
Assessing the Performance of Autoencoders for Particle Density
Estimation in Acoustofluidic Medium: A Visual Analysis Approach
Lucas M. Massa
1 a
, Tiago F. Vieira
1 b
, Allan de M. Martins
2 c
and Bruno G. Ferreira
3 d
1
Institute of Computing, Federal University of Alagoas, Lourival Melo Mota Av., Macei
´
o, Brazil
2
Department of Electrical Engineering, Federal University of Rio Grande do Norte, Natal, Brazil
3
Edge Innovation Center, Federal University of Alagoas, Macei
´
o, Brazil
Keywords:
Particle Density Estimation, Convolutional Autoencoder, Particle Size Estimation, Acoustofluidics.
Abstract:
Micro-particle density is important for understanding different cell types, their growth stages, and how they
respond to external stimuli. In previous work, a Gaussian curve fitting method was used to estimate the
size of particles, in order to later calculate their density. This approach required a long processing time,
making the development of a Point of Care (PoC) device difficult. Current work proposes the application
of a convolutional autoencoder (AE) to estimate single particle density, aiming to develop a PoC device that
overcomes the limitations presented in the previous study. Thus, we used the AE to bottleneck a set of particle
images into a single latent variable to evaluate its ability to represent the particle’s diameter. We employed
an identical physical apparatus involving a microscope to take pictures of particles in a liquid submitted to
ultrasonic waves before the settling process. The AE was initially trained with a set of images for calibration.
The acquired parameters were applied to the test set to estimate the velocity at which the particle falls within
the ultrasonic chamber. This velocity was later used to infer the particle density. Our results demonstrated that
the AE model performed much better, notably exhibiting significantly enhanced computational speed while
concurrently achieving comparable error in density estimation.
1 INTRODUCTION
Density establishes a fundamental relationship be-
tween the mass and volume of a particle, thereby as-
sisting in the determination of cell types and their cor-
responding stage cycles (Bryan et al., 2010). Addi-
tionally, cell volume shows a direct correlation with
the mass and energy requirements of cell division. By
leveraging density measurements, it becomes possi-
ble to estimate fluctuations in the volume of a cell. To
(Zhao et al., 2014), cell mass and density measure-
ments offer a powerful and direct method for monitor-
ing cellular responses to various external stimuli, such
as drug interventions and environmental changes.
On this basis, researchers across various scientific
areas have employed different methods to measure the
density of particles. In (Castiglioni et al., 2021), a
method is proposed to measure the density of porous
particles, specifically activated carbons, by measur-
a
https://orcid.org/0009-0001-6023-9318
b
https://orcid.org/0000-0002-5202-2477
c
https://orcid.org/0000-0002-9486-4509
d
https://orcid.org/0000-0003-1345-5103
ing the volume of pores using gas or solution expo-
sure. The work proposed by (Pl
¨
uisch et al., 2021)
presents a microfluidic device to measure the den-
sity of single cells using a suspended microchannel
resonator (SMR). Some authors propose using cen-
trifuges (Minelli et al., 2018; Uttinger et al., 2020;
Ullmann et al., 2017; Maric et al., 1998) to obtain this
measurement. However, these methods have some
significant downsides. While SMRs have a costly
and challenging fabrication process (De Pastina et al.,
2018), centrifuges may potentially damage the cells
under analysis.
There is a particularly interesting method based on
acoustofluidics where the particle is placed inside a
resonant chamber containing a specific fluid and sub-
mitted to an ultrasonic field used for manipulation and
observation. This consists of a contactless, biocom-
patible approach and has been applied to different ar-
eas of science (Wu et al., 2019; Yazdani and S¸is¸man,
2020; Rasouli et al., 2023; Xie et al., 2019; Li and
Huang, 2018).
To the best of our knowledge, the most recent
work using computer vision in this context was in-
436
Massa, L., Vieira, T., Martins, A. and Ferreira, B.
Assessing the Performance of Autoencoders for Particle Density Estimation in Acoustofluidic Medium: A Visual Analysis Approach.
DOI: 10.5220/0012364500003660
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 19th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2024) - Volume 3: VISAPP, pages
436-443
ISBN: 978-989-758-679-8; ISSN: 2184-4321
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
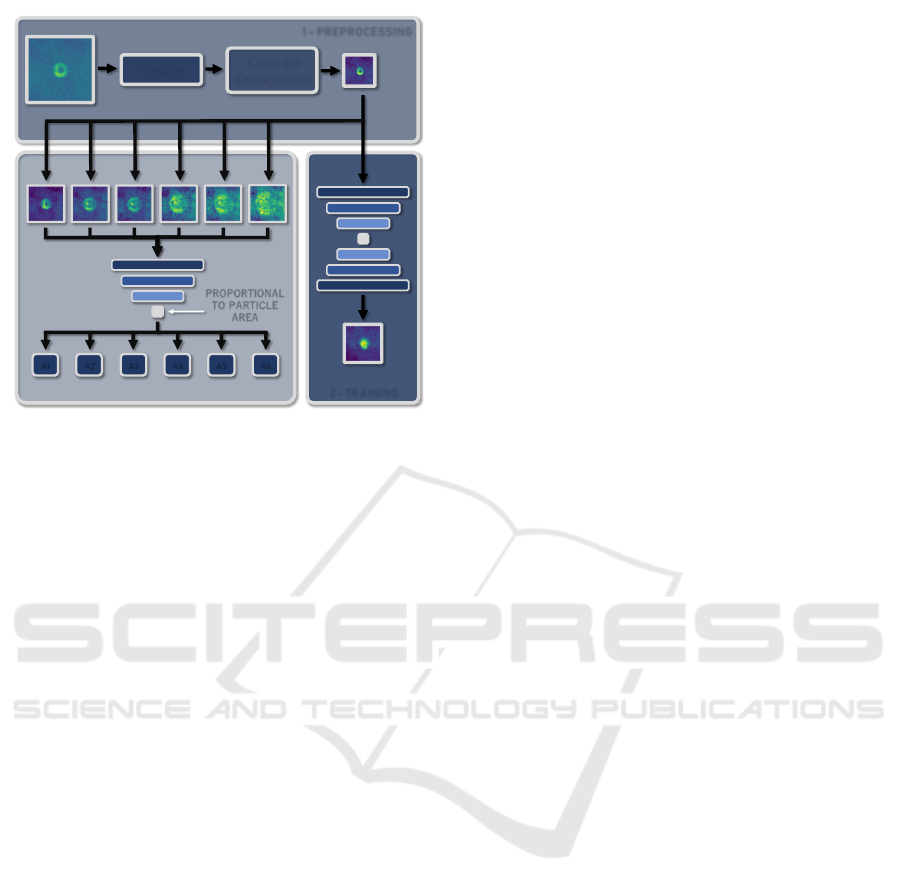
2 - TRAINING
1 - PREPROCESSING
Resize
Contrast
Enhacement
PROPORTIONAL
TO PARTICLE
AREA
A1 A2 A3 A4 A5 A6
3 - INFERENCE
Figure 1: Overview of the proposed approach. Initially,
some particle images collected from a microscope were
used to train a convolutional autoencoder that squeezes a
single latent variable. Hypothetically, this value is expected
to be proportional to the particle area. Lastly, the values
returned by the encoder are used along with the calibration
curve and physical relationship to estimate the particle den-
sity.
troduced in (Massa et al., 2023), where they analyzed
microscopic images acquired during the settling pro-
cess within an acoustofluidic medium. Their method
involves extracting particle areas by fitting 2D Gaus-
sian functions to estimate particle size using a ge-
netic algorithm optimization. These extracted values
are subsequently employed to estimate the particle
fall velocity and its density. While the authors have
demonstrated promising outcomes, some aspects ne-
cessitate improvement. The primary concern is the
requirement of executing the genetic algorithm for ev-
ery new image along a given experiment. In contrast,
we propose using a machine learning pipeline, which
allows the training of the model with a fixed calibra-
tion single particle image dataset. Subsequently, the
same model can be applied to compute particle ar-
eas in new input image sets, removing the need for
retraining and greatly improving inference execution
time.
In the context of deep learning, Autoencoders
(AE) have been widely used to analyze microscopic
scenarios, leveraging their ability to capture intricate
non-linear relationships, reduce dimensionality, and
retain essential features. In (Ignatans et al., 2022)
deep learning, variational autoencoders (VAE), and
matrix factorization were combined to learn latent
representations with rotational equivalence, enabling
the exploration of dynamic data in diverse imag-
ing techniques with improved descriptors. Also, the
approach proposed by (Kalinin et al., 2021a) uti-
lizes VAE to describe dynamic structural changes and
chemical transformations. It focuses on disordered
systems and extracts order parameters to study dy-
namic processes. The work proposed by (Kalinin
et al., 2021b) presents a machine-learning workflow
that combines semantic segmentation and rotationally
invariant VAE. It explores complex ordering systems,
separating rotational dynamics and ordering transi-
tions.
As discussed later in Section 2.5, Autoencoders
tend to compress the inputs into meaningful represen-
tations in the latent space. So, it can be assumed that,
by correctly training an AE with particle images, one
can get the latent space to deliver values representa-
tive of the particle area. In addition, as the AE is
trained in an unsupervised fashion, there is no need
to give the real particle area as input for the model.
Thus, this allows an improvement from the work pre-
sented by (Massa et al., 2023), as they manually ex-
tract particle area values for each image for calibra-
tion.
Given what has been said, the present work pro-
poses using a standard convolutional autoencoder to
evaluate its performance in the particle density esti-
mation process. The AE is specifically applied to the
area calculation task, replacing the Gaussian curves
presented by (Massa et al., 2023). Finally, a compari-
son will be made between the results obtained in this
study and those achieved by (Massa et al., 2023) in or-
der to assess the scenarios in which the autoencoder
solution outperforms the previous one. An overview
of the proposed Autoencoder area estimation can be
seen in Fig. 1. This approach will also be explained
in more detail in Sections 2.2 and 2.5.
2 EXPERIMENTAL
METHODOLOGY
The methodology followed in this paper is similar to
the one presented by (Massa et al., 2023). The uti-
lized acustofluidic device is formed by a disk cast in-
side a cylindrical structure that is sealed with glass.
The disk can be filled with fluidic solutions that con-
tain the studied particle and is also used as an acous-
tic chamber. A piezoelectric actuator was placed at
the bottom of the device. When the actuator is on,
the glass cover acts as an acoustic reflector, causing
the formation of an acoustic standing wave inside the
chamber. As exposed in Fig. 2 (a), the force created
by the standing wave traps the particle present in the
solution into the wave node. A microscope was at-
tached at the top of the device to capture images of
Assessing the Performance of Autoencoders for Particle Density Estimation in Acoustofluidic Medium: A Visual Analysis Approach
437
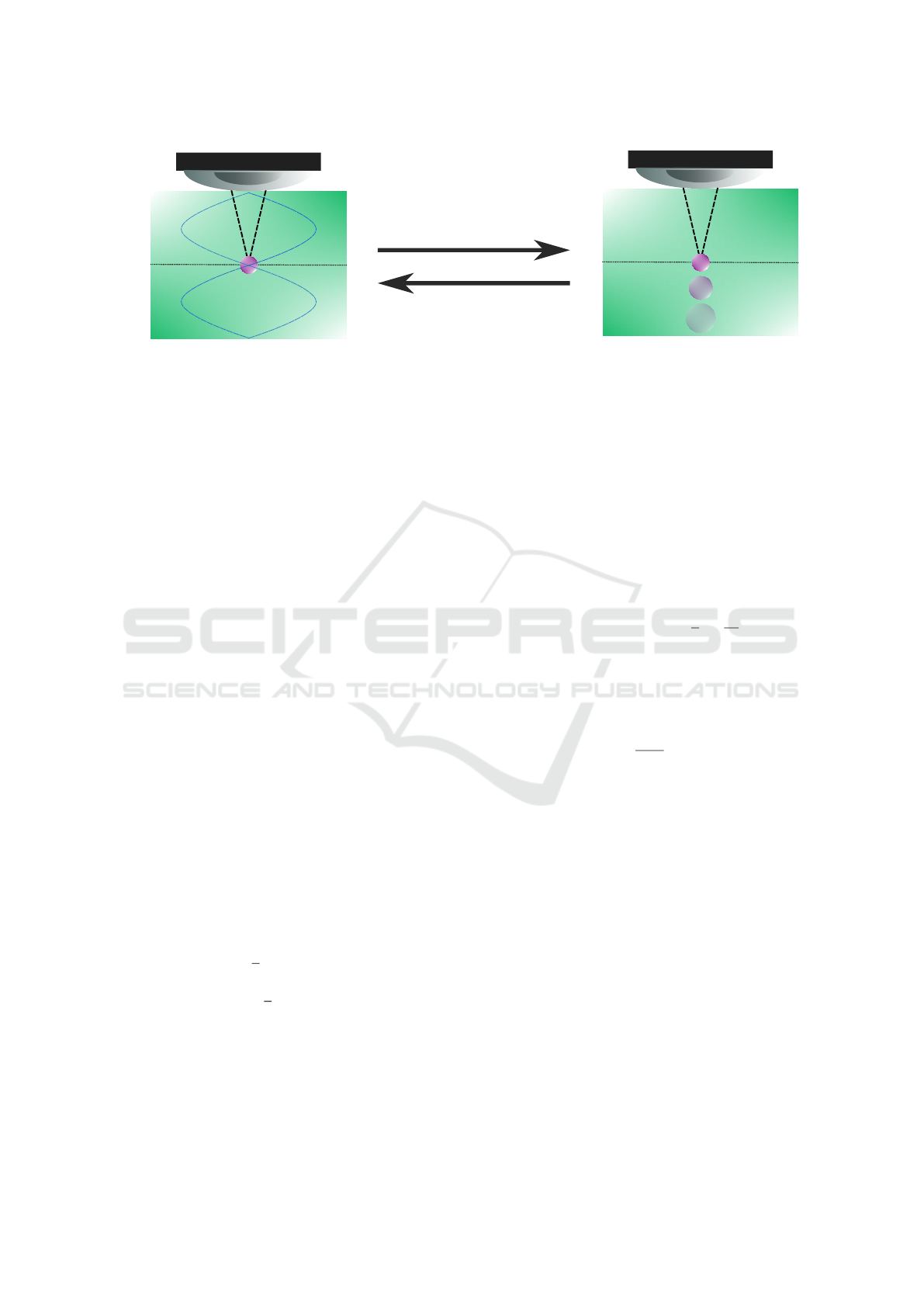
Fluid
medium
Confocal
plane
Levitated
particle
Standing
wave
Confocal
plane
Fluid
medium
Falling
particle
(a)
(b)
Actuator off
Microscope lens
Microscope lens
Actuator on
Figure 2: The hardware follows the same setup used by (Massa et al., 2023). Figure above illustrates the main stages of the
process. (a) When the actuator is on, a generated acoustic standing wave traps the particle in the microscope confocal plane.
(b) When the actuator is turned off, as the acoustic wave vanishes, the particle falls to the bottom of the cavity (perpendicular
to the confocal plane) and presents the physical behaviour explained in Section 2.1.
the particle. The acoustic wave frequency is carefully
defined so that the levitation node plane matches the
confocal plane of the microscope. Once the actuator
is turned off, the particle starts to fall along the fluid,
moving away from the microscope’s confocal plane,
as shown in Fig. 2 (b). Due to this displacement, sub-
sequent images acquired from the particle present in-
creasingly higher amounts of blur, which can be seen
as an increase in their area. Thus, a relationship be-
tween particle area, fall velocity and, consequently,
particle density can be established. During the course
of this work we attempted the usage of a computer
vision approach to measure the density of a 10µm di-
ameter polystyrene bead, which has a known density
of around 1050 kg·m
−3
. We also utilized a solution
with density of ρ
f luid
= 997 kg·m
−3
and a dynamic
viscosity of µ = 0.89 × 10
−1
Pa·s along with a gravi-
tational acceleration of g = 9.82 m·s
−2
.
2.1 Physical Background
The velocity with which a particle falls when embed-
ded in a fluidic medium is directly related to its den-
sity. When the particle is approximated by a perfect
sphere, this relation is mainly controlled by gravita-
tional (Eq. 1), buoyancy (Eq. 2) and viscosity (Eq. 3)
forces (Zhao et al., 2014), i.e.;
F
g
=
4
3
πgr
3
ρ
particle
, (1)
F
b
= −
4
3
πgr
3
ρ
f luid
, (2)
and
F
v
= 6πrµv, (3)
where:
• ρ
particle
: Particle density.
• ρ
f luid
: Fluid density.
• r: Particle spherical approximation radius.
• µ: Fluid dynamic viscosity.
• g: Gravitational acceleration.
• v: Particle velocity.
The resulting force pulls the particle down to the
bottom of the cavity in which it is inserted and can be
found by the application of Newton’s Second Law.
∑
F = F
g
+ F
b
+ F
v
=
4
3
πr
3
dv
dt
(4)
Solving the first order differential Eq. 4 and subse-
quently simplifying the results, it is possible to reach
the equation that describes the physical relation be-
tween fall velocity and particle density:
ρ
particle
=
9µv
2r
2
g
+ ρ
f luid
. (5)
2.2 Dataset
As previously stated, we apply an optical micro-
scope to extract images from particle fall experiments.
The original images obtained by the microscope are
monochromatic (grayscale) and have a resolution of
2448×1920 pixels. Used images are 500×500 pixels
containing a single particle during the settlement pro-
cess. This resolution allows us to capture the whole
particle across the entire size range observed in exper-
iments due to the defocus.
Conducting the discussed experiments typically
presents significant challenges. It is very common for
particles to group together in packs. In this way, the
number of images that enable the analysis of isolated
particles is extremely reduced, leading to a challeng-
ing problem. Only two successful experiments could
deliver calibration and test image sets. The calibration
set was comprised of 18 images acquired during the
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
438
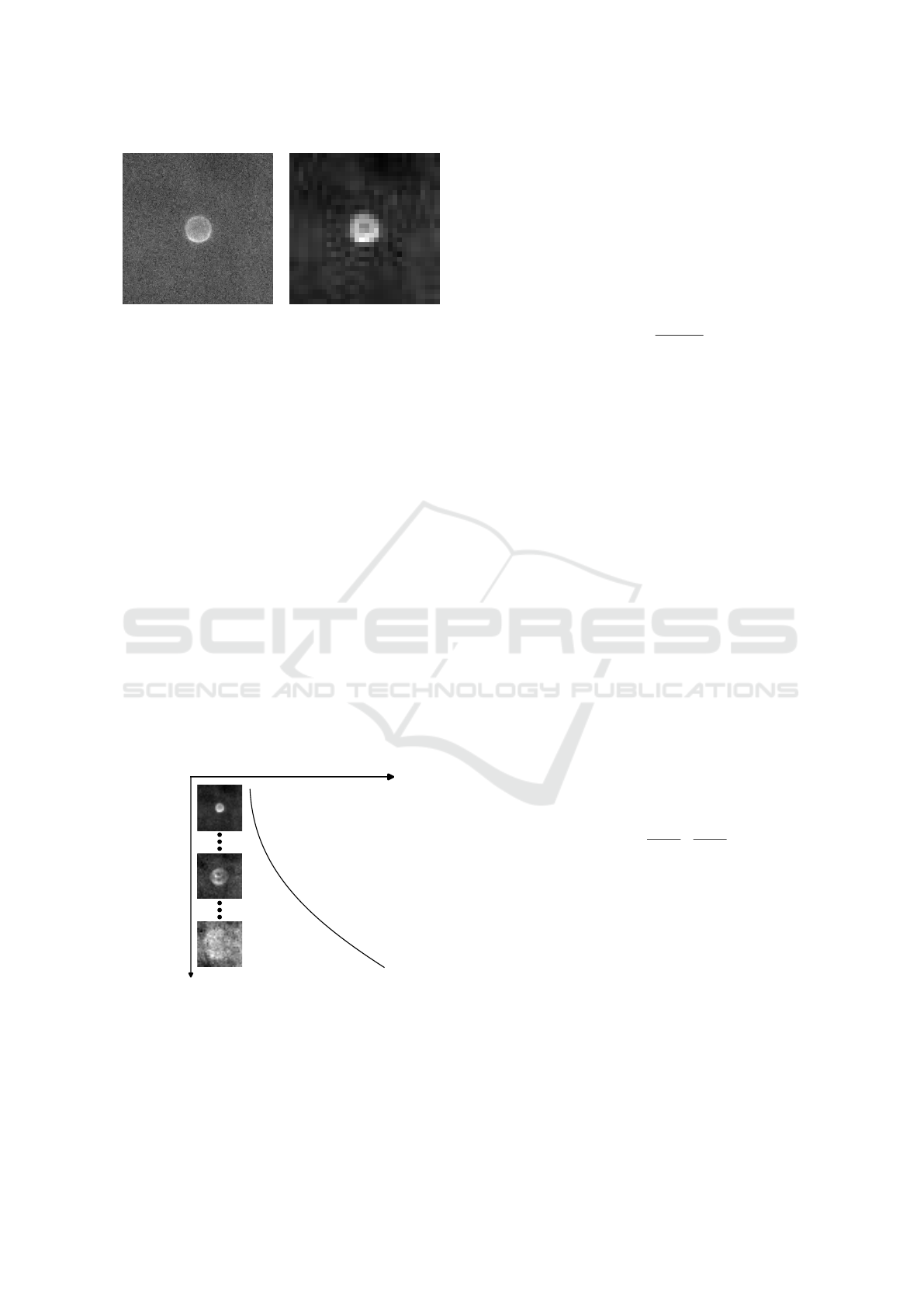
(a) Original image. (b) Preprocessed image.
Figure 3: Example of the images obtained through the ex-
plained pipeline. The left one represents the original image.
The right one represents the result after preprocessing.
particle settlement. The test set consisted of 13 im-
ages acquired in the same manner. With such limited
number of examples, the risk of overfitting was man-
aged with the adjustment of model’s hyperparameters
to reduce its complexity (cf. Sec. 2.5).
For the sake of reproducibility, we applied the
same preprocessing steps used in (Massa et al., 2023)
to each acquired image. The preprocessing stage in
our approach involves cropping, resizing, and enhanc-
ing, as illustrated in Fig. 1. Examples can be observed
in Fig. 3. It is also worth noting that last images in the
sequence, where the particle is close to cavity’s bot-
tom, are very noisy and unfocused, making it hard
to segment the particle via classical image process-
ing approaches. This noisy behaviour can be seen in
the bottom particle image from Fig. 4 and also in the
input image from Fig. 5.
2.3 Calibration
Relative Area
Height
Calibration curve
Figure 4: Illustration of the calibration process. As the par-
ticle falls, the blur causes an increase in the area related to
the particle’s height in each image. This relationship can be
expressed as a calibration curve.
When the particle falls, the distance to the image con-
focal plane becomes larger, resulting in a defocus that
“increases” the particle size in the acquired images.
Therefore, the dynamics of a particle embedded into
a fluidic medium can be analyzed by knowing its area
and height in each image acquired during the fall pro-
cess. To do so, we fit a calibration curve capable
of modeling the correspondence between the relative
area of the particle in each image and the respective
height relative to the bottom of the chamber.
For a set of n acquired images, relative area values
can be calculated with the following equation:
A
i relative
=
A
i
− A
n
A
1
− A
n
(6)
where A
1
, A
i
, and A
n
are, respectively, area values for
the first, current, and last captured images.
The computed values for relative area and their re-
spective height values can fit an exponential function,
as illustrated in Fig. 4, which is used as a calibra-
tion curve. With this curve, it is possible to obtain
the height of a different particle by giving as input
the area value, as long as the studied particle has the
same diameter as the calibrated one. Finally, the fall
velocity can be derived from the height values and
used along with Eq.(5) to calculate the particle den-
sity. This highlights the importance of obtaining an
efficient and automated way to estimate the particle
area, a process which, in general, is done manually.
2.4 Baseline
To serve as a baseline for further investigations, we
applied the Gaussian curve fitting approach proposed
by (Massa et al., 2023) to the task of estimating parti-
cle areas. The main idea is to fit a Gaussian model to
the image by minimizing an error function. As we are
dealing with images, we need to use the 2D version
of the Gaussian curve, which can be generated by the
following expression:
G(x, y) = α e
−
(x−µ
x
)
2
2σ
2
+
(y−µ
y
)
2
2σ
2
4
(7)
where:
• α: Gaussian amplitude.
• µ
x
: Position of the Gaussian’s center in the image
along the horizontal axis.
• µ
y
: Position of the Gaussian’s center in the image
along the vertical axis.
• σ: Standard deviation of the Gaussian.
As discussed in (Massa et al., 2023), defining val-
ues for α, µ
x
, µ
y
and σ
2
which generate a Gaussian
that fits a specific image is not trivial. Furthermore, as
the particle is centered in the image, µ
x
and µ
y
were
always zero, and we only had to optimize α and σ.
Thus, aiming to reproduce the experiment, we also
Assessing the Performance of Autoencoders for Particle Density Estimation in Acoustofluidic Medium: A Visual Analysis Approach
439
delegated this task to a Genetic Algorithm approach.
The evolutionist theory inspires this algorithm and is a
common choice for optimization tasks (Katoch et al.,
2021).
One core concept of this method is fitness, which
is the metric that determines how much an individ-
ual is adapted to the problem it intends to solve. This
metric is defined according to the problem necessi-
ties. As we need to measure the error between a 2D
Gaussian-generated image and the particle image, the
Mean Squared Error was chosen as the fitness metric.
Considering the particle image I(x, y) and the Gaus-
sian G(x, y), where x and y are the pixel coordinates,
the Mean Squared Error can be calculated by the fol-
lowing expression:
MSE =
1
W H
W
∑
x=1
H
∑
y=1
[I(x, y) − G(x, y)]
2
(8)
where W and H are, respectively, the width and height
of the images.
First, we used the calibration image set, which had
previously known area and height values, to calculate
a calibration curve, as discussed in Section 2.3. Then,
the test set was applied to the Genetic Algorithm op-
timization, which was executed for 1000 generations
with a population of 100 individuals. We also applied
an elitist selection by, in each generation, maintaining
the most adapted half of the population and mixing
it with the newly generated individuals. The calcu-
lated Gaussian curves were used to estimate the par-
ticle area in each image, which served as input to the
calibration curve to reach height values for the test
images and, subsequently, particle fall velocity and
density.
2.5 Autoencoder
As stated in Section 1, autoencoders (AEs) are a pow-
erful type of neural network. Their architecture com-
prises two main parts: an encoder and a decoder. The
encoder is formed by a set of layers that consecutively
reduces the dimensionality of data until it reaches the
intermediary layer, also known as bottleneck. On the
other hand, the decoder aims to reconstruct the in-
put data from the values delivered by the intermediary
layer. Both encoder and decoder can be composed of
various combinations of layer types, including convo-
lutional layers, which are ideal for computer vision-
related tasks.
The main goal of autoencoders is to reduce dimen-
sionality so the input data is compressed into mean-
ingful representations. This is done by minimizing
the distance between an input and its respective re-
construction. Encoded data is projected into a so-
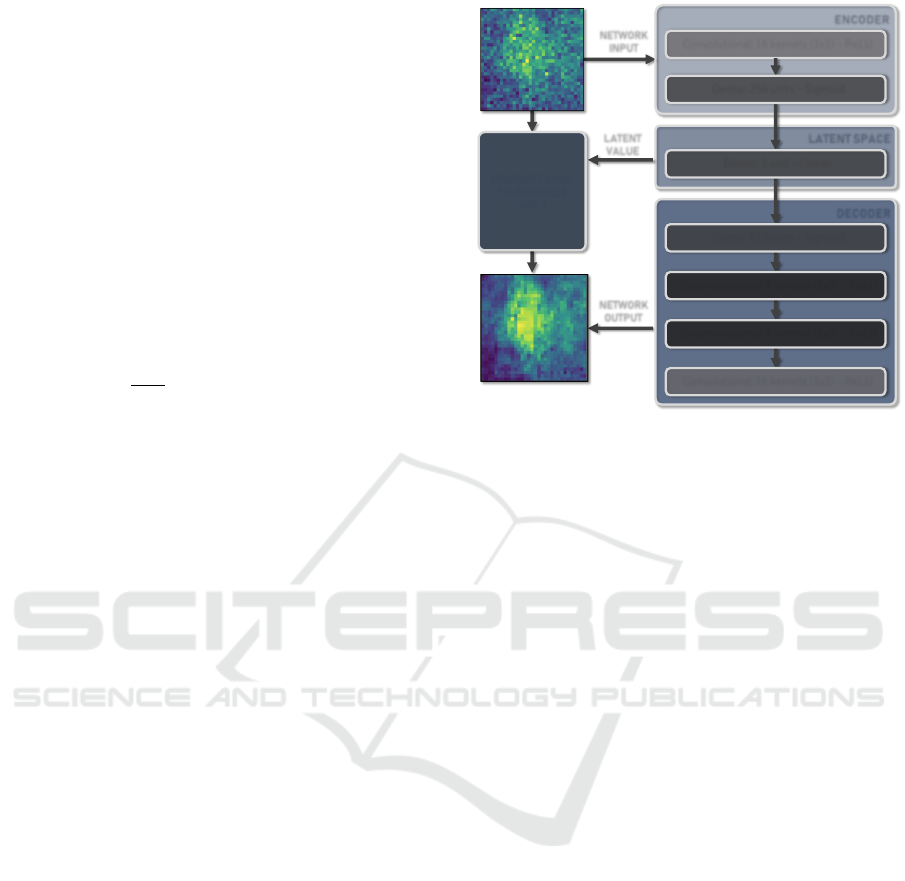
ENCODER
LATENT SPACE
Convolutional: 16 kernels (3x3) - ReLU
DECODER
Dense: 1 unit - Linear
Dense: 256 units - Sigmoid
Convolutional: 16 kernels (3x3) - ReLU
Dense: 512 units - Sigmoid
Deconvolutional: 8 kernels (3x3) - ReLU
Deconvolutional: 8 kernels (3x3) - ReLU
PROPORTIONAL
TO PARTICLE
AREA
NETWORK
INPUT
NETWORK
OUTPUT
LATENT
VALUE
Figure 5: Autoencoder architecture diagram. Images on the
left side show examples of a particle image (upper) given as
input and a reconstructed image (lower) received as output.
The latent space is represented by a single dense unit and
is expected to contain relevant information about the parti-
cle contained in the input image, more specifically about its
area.
called latent representation space. This approach also
forces the model to focus on features with high vari-
ability, being well-suited to denoising tasks (Pratella
et al., 2021). As exposed by (Bank et al., 2020), an
AE can be formally defined as learning the functions
F : R
n
→ R
l
and G : R
l
→ R
n
that satisfy the expres-
sion
argmin
F,G
E[L (x, G ◦ F(X))], (9)
where L is the reconstruction loss function and E is
the expectation over the distribution of the input x.
Furthermore, F, G, n, and l represent the encoder, the
decoder, the input data dimension and the latent space
dimension, respectively.
In this phase of our methodology, we intended to
test the limits of this architecture by squeezing parti-
cle images into a latent vector comprised by a single
real value. The purpose was to validate if the network
could extract a value proportional to the particle area
in each image through the encoder.
To do so, we used the same calibration and test im-
ages, which is a challenge due to the small number of
examples. The AE followed a simple and asymmetric
architecture, as seen in Fig. 5. The encoder was com-
prised of a single convolutional layer followed by a
dense layer that was later squeezed into a single neu-
ron. The decoder, in turn, was formed by a dense layer
followed by two deconvolutional layers.
First, we trained the autoencoder using the cali-
bration set. The target was to reconstruct the original
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
440

image after squeezing it to a single value. The train-
ing loop was executed for 1000 epochs. Was used the
Adam optimizer along with a learning rate of 10
−3
and a batch size of 1. As we are dealing with images,
the MSE was chosen as the loss function. No area tar-
gets were given to the latent space layer. This step is
represented by the training section in Fig. 1.
After the training, we extracted the values deliv-
ered by the latent space for each calibration image and
fit them to an exponential curve. As we are dealing
with relative areas, values used to generate the cali-
bration curve do not need to be the exact areas of each
particle, as long as they are proportional to them.
Lastly, as shown in the inference section of Fig.
1, the test set was also given as input to the trained
network to obtain latent space values expected to be
proportional to the particle area. These values were
later used as input to the calibration curve to estimate
particle height. As in Section 2.4, the resulting height
values for the test images were used to derive the fall
velocity and, consequently, estimate the particle den-
sity.
All experiments were performed on a computer
with the following settings: an Intel Core i7-9700
CPU, 32GB RAM, and RTX 2060 GPU. For a fair
comparison with the baseline work, the GPU is only
used during training, leaving the CPU exclusively for
inference time calculation. Also, we used Python
3 language along with TensorFlow 2.8 (Keras) and
OpenCV 4.7 for training and inference.
3 RESULTS AND DISCUSSION
3.1 Baseline Results
After applying the pipeline proposed in Section 2.4,
we could estimate the area of the test images. The
particle radius was manually extracted for each cali-
bration image and converted via a pixel-nanometer re-
lation. By considering the particles as a perfect circle,
we calculated their areas. These values were used,
along with previously known height, to generate the
calibration curve, as discussed in section 2.3.
Compared to the results obtained by (Massa et al.,
2023), the genetic algorithm showed some lack of
performance. Despite finding good parameters for
the 2D Gaussian functions on most test images, the
curve sometimes did not fit the particle correctly. The
obtained Gaussian curves were then used to compute
particle areas in each image and generate relative area
values by applying Eq. (6). Such values served as in-
put for the calibration curve. By deriving the fall ve-
locity and applying the result to Eq. (5), we could cal-
culate the particle density. The lack of performance
from the Genetic Algorithm was translated in a den-
sity estimation of 999 kg·m
−3
, which is farther from
the real value when compared to the 1059 kg·m
−3
ob-
tained by (Massa et al., 2023).
Even though the resulting density was reasonable,
the baseline approach presented some other limita-
tions. The main problem, as discussed in Section
1, is the lack of replicability, as the Genetic Algo-
rithm must be executed for each new image that will
be applied to the pipeline. Furthermore, the neces-
sity of rerunning the algorithm leads to a time effi-
ciency problem, as the optimization loop takes, on
average, 13 minutes to finish (Table 1). The man-
ual extraction of the particle radius for the calibration
step is also worth mentioning, which can lead to er-
rors more easily when compared to an automated and
well-designed area extraction algorithm.
3.2 Autoencoder Results
By applying the methodology proposed in Section
2.5, we could as well estimate the density of the
polystyrene bead. The values extracted from the au-
toencoder latent space showed a valid proportional-
ity concerning the expected area values, as the ob-
tained relative areas could be successfully fitted to an
exponential function, which is exposed in Figure 6.
Then, we input the test image set to the autoencoder
and used the resulting values to calculate their relative
areas. As in Section 3.1, we applied the calibration
curve to extract a height curve, which can be seen in
Fig. 7. In order to determine the fall velocity we con-
sidered this curve along with the time between each
consecutively captured images, calculating the mean
value of height variations for each interval. The veloc-
ity was finally applied to estimate the particle density.
Figure 6: Calibration curve calculated by fitting height and
relative area values obtained from the autoencoder to an ex-
ponential function.
Using the same experiment configuration as in
Section 3.1, this approach achieved a result of
Assessing the Performance of Autoencoders for Particle Density Estimation in Acoustofluidic Medium: A Visual Analysis Approach
441
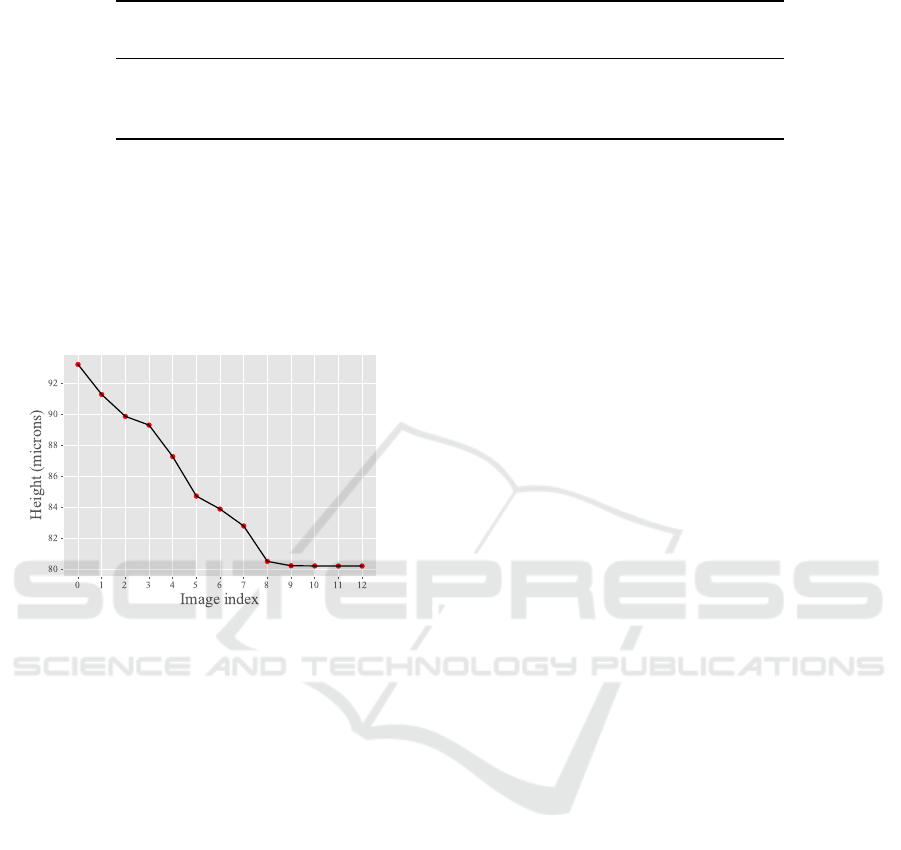
Table 1: Model Performance.
Model
Predicted
density
Absolute
error
Mean
inference time
Previous work (Massa et al., 2023) 1059 kg·m
−3
0.8% -
Baseline experiment 999 kg·m
−3
4.85% 13 minutes
Autoencoder (Ours) 1005 kg·m
−3
4.28% 6.4ms
This table summarizes the experiments discussed in this paper. The best results from the conducted ex-
periments are highlighted in bold.
1005 kg·m
−3
, which represents an absolute error of
4.28% ( Table 1). This represents some reasonable
performance, specially considering the small number
of training images. Compared to the baseline exper-
iment, this method could achieve a better result and
was executed without manual information extraction.
Figure 7: Height curve obtained by giving the relative areas
calculated through the autoencoder for the test set as input
to the calibration curve.
Moreover, after training, the AE can be applied to
any new image, without retraining. This is important
due to the fact that the baseline approach is done by
a classical Genetic Algorithm, which runs an iterative
optimization process. This process does not create a
model capable of generalizing for new inputs. Thus,
there is an increase in the computational cost of the
pipeline, as the optimization process needs to be run
for each new input image.
Lastly, the neural network is more time efficient
and can be trained faster than the Genetic Algorithm.
Indeed, the AE took approximately 2.5 minutes to
train along 1000 epochs in the hardware described
in Section 2.5. Moreover, regarding the Genetic Al-
gorithm (GA) optimization loop time as its inference
time, the AE is also much more efficient, presenting
a mean prediction time of only 6.4ms for each input
image, without using a GPU. The GA, on the other
hand, delivered a much higher prediction time of 13
minutes. Results are summarized in Table 1.
It is also worth mentioning that the resulting curve
obtained with the autoencoder (Fig. 7) exposes satura-
tion of some height values. This can be seen in the last
4 points since, despite the fact that the particle was
still falling, the network predicted similar area values,
resulting in an almost constant curve. We believe that
some limitations of the AE might cause this in dealing
with data outside of its training range (original man-
ifold). In this way, if an image with a larger particle
than the ones presented during the training is given
as input, the value returned by the latent space might
be saturated and lead to wrong predictions. Thus, we
intend to investigate this scenario in future work.
4 CONCLUSIONS
In this paper, we reproduced a previous work and pro-
posed a method to estimate the density of particles on
a microscopic scale using acoustofluidics and com-
puter vision. Aiming at obtaining an automated par-
ticle area extraction, we proposed the use of a simple
convolutional autoencoder (AE) architecture capable
of reconstructing the input image while delivering,
through its latent space layer, a value proportional to
the particle area. The model was trained with a cali-
bration set and the values returned by the latent space
were used to fit a calibration curve. Then, test im-
ages were given as input to the autoencoder and the
resulting latent values were used along with the cali-
bration curve to estimate the fall velocity and, finally,
the particle density.
Our experiments revealed a reasonable perfor-
mance for the AE model, estimating a density of
1005 kg·m
−3
which improves the baseline approach.
The network also vastly outperforms the baseline
model when considering the efficiency and usability,
as it has a much quicker inference time and does not
require retraining for a new experiment.
As future work, we intend to analyze the applica-
bility of VAEs, as they tend to learn continuous latent
spaces that allow the generation of data from values
outside the training range, a property that can help
in mitigating the saturation that was previously dis-
cussed. It is also worth investigating an increase in
the amount of bottleneck neuron units, as it may be
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
442

capable of learning a latent space that, besides parti-
cle area, can describe other important image features.
ACKNOWLEDGEMENTS
This project was supported by the Science, Tech-
nology and Innovations Ministry of Brazil, with re-
sources from law nº 8.2428 of October 23, 1991
within the Softex
1
National Innovation Priority pro-
gram, coordinated by Softex and EDGE Innovation
Center and published by [RESIDENCIA EM TIC ˆ
09(process[01245.005714/2022-18])].
REFERENCES
Bank, D., Koenigstein, N., and Giryes, R. (2020). Autoen-
coders. arXiv preprint arXiv:2003.05991.
Bryan, A. K., Goranov, A., Amon, A., and Manalis, S. R.
(2010). Measurement of mass, density, and volume
during the cell cycle of yeast. Proceedings of the Na-
tional Academy of Sciences, 107(3):999–1004.
Castiglioni, M., Rivoira, L., Ingrando, I., Del Bubba, M.,
and Bruzzoniti, M. C. (2021). Characterization tech-
niques as supporting tools for the interpretation of
biochar adsorption efficiency in water treatment: A
critical review. Molecules, 26(16).
De Pastina, A., Maillard, D., and Villanueva, L. (2018).
Fabrication of suspended microchannel resonators
with integrated piezoelectric transduction. Microelec-
tronic Engineering, 192:83–87.
Ignatans, R., Ziatdinov, M., Vasudevan, R., Valleti, M.,
Tileli, V., and Kalinin, S. V. (2022). Latent mecha-
nisms of polarization switching from in situ electron
microscopy observations. Advanced Functional Ma-
terials, 32(23):2100271.
Kalinin, S. V., Dyck, O., Jesse, S., and Ziatdinov, M.
(2021a). Exploring order parameters and dynamic
processes in disordered systems via variational au-
toencoders. Science Advances, 7(17):eabd5084.
Kalinin, S. V., Zhang, S., Valleti, M., Pyles, H., Baker, D.,
De Yoreo, J. J., and Ziatdinov, M. (2021b). Disentan-
gling rotational dynamics and ordering transitions in
a system of self-organizing protein nanorods via ro-
tationally invariant latent representations. ACS nano,
15(4):6471–6480.
Katoch, S., Chauhan, S. S., and Kumar, V. (2021). A re-
view on genetic algorithm: past, present, and future.
Multimedia Tools and Applications, 80:8091–8126.
Li, P. and Huang, T. J. (2018). Applications of acoustoflu-
idics in bioanalytical chemistry. Analytical chemistry,
91(1):757–767.
Maric, D., Maric, I., and Barker, J. L. (1998). Buoyant den-
sity gradient fractionation and flow cytometric analy-
1
https://softex.br/
sis of embryonic rat cortical neurons and progenitor
cells. Methods, 16(3):247–259.
Massa, L., Vieira, T. F., de Medeiros Martins, A., Ara
´
ujo,
´
I. B. Q., Silva, G. T., and Santos, H. D. A. (2023).
Model fitting on noisy images from an acoustoflu-
idic micro-cavity for particle density measurement. In
VISIGRAPP.
Minelli, C., Sikora, A., Garcia-Diez, R., Sparnacci, K.,
Gollwitzer, C., Krumrey, M., and Shard, A. G. (2018).
Measuring the size and density of nanoparticles by
centrifugal sedimentation and flotation. Analytical
Methods, 10(15):1725–1732.
Pl
¨
uisch, C. S., Stuckert, R., and Wittemann, A. (2021). Di-
rect measurement of sedimentation coefficient distri-
butions in multimodal nanoparticle mixtures. Nano-
materials, 11(4).
Pratella, D., Ait-El-Mkadem Saadi, S., Bannwarth, S.,
Paquis-Fluckinger, V., and Bottini, S. (2021). A sur-
vey of autoencoder algorithms to pave the diagnosis
of rare diseases. International journal of molecular
sciences, 22(19):10891.
Rasouli, R., Villegas, K. M., and Tabrizian, M. (2023).
Acoustofluidics–changing paradigm in tissue engi-
neering, therapeutics development, and biosensing.
Lab on a Chip, 23(5):1300–1338.
Ullmann, C., Babick, F., Koeber, R., and Stintz, M. (2017).
Performance of analytical centrifugation for the parti-
cle size analysis of real-world materials. Powder Tech-
nology, 319:261–270.
Uttinger, M. J., Boldt, S., Wawra, S. E., Freiwald, T. D.,
Damm, C., Walter, J., Lerche, D., and Peukert, W.
(2020). New prospects for particle characteriza-
tion using analytical centrifugation with sector-shaped
centerpieces. Particle & Particle Systems Characteri-
zation, 37(7):2000108.
Wu, M., Ozcelik, A., Rufo, J., Wang, Z., Fang, R., and
Jun Huang, T. (2019). Acoustofluidic separation of
cells and particles. Microsystems & nanoengineering,
5(1):32.
Xie, Y., Bachman, H., and Huang, T. J. (2019). Acoustoflu-
idic methods in cell analysis. TrAC Trends in Analyti-
cal Chemistry, 117:280–290.
Yazdani, A. M. and S¸ is¸man, A. (2020). A novel numeri-
cal model to simulate acoustofluidic particle manipu-
lation. Physica Scripta, 95(9):095002.
Zhao, Y., Lai, H. S. S., Zhang, G., Lee, G.-B., and Li, W. J.
(2014). Rapid determination of cell mass and density
using digitally controlled electric field in a microflu-
idic chip. Lab on a Chip, 14(22):4426–4434.
Assessing the Performance of Autoencoders for Particle Density Estimation in Acoustofluidic Medium: A Visual Analysis Approach
443
