
based prediction of human forearm movements
utilizing a biomechanical model based on individual
anatomical/ physiological measures and a reduced set
of optimization parameters. PLOS ONE, 18(8):1–28.
Hill, A. V. (1964). The effect of load on the heat of
shortening of muscle. Proceedings of the Royal
Society of London. Series B. Biological Sciences.
Publisher: The Royal Society London.
Kim, M., Chung, W. K., and Kim, K. (2019). Preliminary
Study of Virtual sEMG Signal-Assisted Classification.
In 2019 IEEE 16th International Conference on
Rehabilitation Robotics (ICORR), pages 1133–1138.
BIOSIGNALS 2024 - 17th International Conference on Bio-inspired Systems and Signal Processing
620

Kingma, D. and Ba, J. (2015). Adam: A method for
stochastic optimization. In International Conference
on Learning Representations (ICLR), San Diega, CA,
USA.
Koo, T. K. and Mak, A. F. (2005). Feasibility of using emg
driven neuromusculoskeletal model for prediction
of dynamic movement of the elbow. Journal of
Electromyography and Kinesiology, 15(1):12–26.
Leserri, D., Grimmelsmann, N., Mechtenberg, M., Meyer,
H. G., and Schneider, A. (2022). Evaluation of
semg signal features and segmentation parameters for
limb movement prediction using a feedforward neural
network. Mathematics, 10(6).
Machado, J. C., Cene, V. H., and Balbinot, A. (2019).
Recurrent Neural Network as Estimator for a Virtual
sEMG Channel. In 2019 41st Annual International
Conference of the IEEE Engineering in Medicine and
Biology Society (EMBC), pages 3620–3623. IEEE.
Mechtenberg, M., Grimmelsmann, N., Meyer, H. G., and
Schneider, A. (2023). Surface electromyographic
recordings of the biceps and triceps brachii for various
postures, motion velocities and load conditions. FH
Bielefeld.
Merletti, R. and Farina, D. (2008). Analysis of
intramuscular electromyogram signals. Philosophical
Transactions of the Royal Society A: Mathematical,
Physical and Engineering Sciences, 367(1887):357–
368.
Merletti, R., Farina, D., and Holobar, A. (2018). Surface
Electromyography (sEMG), pages 1–22. John Wiley
& Sons, Ltd.
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V.,
Thirion, B., Grisel, O., Blondel, M., Prettenhofer,
P., Weiss, R., Dubourg, V., Vanderplas, J., Passos,
A., Cournapeau, D., Brucher, M., Perrot, M., and
Duchesnay, E. (2011). Scikit-learn: Machine learning
in Python. Journal of Machine Learning Research,
12:2825–2830.
Sch
¨
unke, M., Schulte, E., Schumacher, U., Voll, M., and
Sch
¨
unke, M. (2010). Thieme atlas of anatomy:
general anatomy and musculoskeletal system; 100
tables. Thieme, corr. repr., plus version edition.
Zajac, F. E. (1989). Muscle and tendon: properties, models,
scaling, and application to biomechanics and motor
control. Crit Rev Biomed Eng, 17(4):359–411.
Predicting the Level of Co-Activation of One Muscle Head from the Other Muscle Head of the Biceps Brachii Muscle by Linear Regression
and Shallow Feedforward Neural Networks
621
