
Cardiac Arrhythmia Detection in Electrocardiogram Signals with
CNN-LSTM
Igor Lopes Souza
a
and Daniel Oliveira Dantas
b
Departamento de Computac¸
˜
ao, Universidade Federal de Sergipe, S
˜
ao Crist
´
ov
˜
ao, SE, Brazil
Keywords:
CNN, Electrocardiography, ECG, Atrial Fibrillation.
Abstract:
Sudden cardiac death and arrhythmia account for a large percentage of all deaths worldwide. Electrocardiog-
raphy is essential in the clinical evaluation of patients who have heart disease. Through the electrocardiogram
(ECG), medical doctors can identify whether the cardiac muscle dysfunctions presented by the patient have
an inflammatory origin and diagnose early serious diseases that primarily affect the blood vessels and the
brain. The basis of arrhythmia diagnosis is the identification of normal and abnormal heartbeats and their
classification into different diagnoses based on ECG morphology. Traditionally, ECG signals are classified
manually, requiring experience and great skill, while being time-consuming and prone to error. Thus, machine
learning algorithms have been widely adopted because of their ability to perform complex data analysis. The
objective of this study is to develop a classifier capable of classifying a patient’s ECG signals for the detection
of arrhythmia in clinical patients. We developed a convolutional neural network (CNN) with long short mem-
ory (LSTM) to identify five classes of heartbeats in ECG signals. Our experiment was conducted with ECG
signals obtained from a publicly available MIT-BIH database. The number of instances was even out to five
classes of heartbeats. The proposed model achieved an accuracy of 98.12% and an F1-score of 99.72% in the
classification of ventricular ectopic beats (V), and an accuracy of 97.39% and an F1-score of 95.25% in the
classification of supraventricular ectopic beats (S).
1 INTRODUCTION
According to the World Health Organization, cardio-
vascular diseases (CVDs) are the leading cause of
death in the world (Wold Health Organization, 2023).
Arrhythmia, a heart rhythm disorder, is considered
one of the most common disorders of the heart. Ar-
rhythmias can lead to tachycardia or even sudden car-
diac arrest. Heartbeat classification based on ECG
signal has become a valuable and promising tech-
nique for early warning of arrhythmias. However,
variations in ECG signals can be significant among
different subjects. Under different circumstances, the
waveform and rhythms produced by the arrhythmia
symptoms can be quite different as well. Experi-
enced cardiologists can easily distinguish abnormal
heartbeats from normal sinus rhythms by observing
the ECG. However, this is still challenging for pa-
tients who wish to accompany their clinical symp-
toms. Computer-driven signal processing is an im-
portant tool to diagnose arrhythmia in the field of
a
https://orcid.org/0000-0002-2499-4607
b
https://orcid.org/0000-0002-0142-891X
biomedical engineering. Today, biomedicine has ad-
vanced to the stage of the practical application of sig-
nal processing and pattern analysis techniques.
Many approaches to arrhythmia heartbeat classi-
fication with convolutional neural networks (CNN)
have been proposed. Ozaltin (Ozaltin and Yeniay,
2022) proposed a novel CNN architecture to detect
ECG types. In addition, the proposed CNN can au-
tomatically extract features from images. He clas-
sifies an ECG dataset using a CNN with 34 layers.
While this dataset is composed of 1D signals, these
are transformed into images using continuous wavelet
transform (CWT). In addition, the proposed CNN
is compared to known architectures, AlexNet and
SqueezeNet, for classifying ECG images. Ozaltin not
only performed CWT but also implemented a short-
time Fourier transform. Ozaltin obtained an accuracy
of 99.21% from the proposed CNN–SVM when using
CWT.
Li (Li et al., 2018) proposed a generic convo-
lutional neural network (GCNN) trained first using
heartbeats without distinguishing patients. Based on
the GCNN, the fine-tuning technique is applied to
304
Souza, I. and Dantas, D.
Cardiac Arrhythmia Detection in Electrocardiogram Signals with CNN-LSTM.
DOI: 10.5220/0012362600003654
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 13th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2024), pages 304-310
ISBN: 978-989-758-684-2; ISSN: 2184-4313
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

modify the GCNN to a tuned dedicated CNN (TD-
CNN) for the corresponding individual. Fine-tuning
is done in several seconds rather than dozens of min-
utes, necessary for the TDCNN to be used to monitor
the long-term ECG signals in clinics. To accelerate
the ECG classification, only the original ECG heart-
beat is input to the CNN without other extended in-
formation from the adjacent heartbeats or FFT repre-
sentation.
Cao (Cao et al., 2022) proposed a novel nonlinear
adaptive noise-canceling framework (ANC) based on
a temporal CNN to effectively extract fetal ECG sig-
nals from mothers’ abdominal ECG recordings. The
proposed framework consists of a two-step network,
using the ANC architecture; one network is for the
maternal ECG component elimination and the other
is for the residual noise component removal of the
extracted fetal ECG signal. Then, joint approxima-
tion diagonalization of eigenmatrices (JADE), one of
the blind source separation algorithms, is applied as
a post-processing step to produce a clean fetal ECG
signal.
Kamozawa (Kamozawa et al., 2023) proposed
a method for detecting atrial fibrillation (AF) from
an electrocardiogram (ECG) measured by a 24-hour
Holter electrocardiograph (Holter-ECG) using CNN.
In the preprocessing step, artifacts and noises on
Holter-ECG are removed by a bandpass filter. The
detection method consists of extraction of abnormal
waveforms using 1D CNN trained with segmented
ECG waveform, spectral entropy, and identification
of AF.
Xiong (Xiong et al., 2017) created a data-driven
deep learning pipeline using a 16-layer CNN for the
automatic classification of ECG signals. Xiong used
a large dataset recorded from patients and labeled by
medical experts in ECG for developing and validating
his approach.
Kiranyaz (Kiranyaz et al., 2017) created a sys-
tem that can detect early occurrences of arrhythmias,
by modeling common causes of arrhythmias in a pa-
tient’s ECG signal. The causes of arrhythmia are
modeled as a degradation from normal ECG beats.
The CNN was trained using real normal beats and
synthesized abnormal beats.
Han (Han and Shi, 2020) proposed a multi-lead
attention (MLA) mechanism integrated with a CNN
to detect myocardial infarction (MI) in 12-lead ECG
recordings. The MLA automatically assigns weights
to different leads and a 2D CNN extracts discrimina-
tive features from all the 12 leads. The robustness of
the MI detection was tested in both intra-patient and
inter patient schemes.
Martis (Martis et al., 2014) investigated four dif-
ferent methods for atrial fibrillation and atrial flut-
ter feature extraction: the principal components
of discrete wavelet transform coefficients, indepen-
dent components of discrete wavelet transform co-
efficients, principal components of discrete cosine
transform coefficients, and independent components
of discrete wavelet transform coefficients methods.
Martis explored three different classification tech-
niques: K-nearest neighbor, decision tree, and artifi-
cial neural network. The methodology used data from
MIT-BIH arrhythmia and atrial fibrillation databases.
Discrete cosine transform coupled with independent
component analysis and K-nearest neighbor yielded
the highest average sensitivity of 99.61%, average
specificity of 99.99%, and classification accuracy of
99.45% using tenfold cross-validation.
We believe that it is possible to further improve
the accuracy, sensitivity, specificity, precision, and
F1-score of CNN heartbeat classifiers of our previous
works. Our study aims to improve the classification
metrics of our previous study by using LSTM com-
bined with CNN models (Souza and Dantas, 2023).
We used a fine-tuning step for further optimization
of the classification of arrhythmia. The improved
classification of ECG signals will generate more ac-
curate responses in the detection of cardiac arrhyth-
mias, facilitating the health care of patients. The
proposed neural network architecture provides a bet-
ter F1-score when compared to the previously listed
works, and provides a higher F1-score and less train-
ing time when compared to our previous work.
2 METHODOLOGY
In this study, we created classifiers based on CNN-
LSTM and AlexNet capable of distinguishing the dif-
ferent types of heartbeats and detecting cardiac ar-
rhythmia. Their architecture was fine-tuned so that
the models achieved the highest validation accuracy
possible and were compared to the decision tree, ran-
dom forest and extra trees classifiers (Kumar, 2022).
Our models were based on a previous study (Souza
and Dantas, 2023) and evaluated in the test set. The
ECG heartbeat classifier is composed of two main
steps: preprocessing and classification. The CNN-
LSTM and AlexNet network architectures are shown
in Figure 1 and Figure 2 respectively. The implemen-
tation of this methodology is publicly available
1
and
was coded in Python using Tensorflow, Keras, and
Numpy.
1
https://github.com/Igor-Lopes-Souza/2023-CNN-
LSTM
Cardiac Arrhythmia Detection in Electrocardiogram Signals with CNN-LSTM
305
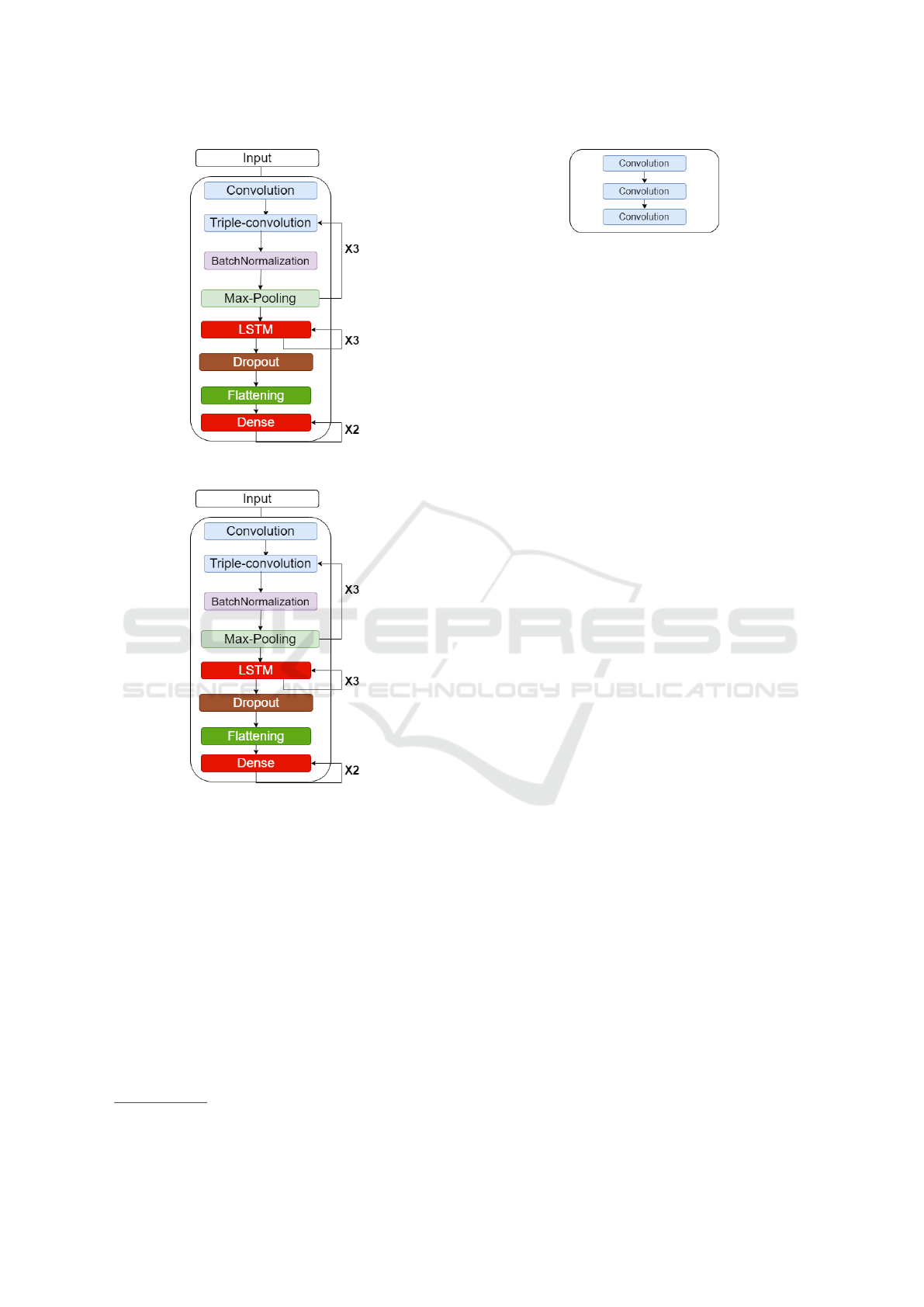
Figure 1: CNN-LSTM architecture.
Figure 2: AlexNet architecture.
In the following subsections we will describe the
dataset used, the preprocessing step, which includes
filtering and data augmentation, the classifier archi-
tecture, and classifier training.
2.1 Dataset
In this study, we used the ECG Heartbeat Catego-
rization Dataset, freely available in the Internet
2
. We
used only the portion of the dataset derived from the
Physio Bank MIT-BIH Arrhythmia database (Mark
and Moody, 1988). This database consists of a 48
half-hour long ECG recordings from 47 subjects—
obtained with a Lead II ECG configuration—that
2
https://www.kaggle.com/datasets/shayanfazeli/heartbeat
Figure 3: Triple-convolution.
were band-pass filtered over the frequency range from
0.1 to 100Hz and digitized at 360 samples per second.
Furthermore, these recordings were interpreted and
validated by at least two cardiologists. The database
consists of annotations for both heartbeat class in-
formation and R-peak position information verified
by two or more expert cardiologists. The 17 beat
types can be grouped into five beat classes defined by
the Association of Advancement for Medical Instru-
mentation (AAMI) which follows the American Na-
tional Standard for Ambulatory ECGs (ANSI/AAMI
EC38:2007) recommendations. The five beat types
are the non-ectopic beat (N), supraventricular ectopic
beat (S), ventricular ectopic beat (V), fusion beat (F),
and unknown (Q).
2.2 Preprocessing
The MIT-BIH dataset is unbalanced, difficulting the
analysis of the signal. The original dataset contains
a total of 109,446 data rows. Each data row contains
a fraction of the ECG signal with duration of 10 sec-
onds and its class, specified in the last column by a
number from 0 to 4 representing N, S, Q, F, and Q.
There are 70,043 data rows for training, 17,511 for
validation and 21,892 for testing, making the propor-
tions 65/15/20. We augmented the training and vali-
dation datasets to match the value of the biggest class
from the five types of heartbeats.
The raw MIT-BIH signal is corrupted by myo-
electric interference, power line interference, and line
drift. We added noise through the Gaussian func-
tion, by creating a random variable and adding it to
our train dataset in order to simulate the myoelec-
tric interferences and better train our model (Yong
et al., 2011). The noise serves to better train our
model by adding a varying variable to our dataset,
acting as a probability distribution. To remove the
noise, the raw ECG signal is filtered using wavelet
filters (Audhkhasi et al., 2016). The raw signal is de-
composed by Daubechies wavelet 6(db6) at six levels,
and wavelet coefficients from the third to the sixth
level were retained and used for signal reconstruc-
tion (Shi et al., 2019).
ICPRAM 2024 - 13th International Conference on Pattern Recognition Applications and Methods
306
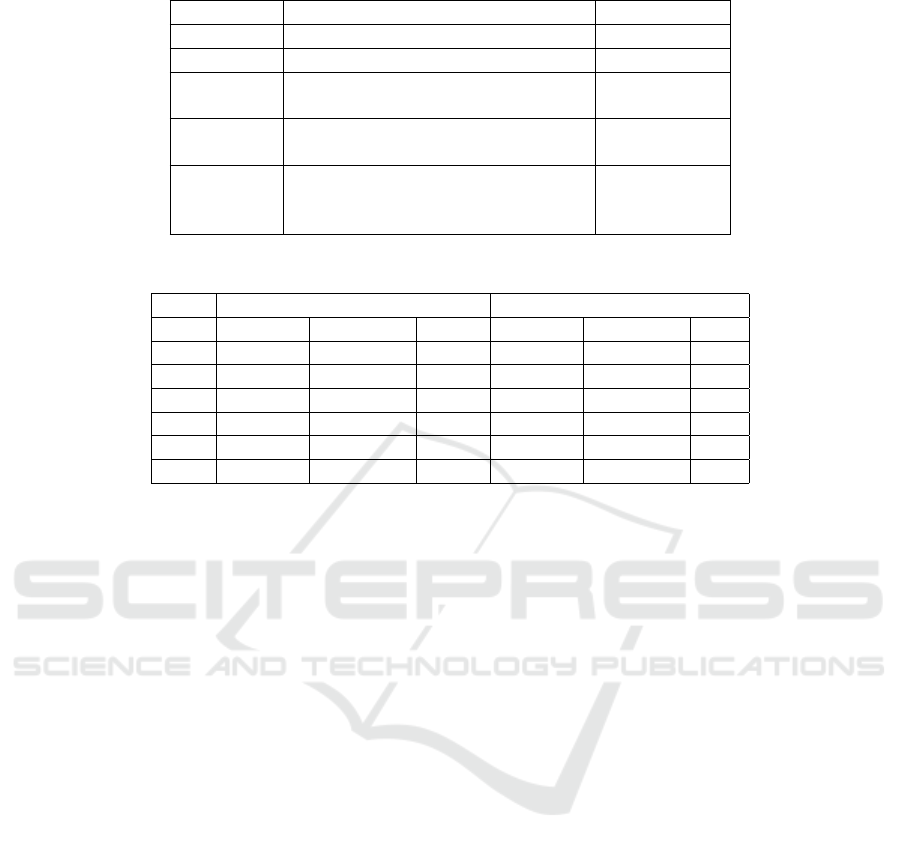
Table 1: Hyperparameter values chosen in classifier fine-tuning.
Parameters Values Chosen Value
Dropout 0.10, 0.20, 0.30, 0.40, 0.50 0.50
Optimizer Adam, Adamax, SGD Adam
Activation
function
Relu, Softmax, Softplus Relu
Batch
size
10, 32, 54, 76, 98 98
Loss
function
Binary cross-entropy, Categorical
cross-entropy, Poisson,
Kullback-Leibler divergence, Huber
Categorical
cross-entropy
Table 2: Number of samples in the training, validation, and test sets.
Before data augmentation After data augmentation
Training Validation Test Training Validation Test
N 57974 14493 18118 57974 14493 N/A
S 1778 450 556 57974 14493 N/A
V 4630 1155 1448 57974 14493 N/A
F 520 127 162 57974 14493 N/A
Q 5141 1286 1608 57974 14493 N/A
Total 70043 17511 21892 289870 72465 N/A
2.3 Classifier Architecture
Figure 1 shows the CNN-LSTM classifier architec-
ture, which comprises convolutional layers, subsam-
pling layers, fully connected layers, batch normaliza-
tion layers, LSTM layers and a dropout layer. Usu-
ally, each convolution layer is followed by a subsam-
pling layer. In order to facilitate mapping between
the heartbeat category and its waveform, we use a
triple-convolution structure to achieve a more pow-
erful fitting capability (Uchida et al., 2018) in our
CNN model. Figure 3 shows the structure of a triple-
convolution layer sequence.
Figure 2 show the schematic of our AlexNet clas-
sifier. The standard AlexNet classifier (Krizhevsky
et al., 2012) is used for 2D image classification,
while we modify its architecture for the analysis of
ECG signals, which are 1D. The AlexNet classifier
comprises convolutional layers, subsampling layers,
fully connected layers, batch normalization layers,
and dropout layers. We altered the convolution, max-
pooling, and dropout layers to use their 1D versions.
In our AlexNet architecture when compared to the
standard format, we doubled the number of triple-
convolutions to obtain better accuracy and used the
batch-normalization layer to normalize the interlayer
outputs into a standard format.
In order to compare the performance of the pro-
posed models with standard classification algorithms,
our CNN-LSTM and AlexNet models are compared
with extra trees, random forest and decision tree clas-
sifiers (Alom et al., 2018; Yu et al., 2019) that were
trained with the sklearn default configuration and our
training dataset (Kramer and Kramer, 2016). The ex-
tra trees, decision tree, and random forest classifiers
architecture use the following parameters in their de-
fault form:
• Minimal Number of Leaves: 1
• Minimal Number of Samples Split: 2
• Criterion: gini,
• Maximum Depth: None,
• Maximum Number of Features: sqrt,
• Maximum Number of Leaf Nodes: None,
• Maximum Number of Samples: None,
• Number of Estimators: 100
In this study, we use the ReLu activation function
in both convolutional layers and fully connected lay-
ers (Nair and Hinton, 2010; Girosi et al., 1995). In the
output layer, we use the softmax activation function to
obtain the five heartbeat classes.
2.4 Training Method
The goal of training is to reduce the value of the
loss function L, i.e., to decrease the CNN-LSTM and
AlexNet models loss and adjust the weights and bi-
ases so that Equation 1 fits the model training set. The
cross-entropy function is used as the loss function (Xu
and Liu, 2020):
Cardiac Arrhythmia Detection in Electrocardiogram Signals with CNN-LSTM
307
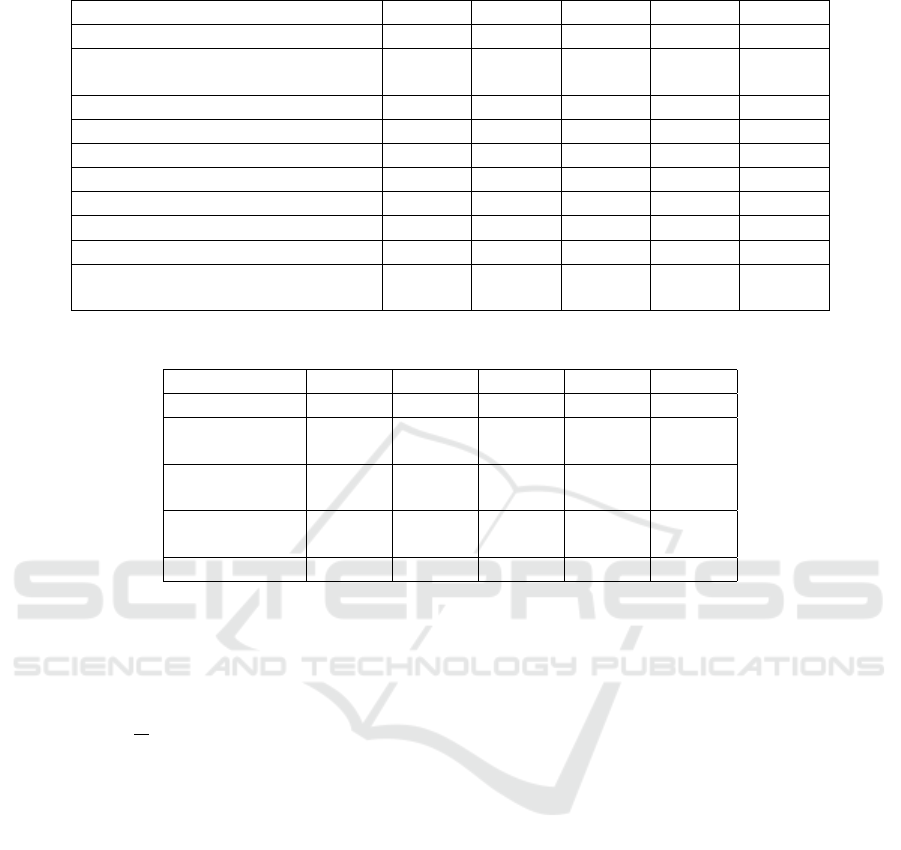
Table 3: Comparison of the proposed algorithm classification using ventricular ectopic beats (V).
ACC SEN SPE PRE F1S
Martis (Martis et al., 2014)
99.45% 99.61% 99.99% 99.99% 99.80%
Proposed classifier:
CNN-LSTM
98.12% 99.00% 98.85% 99.39% 99.72%
Souza (Souza and Dantas, 2023)
99.33% 99.59% 99.30% 99.12% 99.44%
Sellami (Sellami and Hwang, 2019)
99.48% 96.97% 99.87% 98.83% 97.80%
Acharya (Acharya et al., 2017)
94.03% 96.71% 91.54% 97.85% 97.27%
Zhai (Zhai and Tin, 2018)
99.10% 96.40% 99.50% 96.40% 96.40%
Jiang (Jiang and Kong, 2007)
98.80% 94.30% 99.40% 95.30% 94.70%
Xiang (Xiang et al., 2018)
99.20% 93.70% 99.60% 94.80% 94.20%
Ince (Ince et al., 2009)
97.60% 83.60% 98.10% 87.40% 85.40%
Proposed classifier:
AlexNet
96.66% 99.45% 99.10% 96.85% 80.07%
Table 4: Comparison of proposed implementations using ventricular ectopic beats (V).
ACC SEN SPE PRE F1S
CNN-LSTM
98.12% 99.00% 98.85% 99.39% 99.72%
Decision tree
classifier
99.11% 99.22% 99.16% 99.00% 99.36%
Random forest
classifier
95.32% 95.73% 98.83% 96.39% 95.40%
Extra trees
classifier
95.40% 95.27% 94.70% 95.25% 95.35%
AlexNet
96.66% 99.45% 99.10% 96.85% 80.07%
We update the weights and offsets using the Adam
optimizer (Kingma and Ba, 2014). First, a batch
of samples was sent to calculate the gradient of the
Equation 1, and we set the batch size to 98:
g =
1
m
∇
θ
∑
i
L( f (x
(i)
;θ), y
(i)
)
!
. (1)
The g is the gradient value, m is the batch size, θ is
the parameter to be updated, f (x
(i)
;θ) is the heartbeat
type predicted by the i-th sample, y
(i)
is the actual type
of the i-th sample, and L is the loss function.
After defining the architecture, fine-tuning was
performed to obtain the best values of dropout, op-
timizer, activation function, loss function, and batch
size. A grid search in the hyperparameter space tested
each possible combination with 20 epochs. Table 1
shows the tested hyperparameter values and the ones
that maximized accuracy.
3 RESULTS AND DISCUSSION
We performed classification experiments on 44
recordings from the MIT-BIH arrhythmia database,
among the 48 recordings obtained from 47 patients
studied by the BIH arrhythmia laboratory, and the
heartbeats were classified according to the recom-
mendation of the AAMI.
The training dataset contains a total of 109,466
data rows of representative beats from all classes:
type-N, non-ectopic beats; type-S, supraventricular
ectopic beats; type-V, ventricular ectopic beats; type-
F, fusion beats; and type-Q, unknown beats. Clas-
sification performance is measured using the statisti-
cal error metrics found in the literature (Chen et al.,
2022): accuracy (ACC), sensitivity (SEN), specificity
(SPE), precision (PRE), and F1-score (F1S). The F1-
score measures the overall performance of the beat
classification, as shown in Table 3.
Our CNN-LSTM and AlexNet models were im-
plemented using the TensorFlow framework. The
CNN-LSTM and AlexNet models training time of
each epoch was approximately 20s, and the maximum
epoch number was set to 100. Table 3 shows that the
CNN-LSTM model has an F1-score value compara-
ble to those of other studies, presenting the second
best results. Table 4 shows the results of the differ-
ent proposed architectures. In this study, our CNN-
LSTM model achieved an accuracy of 98.12%, sen-
sitivity of 99.00%, specificity of 98.85%, precision
of 99.39%, and F1-score of 99.72% and our AlexNet
model achieved an accuracy of 96.66%, sensitivity of
99.45%, specificity of 99.10%, precision of 96.85%,
ICPRAM 2024 - 13th International Conference on Pattern Recognition Applications and Methods
308
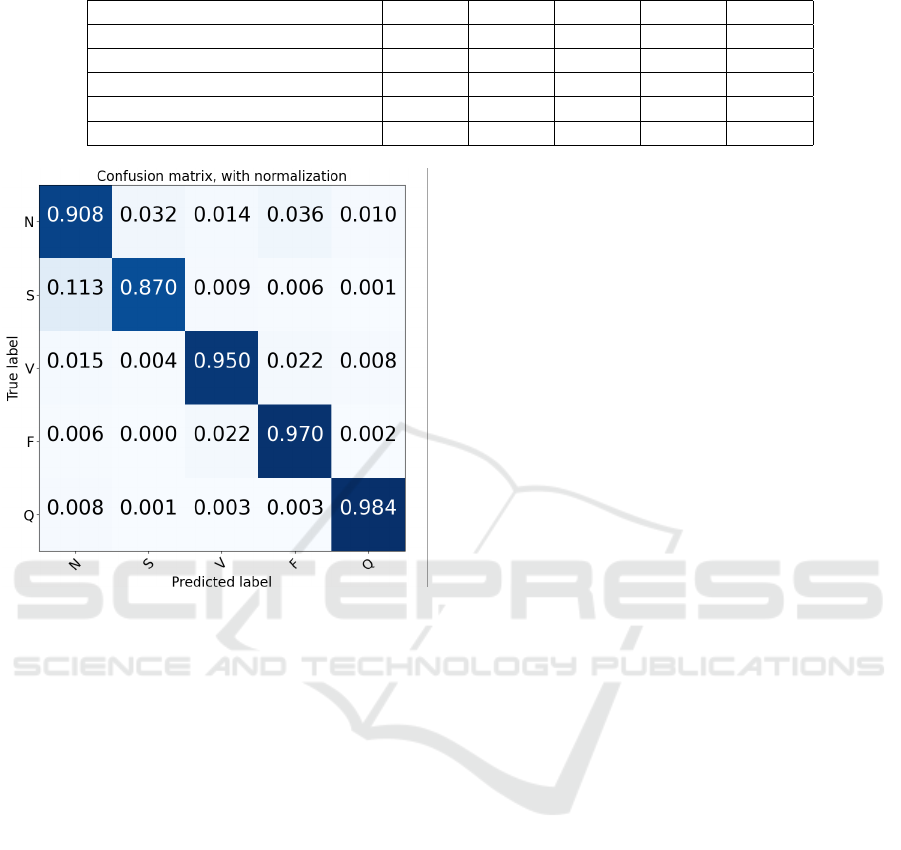
Table 5: Comparison of the types of heartbeats.
ACC SEN SPE PRE F1S
Normal (N)
99.45% 99.98% 92.83% 90.89% 99.44%
Supraventricular ectopic beats (S)
97.39% 88.61% 98.92% 88.92% 95.25%
Ventricular ectopic beats (V)
98.12% 99.00% 98.85% 99.39% 99.72%
Fusion Beats (F)
87.40% 77.27% 84.70% 95.25% 82.35%
Unknown Beats (Q)
99.32% 99.65% 98.72% 97.20% 99.10%
Figure 4: Confusion matrix for heartbeat classification on
the test set.
and F1-score of 80.07%.
Figure 4 shows the confusion matrix of the clas-
sification results of the CNN-LSTM test set. The
model is able to make accurate predictions and distin-
guish different classes. The main reason behind this
might be that we have fine-tuned our model, as unre-
fined tests with ventricular ectopic beats (V) obtained
an average accuracy of 89.99% and an F1-score of
88.54%.
4 CONCLUSIONS
In this study, we designed an ECG signals classifier
for cardiac arrhythmia detection using CNN-LSTMs.
The proposed model achieved an accuracy of 98.12%
and an F1-score of 99.72% in the classification of ven-
tricular ectopic beats (V), and an accuracy of 97.39%
and an F1-score of 95.25% in the classification of
supraventricular ectopic beats (S) as shown in Table 5.
In order to optimize our model, we fine-tuned the hy-
perparameters. The selected values compose our fi-
nal version of the classifier and are displayed in Ta-
ble 1. Compared with the methods in the literature,
our model performed better in terms of ventricular ec-
topic beat classification precision and F1-score, only
being surpassed by Martis (Martis et al., 2014).
Our trained CNN heartbeat classifier model can
be used for real-life and real-time applications. It
can also be used to analyze other 1D biosignals. Fu-
ture work may refine this approach with a better set
of hyperparameter values and different augmentation
strategies and training methods.
REFERENCES
Acharya, U. R., Oh, S. L., Hagiwara, Y., Tan, J. H., Adam,
M., Gertych, A., and San Tan, R. (2017). A deep
convolutional neural network model to classify heart-
beats. Computers in biology and medicine, 89:389–
396.
Alom, M. Z., Taha, T. M., Yakopcic, C., Westberg, S.,
Sidike, P., Nasrin, M. S., Van Esesn, B. C., Awwal, A.
A. S., and Asari, V. K. (2018). The history began from
AlexNet: A comprehensive survey on deep learning
approaches. arXiv preprint arXiv:1803.01164.
Audhkhasi, K., Osoba, O., and Kosko, B. (2016). Noise-
enhanced convolutional neural networks. Neural Net-
works, 78:15–23.
Cao, S., Xiao, H., Gong, G., Fang, W., and Chen, C. (2022).
Morphology extraction of fetal ECG using temporal
CNN-based nonlinear adaptive noise cancelling. Plos
one, 17(12):e0278917.
Chen, S. W., Wang, S. L., Qi, X. Z., Samuri, S. M., and
Yang, C. (2022). Review of ECG detection and clas-
sification based on deep learning: Coherent taxon-
omy, motivation, open challenges and recommenda-
tions. Biomedical Signal Processing and Control,
74:103493.
Girosi, F., Jones, M., and Poggio, T. (1995). Regulariza-
tion theory and neural networks architectures. Neural
computation, 7(2):219–269.
Han, C. and Shi, L. (2020). ML–ResNet: A novel net-
work to detect and locate myocardial infarction using
12 leads ECG. Computer methods and programs in
biomedicine, 185:105138.
Ince, T., Kiranyaz, S., and Gabbouj, M. (2009). A generic
and robust system for automated patient-specific clas-
sification of ECG signals. IEEE Transactions on
Biomedical Engineering, 56(5):1415–1426.
Jiang, W. and Kong, S. G. (2007). Block-based neural
networks for personalized ECG signal classification.
Cardiac Arrhythmia Detection in Electrocardiogram Signals with CNN-LSTM
309

IEEE Transactions on Neural Networks, 18(6):1750–
1761.
Kamozawa, H., Muroga, S., and Tanaka, M. (2023). A
detection method of atrial fibrillation from 24-hour
Holter-ECG using CNN. IEEJ Transactions on Elec-
trical and Electronic Engineering.
Kingma, D. P. and Ba, J. (2014). Adam: A
method for stochastic optimization. arXiv preprint
arXiv:1412.6980.
Kiranyaz, S., Ince, T., and Gabbouj, M. (2017). Personal-
ized monitoring and advance warning system for car-
diac arrhythmias. Scientific reports, 7(1):1–8.
Kramer, O. and Kramer, O. (2016). Scikit-learn. Machine
learning for evolution strategies, pages 45–53.
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2012). Im-
agenet classification with deep convolutional neural
networks. Advances in neural information processing
systems, 25.
Kumar, V. (2022). Analysis of CNN features with multiple
machine learning classifiers in diagnosis of monkey-
pox from digital skin images. medRxiv, pages 2022–
09.
Li, J., Si, Y., Xu, T., and Jiang, S. (2018). Deep convolu-
tional neural network based ECG classification system
using information fusion and one-hot encoding tech-
niques. Mathematical problems in engineering, 2018.
Mark, R. and Moody, G. (1988). MIT-BIH arrhythmia
database directory. Cambridge: Massachusetts Insti-
tute of Technology.
Martis, R. J., Acharya, U. R., Adeli, H., Prasad, H., Tan,
J. H., Chua, K. C., Too, C. L., Yeo, S. W. J., and Tong,
L. (2014). Computer aided diagnosis of atrial arrhyth-
mia using dimensionality reduction methods on trans-
form domain representation. Biomedical signal pro-
cessing and control, 13:295–305.
Nair, V. and Hinton, G. E. (2010). Rectified linear units
improve restricted boltzmann machines. In Icml.
Ozaltin, O. and Yeniay, O. (2022). A novel proposed CNN–
SVM architecture for ECG scalograms classification.
Soft Computing, pages 1–20.
Sellami, A. and Hwang, H. (2019). A robust deep convo-
lutional neural network with batch-weighted loss for
heartbeat classification. Expert Systems with Applica-
tions, 122:75–84.
Shi, H., Wang, H., Zhang, F., Huang, Y., Zhao, L., and Liu,
C. (2019). Inter-patient heartbeat classification based
on region feature extraction and ensemble classifier.
Biomedical Signal Processing and Control, 51:97–
105.
Souza, I. and Dantas, D. (2023). Cardiac arrhythmia clas-
sification in electrocardiogram signals with convolu-
tional neural networks. In Proceedings of the 12th
International Conference on Pattern Recognition Ap-
plications and Methods. SCITEPRESS - Science and
Technology Publications.
Uchida, K., Tanaka, M., and Okutomi, M. (2018). Cou-
pled convolution layer for convolutional neural net-
work. Neural Networks, 105:197–205.
Wold Health Organization (2023). The top 10 causes of
death. Available at: https://www.who.int/news
-room/fact-sheets/detail/the-top-10-cause
s-of-death. Last accessed: 03 September.
Xiang, Y., Luo, J., Zhu, T., Wang, S., Xiang, X., and Meng,
J. (2018). ECG-based heartbeat classification using
two-level convolutional neural network and rr interval
difference. IEICE TRANSACTIONS on Information
and Systems, 101(4):1189–1198.
Xiong, Z., Stiles, M. K., and Zhao, J. (2017). Robust ECG
signal classification for detection of atrial fibrillation
using a novel neural network. In 2017 Computing in
Cardiology (CinC), pages 1–4. IEEE.
Xu, X. and Liu, H. (2020). ECG heartbeat classification
using convolutional neural networks. IEEE Access,
8:8614–8619.
Yong, P. C., Nordholm, S., Dam, H. H., and Low, S. Y.
(2011). On the optimization of sigmoid function for
speech enhancement. In 2011 19th European Signal
Processing Conference, pages 211–215.
Yu, Y., Si, X., Hu, C., and Zhang, J. (2019). A review of
recurrent neural networks: LSTM cells and network
architectures. Neural computation, 31(7):1235–1270.
Zhai, X. and Tin, C. (2018). Automated ECG classification
using dual heartbeat coupling based on convolutional
neural network. IEEE Access, 6:27465–27472.
ICPRAM 2024 - 13th International Conference on Pattern Recognition Applications and Methods
310
