
Association of Grad-CAM, LIME and Multidimensional Fractal
Techniques for the Classification of H&E Images
Thales R. S. Lopes
1
, Guilherme F. Roberto
2 a
, Carlos Soares
2 b
, Tha
´
ına A. A. Tosta
3 c
,
Adriano B. Silva
4 d
, Adriano M. Loyola
5
, S
´
ergio V. Cardoso
5 e
, Paulo R. de Faria
6 f
,
Marcelo Z. do Nascimento
4 g
and Leandro A. Neves
1 h
1
Department of Computer Science and Statistics, S
˜
ao Paulo State University, S
˜
ao Jos
´
e do Rio Preto-SP, Brazil
2
Faculty of Engineering, University of Porto, Porto, Portugal
3
Institute of Science and Technology, Federal University of S
˜
ao Paulo, S
˜
ao Jos
´
e dos Campos-SP, Brazil
4
Faculty of Computer Science, Federal University of Uberl
ˆ
andia, Uberl
ˆ
andia-MG, Brazil
5
Area of Oral Pathology, School of Dentistry, Federal University of Uberl
ˆ
andia, Uberl
ˆ
andia-MG, Brazil
6
Department of Histology and Morphology, Institute of Biomedical Science,
Federal University of Uberl
ˆ
andia, Uberl
ˆ
andia-MG, Brazil
Keywords:
Deep Learning, Fractal Features, Explainable Artificial Intelligence, Histological Images.
Abstract:
In this work, a method based on the use of explainable artificial intelligence techniques with multiscale
and multidimensional fractal techniques is presented in order to investigate histological images stained with
Hematoxylin-Eosin. The CNN GoogLeNet neural activation patterns were explored, obtained from the
gradient-weighted class activation mapping and locally-interpretable model-agnostic explanation techniques.
The feature vectors were generated with multiscale and multidimensional fractal techniques, specifically frac-
tal dimension, lacunarity and percolation. The features were evaluated by ranking each entry, using the ReliefF
algorithm. The discriminative power of each solution was defined via classifiers with different heuristics. The
best results were obtained from LIME, with a significant increase in accuracy and AUC rates when compared
to those provided by GoogLeNet. The details presented here can contribute to the development of models
aimed at the classification of histological images.
1 INTRODUCTION
Clinical diagnoses and studies in the medical field are
commonly based on biomedical images, a fact that
has motivated applications and research in the fields
of computer vision and pattern recognition (Zerdoumi
et al., 2018). Thus, it is possible to exploit a series
of characteristics present in this category of images,
such as microscopic information (texture, colour, and
morphology) of tissues and cells (Cruz-Roa et al.,
2011).
a
https://orcid.org/0000-0001-5883-2983
b
https://orcid.org/0000-0003-4549-8917
c
https://orcid.org/0000-0002-9291-8892
d
https://orcid.org/0000-0001-8999-1135
e
https://orcid.org/0000-0003-1809-0617
f
https://orcid.org/0000-0003-2650-3960
g
https://orcid.org/0000-0003-3537-0178
h
https://orcid.org/0000-0001-8580-7054
In this context, representation learning encom-
passes a set of techniques for automatically trans-
forming data, such as pixels in a digital image, into
a feature vector, with the aim of recognising existing
patterns in the domain under analysis. In this context,
models based on deep learning stand out among the
different representation learning approaches, as they
have multiple levels and can therefore learn highly
complex functions (LeCun et al., 2015). The increase
in computer processing capacity has made it possible
to exploit a specific type of deep learning approach,
known as a Convolutional Neural Network (CNN),
which has provided important advances in image pro-
cessing and pattern recognition (LeCun et al., 2015).
Computational models based on the deep learning
paradigm are proving effective in many applications,
such as the categorisation of medical images. The in-
terpretability of the results provided by deep neural
networks is still a challenge due to the ”black box”
Lopes, T., Roberto, G., Soares, C., Tosta, T., Silva, A., Loyola, A., Cardoso, S., R. de Faria, P., Z. do Nascimento, M. and Neves, L.
Association of Grad-CAM, LIME and Multidimensional Fractal Techniques for the Classification of H&E Images.
DOI: 10.5220/0012358200003660
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 19th Inter national Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2024) - Volume 2: VISAPP, pages
441-447
ISBN: 978-989-758-679-8; ISSN: 2184-4321
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
441

nature of CNNs (Adadi and Berrada, 2018; Doshi-
Velez and Kim, 2017). The black box problem refers
to the level of concealment of the internal compo-
nents of a system (Suman et al., 2010). In the context
of artificial intelligence and deep learning, the sys-
tem’s difficulty in generating an explanation of how
it reached a decision defines the black box problem.
In critical decisions, such as the indication of a di-
agnosis, it is important to know the reasons behind
them. Therefore, the concept of explainable artifi-
cial intelligence (XAI) offers interesting solutions for
making the knowledge produced by a computational
model that exploits black box artificial intelligence
techniques more comprehensible.
To this end, specific techniques are being explored
to evaluate neural activation patterns in a CNN (Ma-
hendran and Vedaldi, 2016; Yosinski et al., 2015). For
instance, the values present in the ”average pooling”
layer of a CNN, applied as a structural regularising
strategy, can define the image regions commonly used
in the classification process through the use of the
gradient-weighted class activation mapping (Grad-
CAM) techniques (Rajaraman et al., 2018; Reyes
et al., 2020) and locally-interpretable model-agnostic
explanation (LIME) (Rajaraman et al., 2018; Reyes
et al., 2020; Iam Palatnik de Sousa, 2019). On
the other hand, the literature also shows the suc-
cess of computer models developed using consol-
idated image processing techniques, such as those
used for handcrafted features extraction (Ivanovici
et al., 2009b; M. Sahini, 2014). These studies have
explored strategies based on first and second order
analyses, such as fractal techniques called fractal di-
mension, lacunarity and percolation.
Finally, it is important to note that despite
the advances involving CNN models for investi-
gating diseases in histological images stained with
Haematoxylin-Eosin (H&E), there has not yet been
a proposal aimed at investigating the discriminative
power of regions provided by Grad-CAM and LIME
techniques via multidimensional fractal approaches.
Fractal techniques can be applied to quantify the neu-
ral activation representations of a CNN and provide
new classifications and interpretations of the results.
These associations are relevant contributions to the
classification and pattern recognition of diseases com-
monly investigated from histological images, as well
as to the field of machine learning. The knowledge
obtained can be used to support more comprehensible
computer systems.
In this project, H&E histological images were ex-
plored through quantification with fractal strategies of
the representations provided by the Grad-CAM and
LIME techniques. Specifically, the aim was to: ob-
tain neural activation patterns using the Grad-CAM
and LIME techniques; quantify the representations
of neural activation with multiscale and multidimen-
sional fractal techniques; and define the discrimina-
tive power of the features obtained using recognised
classifiers in the field of artificial intelligence.
2 MATERIALS AND METHODS
The method was structured into four main stages: ob-
taining the CNN’s activation patterns; generating fea-
ture vectors; ranking and selecting features; classify-
ing and identifying the best combinations. The first
stage aimed to organise and define a set of images
representative of the original image dataset, but only
with the images obtained via LIME and Guided Grad-
CAM. The second stage was dedicated to quantising
the images with fractal techniques and composing the
feature vectors. The third stage explored the use of
the ReliefF algorithm to identify the most relevant de-
scriptors for the histological image classification pro-
cess (Robnik-Sikonja and Kononenko, 2003). Finally,
in the fourth stage, each set of features was analysed
by collecting the performances achieved with the rele-
vant classification algorithms. A summary of the pro-
posed method is shown in Figure 1 and the details of
each stage are in the following sections.
The method was applied to six public H&E his-
tological images datasets. The CR dataset consists
of histological images derived from 16 H&E stained
sections of stage T3 or T4 colorectal cancer (Sir-
inukunwattana et al., 2017). The histological sec-
tions were digitised into whole-slide images (WSI)
using a Zeiss MIRAX MIDI digitiser with a pixel
resolution of 0.465µm. The images were catego-
rized into benign or malignant groups. This study
used 151 images measuring 775 x 522 pixels, divided
into 67 benign cases and 84 malignant cases. The
OD dataset was built from 30 tongue tissue sections
from mice stained with H&E previously subjected to
a carcinogen during two experiments carried out in
2009 and 2010, duly approved by the Ethics Com-
mittee on the Use of Animals, under protocol num-
ber 038/39 at the Federal University of Uberl
ˆ
andia.
A total of 66 histological images were obtained us-
ing a LeicaDM500 optical microscope at 400 magni-
fication, using the RGB colour model with a resolu-
tion of 2048 × 1536 pixels. This dataset consists of
74 healthy and 222 severe dysplasia samples (Silva
et al., 2022). The resolution of each image is 452
x 250 pixels. The LA dataset is composed of liver
tissue obtained from mice and consists of 521 im-
ages divided into four classes where each represents
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
442
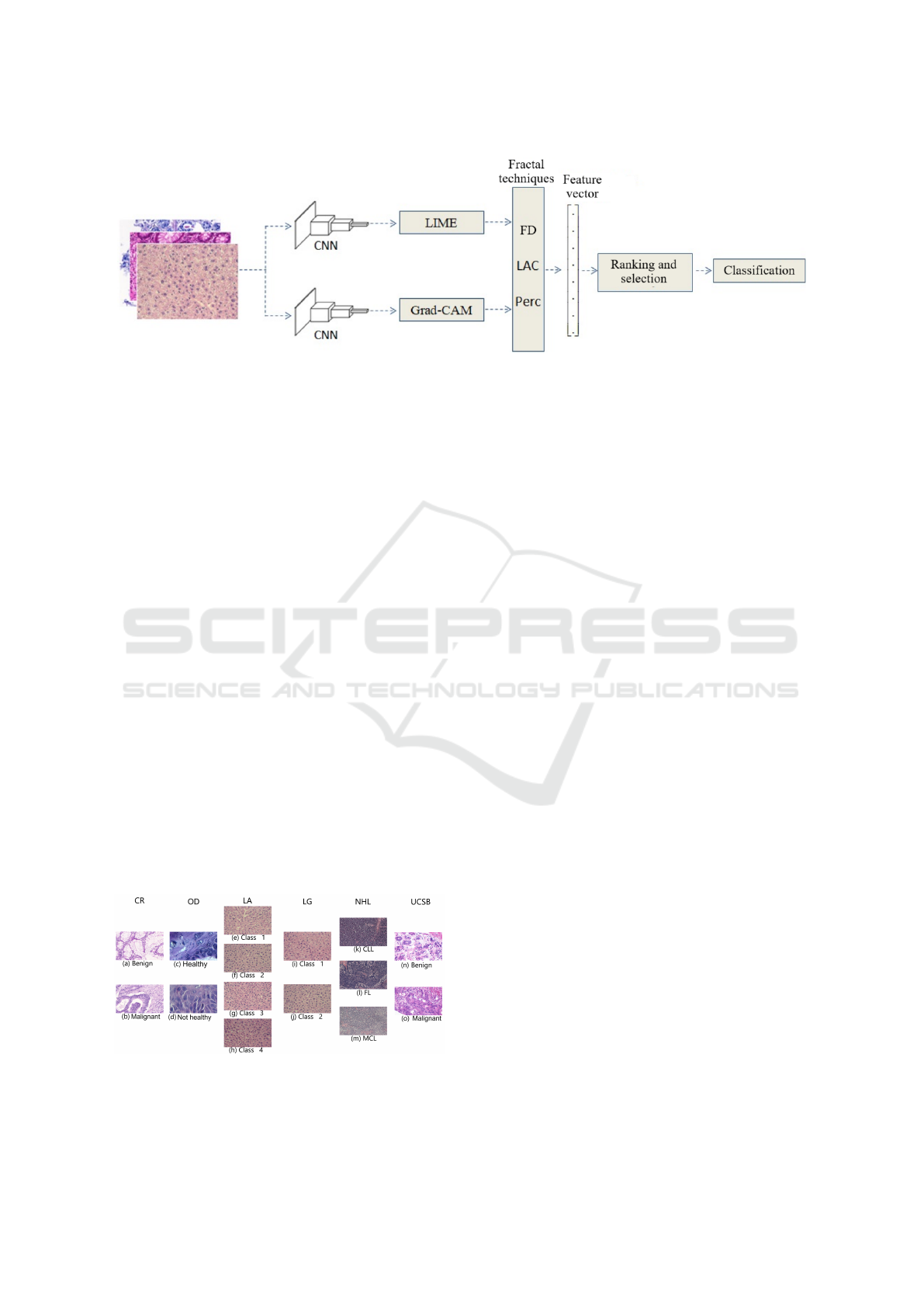
Figure 1: Illustration of the proposed method covering the phases: 1) Obtaining neural activation patterns; 2) Generating
feature vectors; 3) Ranking and selecting features; and 4) Classification.
a group of female mice of different ages with ad li-
bitum diets: one (100), six (115), 16 (162) and 24
(152) months old. All the images have the same reso-
lution of 417 x 312 pixels. The LG dataset also con-
sists of images with dimensions of 417 x 312 pixels
representing liver tissue from mice. The two classes
represent the gender of the sample collected, totalling
265 examples: male with 150 images and female with
115 samples. Both LA and LG datasets were pro-
vided by the Atlas of Gene Expression in Mouse Ag-
ing Project (AGEMAP) (o. A. AGEMAP, 2020). The
NHL dataset consists of representative histological
images of three classes of non-Hodgkin’s lymphoma,
CLL, FL and MCL (do Nascimento et al., 2018). The
images were photographed and stored digitally with-
out compression in tif format, RGB colour model,
1388 x 1040 resolution and 24-bit quantization. A
total of 375 images were used, containing 113, 139
and 122 CLL, FL and MCL regions, respectively.
The UCSB dataset consists of 58 histological images
obtained from biopsies stained with Haematoxylin
and Eosin (H&E). All the images were made avail-
able by the University of California Santa Barbara
(Drelie Gelasca et al., 2008). The images have di-
mensions of 768 x 896 pixels, RGB colour standard
and 24-bit quantization rate. In Figure 2 samples of
the H&E datasets used in this work are shown.
Figure 2: Samples of the image datasets explored in this
paper.
The first stage involves extracting neural activa-
tion patterns from a CNN. To this end, histological
images were analysed by applying the GoogLeNet
(Szegedy et al., 2015) model, pre-trained on the Im-
ageNet (He et al., 2016) dataset. This model was
chosen based on its performance in relation to that
achieved by different CNN architectures for classi-
fying histological H&E images. The model cho-
sen was the one with the lowest distinction rate, in
order to verify whether the proposed methodology
provides relevant gains in the image distinctions ex-
plored. Other models have been tested and could be
applied to define higher classification rates. However,
the choice of the architecture with the lowest perfor-
mance effectively illustrates the worst-case gains via
the proposed model. This fact can guarantee new
strategies through less deep and complex models. The
tested models and the metrics applied are shown in
Table 1.
For each dataset, the fine-tuning method was used
to adjust the output layer of the GooLeNet model to
the number of classes, with a learning rate of 0.001, a
momentum of 0.9 and the dataset was split in 80% to
train and 20% to validate the model.
After running the GoogLeNet model, the neural
activation patterns of the histological images were ob-
tained using the Guided Grad-CAM and LIME tech-
niques. Examples of representations obtained using
these techniques are shown in Figure 3.
The feature vectors were generated by applying
multiscale and multidimensional fractal techniques,
which can quantify self-similarity properties in im-
ages: these properties can be observed in histologi-
cal images and in the growth pattern of some tumours
(Baish and Jain, 2000). Fractal techniques have been
applied to quantify different types of lesions in histo-
logical images, especially multiscale and multidimen-
sional approaches such as fractal dimension, lacunar-
ity and percolation (Ivanovici et al., 2009a; Sahini and
Sahimi, 2014). The multiscale and multidimensional
Association of Grad-CAM, LIME and Multidimensional Fractal Techniques for the Classification of H&E Images
443
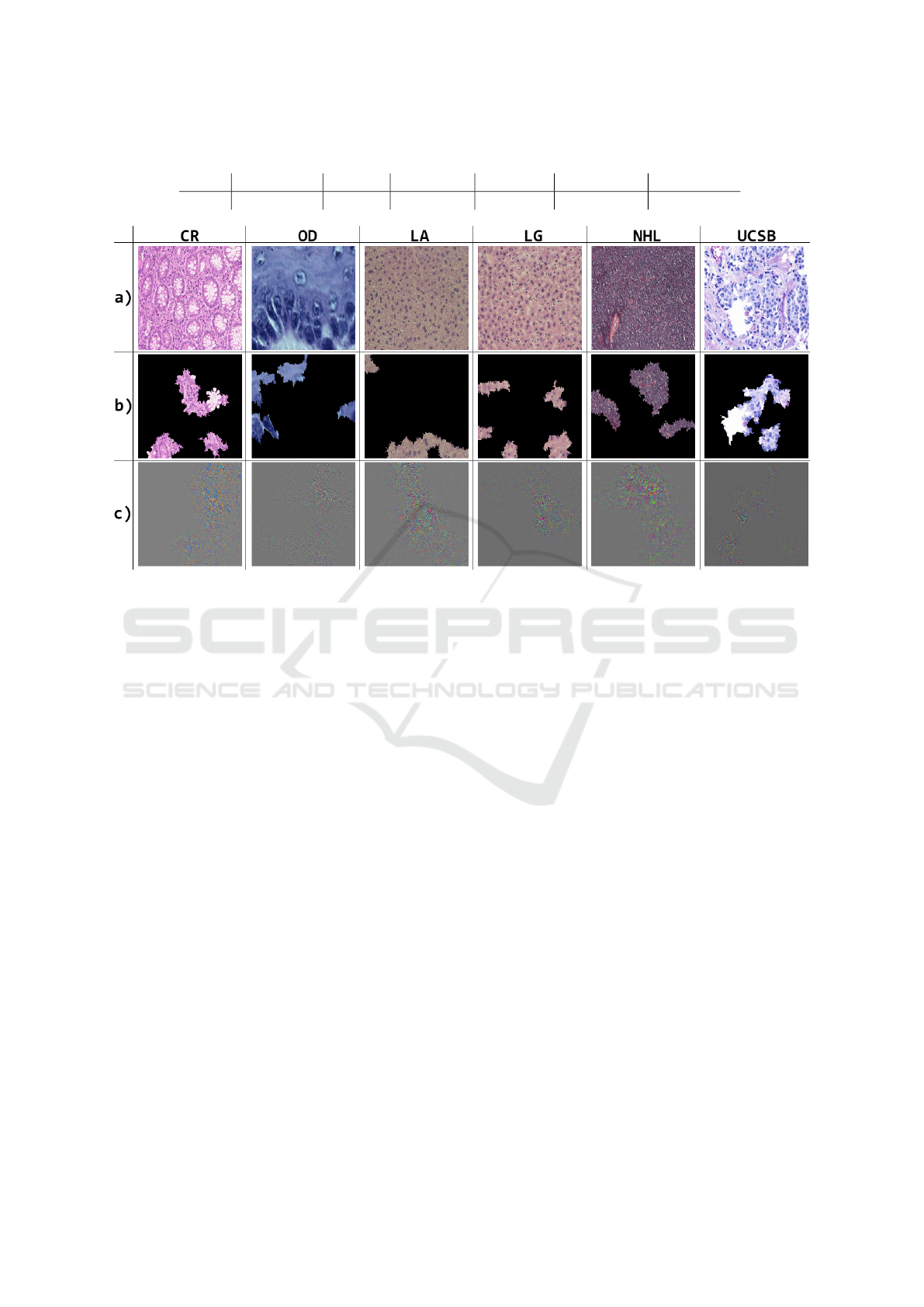
Table 1: Average accuracy of the tested models on the H&E histological image datasets used.
CNN GoogLeNet VGG16 ResNet-50 DenseNet EfficientNet SqueezeNet
ACC 67% 99% 77% 69% 68% 91%
Figure 3: Examples of H&E histological image samples from each base and neural activation patterns, where: (a) original
images; (b) LIME results; (c) results via Guided Grad-CAM.
DF, LAC and PERC techniques available in (Ribeiro
et al., 2019) were applied to quantify the results pro-
vided by the LIME and Grad-CAM techniques. The
attributes obtained were used to compose feature vec-
tors representative of each type of image. Each vector
will be analysed using the strategies indicated in stage
3.
The feature vectors obtained in stage 2 were anal-
ysed by applying a feature selection process in or-
der to reduce the dimensionality and identify the
best combination among the descriptors available
in each subset (Hsu et al., 2011; Mengdi et al.,
2018; Candelero et al., 2020). The process consisted
of ranking each entry using the ReliefF algorithm
(Robnik-Sikonja and Kononenko, 2003) and applying
a threshold to reduce the number of possible combi-
nations. The tested thresholds were defined from the
10 best-ranked features, with increments of 10 fea-
tures (Ribeiro et al., 2019). The largest set was lim-
ited to 116 features, the maximum number of features
obtained at stage 2.
The discriminative power of each solution was ob-
tained by exploring eight classifiers obtained from
the Weka package (Frank et al., 2016): Random
Tree (RT), Random Forest (RaF), IBk, K*, Logit
Boost (LB), Rotation Forest (RoF), Simple Logistic
(SL) and Logistic (L). The evaluation process with
each classification algorithm was carried out using
the cross-validation k-fold strategy, with k=10, and
metrics commonly available in the literature, such as
area under the ROC curve (AUC) and accuracy (Chiu,
2012). The GoogLeNet model was implemented in
the Python language, via the Pytorch library. The
Weka platform, version 3.8.5, was used to collect the
results of the accuracy and AUC measures from the
execution of stage 4. The proposal was carried out on
a notebook with a 2.4GHz Intel Core i5-9300H pro-
cessor, 8GB of RAM and a NVIDIA GeForce 1050
graphics card.
3 RESULTS
The proposed methodology was then applied to the set
of histological images, as described in section 2. The
best results were defined by combining the classifier
and the method for ranking the most relevant descrip-
tors, applying thresholds in the feature space. The
feature rankings were determined using the ReliefF
algorithm. Table 2 shows the highest average accu-
racy rates and AUC, with the total number of features
required to achieve the corresponding results on each
H&E dataset. Figure 4 shows a comparison between
the best average performance obtained by applying
the method using LIME and Guided Grad-CAM.
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
444
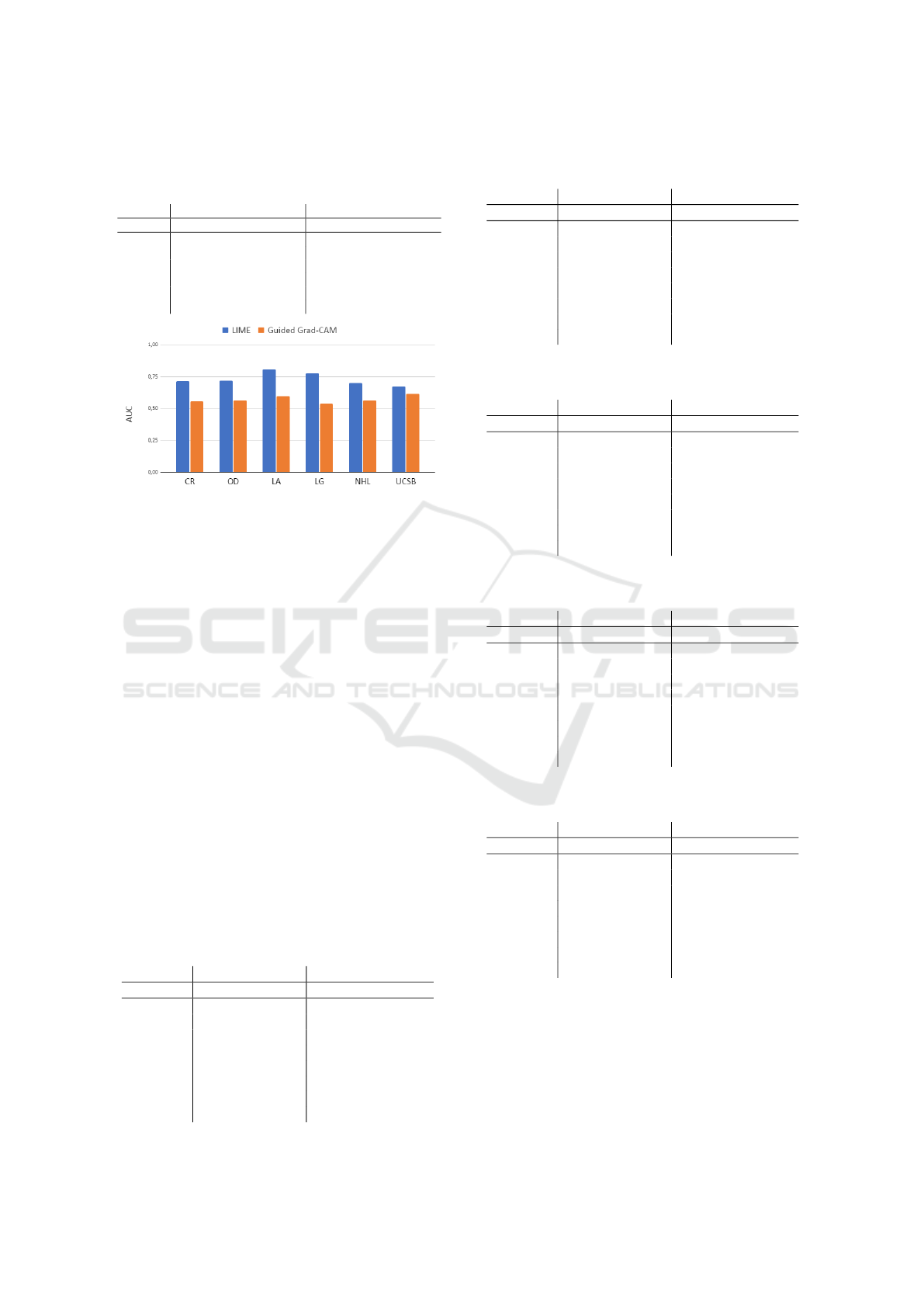
Table 2: Average performances obtained by applying the
proposed method, as well as an indication of the number of
features used (NoF) in each experiment.
LIME Guided Grad-CAM
Dataset AUC ACC NoF AUC ACC NoF
CR 0.714 66.74% 20 0.554 55.96% 110
DO 0.717 73.86% 20 0.562 69.22% 70
LA 0.804 60.80% 20 0.595 36.10% 70
LG 0.775 71.46% 30 0.535 54.76% 70
NHL 0.698 53.24% 40 0.560 38.60% 110
UCSB 0.672 63.15% 30 0.612 57.98% 10
Figure 4: Average AUC rates obtained from eight different
classifiers for each dataset.
From Figure 4, it can be seen that the LIME
method provided the best performance when com-
bined with fractal techniques to classify histological
H&E images. In all datasets, the best combinations
were obtained via LIME, with the highest average
AUC being provided by the LA dataset, using only
20 features.
Finally, to identify the classifier that stood out in
each best result, Tables 3 to 8 show the AUC and ac-
curacy rates. The cases with the highest AUC have
been highlighted in bold. Note that the Logistic,
Simple Logistic, Rotation Forest and Random Forest
classifiers provided the highest AUC values, with the
highest value being 0.896 (30 features obtained via
LIME with the Logistic classifier for the LG dataset).
In this case, the observed accuracy was 82.26%. For
this same dataset, the GoogLeNet model provided a
lower performance, with an AUC of 0.636 and an ac-
curacy of 62.26%, which allows us to verify the con-
tributions of our methodology in terms of classifica-
tion.
Table 3: Results provided by each classifier for the solution
with the highest average AUC for the CR dataset.
LIME Guided Grad-CAM
Classifier AUC ACC AUC ACC
RT 0.605 61.21% 0.550 55.15%
RaF 0.752 69.09% 0.580 55.55%
IBk 0.559 58.18% 0.518 53.33%
K* 0.698 61.82% 0.611 61.21%
LB 0.774 69.09% 0.600 59.39%
RoF 0.768 72.12% 0.584 56.36%
SL 0.771 71.52% 0.504 54.55%
L 0.786 70.91% 0.486 52.12%
Table 4: Results provided by each classifier for the solution
with the highest average AUC for the OD dataset.
LIME Guided Grad-CAM
Classifier AUC ACC AUC ACC
RT 0.610 70.61% 0.556 65.88%
RaF 0.741 74.66% 0.587 70.61%
IBk 0.614 68.24% 0.500 63.18%
K* 0.657 68.92% 0.563 66.55%
LB 0.736 75.00% 0.547 69.26%
RoF 0.789 77.03% 0.645 76.01%
SL 0.798 79.05% 0.481 75.00%
L 0.788 77.36% 0.620 67.23%
Table 5: Results provided by each classifier for the solution
with the highest average AUC for the LA dataset.
LIME Guided Grad-CAM
Classifier AUC ACC AUC ACC
RT 0.672 51.52% 0.544 32.58%
RaF 0.840 63.64% 0.598 37.69%
IBk 0.726 60.42% 0.543 32.20%
K* 0.818 58.71% 0.549 32.00%
LB 0.779 54.55% 0.556 32.20%
RoF 0.875 67.80% 0.634 39.20%
SL 0.857 63.83% 0.651 40.72%
L 0.863 65.91% 0.684 42.23%
Table 6: Results provided by each classifier for the solution
with the highest average AUC for the LG dataset.
LIME Guided Grad-CAM
Classifier AUC ACC AUC ACC
RT 0.719 72.08% 0.547 56.23%
RaF 0.798 70.94% 0.517 54.34%
IBk 0.667 66.04% 0.526 52.83%
K* 0.731 64.91% 0.502 55.85%
LB 0.717 67.17% 0.459 52.45%
RoF 0.844 76.23% 0.577 56.98%
SL 0.829 72.08% 0.533 52.83%
L 0.896 82.26% 0.620 56.60%
Table 7: Results provided by each classifier for the solution
with the highest average AUC for the NHL dataset.
LIME Guided Grad-CAM
Classifier AUC ACC AUC ACC
RT 0.610 47.86% 0.533 37.70%
RaF 0.714 53.21% 0.610 41.18%
IBk 0.592 47.33% 0.545 39.04%
K* 0.631 44.65% 0.569 37.97%
LB 0.729 56.68% 0.526 36.36%
RoF 0.729 53.48% 0.582 41.18%
SL 0.771 59.36% 0.542 37.43%
L 0.807 63.37% 0.569 37.97%
To verify the relevance of the best solution ob-
tained with our proposal, the GoogLeNet model was
applied to the H&E histological image dataset to iden-
tify its discriminative capacity. Table 9 shows the best
results obtained via LIME and Grad-CAM, as well as
the performance obtained by the GoogLeNet network
when classifying the image datasets investigated here
Association of Grad-CAM, LIME and Multidimensional Fractal Techniques for the Classification of H&E Images
445

Table 8: Results provided by each classifier for the solution
with the highest average AUC for the UCSB dataset.
LIME Guided Grad-CAM
Classifier AUC ACC AUC ACC
RT 0.605 60.34% 0.651 65.52%
RaF 0.736 68.97% 0.624 58.62%
IBk 0.691 67.24% 0.584 58.62%
K* 0.714 63.79% 0.627 55.17%
LB 0.653 63.79% 0.599 53.45%
RoF 0.644 55.17% 0.582 53.45%
SL 0.704 67.24% 0.591 53.45%
L 0.626 58.62% 0.641 65.52%
with the number of training epochs set to 30. Thus,
using Grad-CAM, a performance gain is observed in
four of the six datasets, with the highest gain being
17.67% for UCSB. On the other hand, the proposal
provided lower performance in the CR dataset, with
a difference of 22.16%. When the combinations are
made using LIME representations, the methodology
proved to be interesting as it indicated performance
gains in all datasets, with differences ranging from
0.08% to 40.24% in AUC rates. This indicates an-
other important contribution of this study, wherein the
combined use of fractal techniques, LIME representa-
tions and transfer learning can contribute to the foun-
dation of methods aimed at studying and recognising
patterns in the contexts investigated here of histologi-
cal H&E images.
Table 9: Performance rates (AUC) obtained with the model
and differences (Dif.) in each set of H&E images.
Dataset GoogLeNet LIME Grad-CAM
CR 0.785 0.786 0.611
DO 0.569 0.798 0.645
LA 0.612 0.875 0.684
LG 0.636 0.896 0.620
NHL 0.565 0.807 0.610
UCSB 0.536 0.736 0.651
4 CONCLUSION
In this paper. an approach involving the combined
use of XAI strategies to classify histological images
stained with Haematoxylin-Eosin (H&E) was pro-
posed. The neural activation patterns were obtained
from the GoogLeNet network. The feature vectors
were obtained by applying multiscale and multidi-
mensional fractal techniques. Each set of features was
evaluated using the ReliefF algorithm. The results
consisted of vectors with a reduced number of fea-
tures, which were tested by different classifiers. The
best combinations were then identified using LIME.
The results using the proposed method provided gains
in classification rates, ranging from 0.08% to 40.24%.
The best combination occurred when using 30 fea-
tures with the Logistic classifier, which provided an
AUC rate of 0.896 and an accuracy of 82.26% for
the LG dataset. In this same dataset, the GoogLeNet
network achieved a much lower performance than the
proposal, with an AUC of 0.636 and an accuracy of
62.26%. It is important to note that the best solu-
tion exploited a relevant set of features, using only
25.86% of the total descriptors available. This is an-
other important contribution of the proposed method,
as shown in the comparison with the metrics provided
by the GoogLeNet network. We believe that the main
contribution lies in providing detailed information on
combinations of techniques and feature subsets, as
well as an acceptable solution with few features.
For future work, we propose to study other CNN
models, include more feature selection algorithms
and explore other methodologies for extracting fea-
tures from the regions generated via Grad-CAM and
LIME. It would also be interesting to explore solu-
tions using a classifier ensemble.
ACKNOWLEDGEMENTS
We thank the financial support of Coordenac¸
˜
ao
de Aperfeic¸oamento de Pessoal de N
´
ıvel Su-
perior - Brasil (CAPES), the National Coun-
cil for Scientific and Technological Development
CNPq (Grants #132940/2019-1, #313643/2021-0 and
#311404/2021-9); the State of Minas Gerais Research
Foundation - FAPEMIG (Grant #APQ-00578-18); the
State of S
˜
ao Paulo Research Foundation - FAPESP
(Grant #2022/03020-1) and the project NextGenAI
- Center for Responsible AI (2022-C05i0102-02),
supported by IAPMEI, and also by FCT plurianual
funding for 2020-2023 of LIACC (UIDB/00027/2020
UIDP/00027/2020.
REFERENCES
Adadi, A. and Berrada, M. (2018). Peeking inside the black-
box: A survey on explainable artificial intelligence
(xai). IEEE Access, PP:1–1.
Baish, J. W. and Jain, R. K. (2000). Fractals and cancer.
Cancer research, 60(14):3683–3688.
Candelero, D., Roberto, G. F., do Nascimento, M. Z.,
Rozendo, G. B., and Neves, L. A. (2020). Selec-
tion of cnn, haralick and fractal features based on evo-
lutionary algorithms for classification of histological
images. In 2020 IEEE International Conference on
Bioinformatics and Biomedicine (BIBM), pages 2709–
2716. IEEE.
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
446

Chiu, D. (2012). Book review: ”pattern classification”, r.
o. duda, p. e. hart and d. g. stork, second edition. In-
ternational Journal of Computational Intelligence and
Applications, 01.
Cruz-Roa, A., Caicedo, J. C., and Gonz
´
alez, F. A. (2011).
Visual pattern mining in histology image collec-
tions using bag of features. Artificial intelligence in
medicine, 52(2):91–106.
do Nascimento, M. Z., Martins, A. S., Azevedo Tosta, T. A.,
and Neves, L. A. (2018). Lymphoma images analysis
using morphological and non-morphological descrip-
tors for classification. Computer Methods and Pro-
grams in Biomedicine, 163:65–77.
Doshi-Velez, F. and Kim, B. (2017). Towards a rigorous
science of interpretable machine learning.
Drelie Gelasca, E., Byun, J., Obara, B., and Manjunath, B.
(2008). Evaluation and benchmark for biological im-
age segmentation. In 2008 15th IEEE International
Conference on Image Processing, pages 1816–1819.
Frank, E., Hall, M. A., and Witten, I. H. (2016). The weka
workbench. online appendix for ”data mining: Practi-
cal machine learning tools and techniques”. Morgan
Kaufmann, Fourth Edition.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resid-
ual learning for image recognition. In 2016 IEEE Con-
ference on Computer Vision and Pattern Recognition
(CVPR), pages 770–778.
Hsu, H., Hsieh, C. W., and Lu, M. (2011). Hybrid feature
selection by combining filters and wrappers. Expert
Systems with Applications, 38(7):8144–8150.
Iam Palatnik de Sousa, Marley Maria Bernardes Re-
buzzi Vellasco, E. C. d. S. (2019). Local inter-
pretable model-agnostic explanations for classifica-
tion of lymph node metastases. Sensors (Basel,
Switzerland), 19, no. 13.
Ivanovici, M., Richard, N., and Decean, H. (2009a). Fractal
dimension and lacunarity of psoriatic lesions-a colour
approach. medicine, 6(4):7.
Ivanovici, M., Richard, N., and Decean, H. (2009b). Frac-
tal dimension and lacunarity of psoriatic lesions—a
colour approach—. Proceedings of the 2nd WSEAS
International Conference on Biomedical Electronics
and Biomedical Informatics, BEBI ’09, 6.
LeCun, Y., Bengio, Y., and Hinton, G. (2015). Deep learn-
ing. Nature, 521:436–44.
M. Sahini, M. S. (2014). Applications of percolation theory.
CRC Press.
Mahendran, A. and Vedaldi, A. (2016). Visualizing deep
convolutional neural networks using natural preim-
ages. International Journal of Computer Vision.
Mengdi, L., Liancheng, X., Jing, Y., and Jie, H. (2018).
A feature gene selection method based on relieff
and pso. In 2018 10th International Conference on
Measuring Technology and Mechatronics Automation
(ICMTMA), pages 298–301. IEEE.
o. A. AGEMAP, N. I. (2020). The atlas of gene expres-
sion in mouse aging project (agemap). Acesso em:
04/05/2020.
Rajaraman, S., Candemir, S., Kim, I., Thoma, G., and An-
tani, S. (2018). Visualization and interpretation of
convolutional neural network predictions in detecting
pneumonia in pediatric chest radiographs. Applied
Sciences, 8:1715.
Reyes, M., Meier, R., Pereira, S., Silva, C., Dahlweid,
M., Tengg-Kobligk, H., Summers, R., and Wiest, R.
(2020). On the interpretability of artificial intelligence
in radiology: Challenges and opportunities. Radiol-
ogy: Artificial Intelligence, 2:e190043.
Ribeiro, M. G., Neves, L. A., Nascimento, M. Z. d.,
Roberto, G. F., Martins, A. M., and Tosta, T. A. A.
(2019). Classification of colorectal cancer based on
the association of multidimensional and multireso-
lution features. Expert Systems With Applications,
120:262––278.
Robnik-Sikonja, M. and Kononenko, I. (2003). Theoretical
and empirical analysis of relieff and rrelieff. Machine
Learning, 53:23–69.
Sahini, M. and Sahimi, M. (2014). Applications of percola-
tion theory. CRC Press.
Silva, A. B., Martins, A. S., Tosta, T. A. A., Neves, L. A.,
Servato, J. P. S., de Ara
´
ujo, M. S., de Faria, P. R., and
do Nascimento, M. Z. (2022). Computational analysis
of histological images from hematoxylin and eosin-
stained oral epithelial dysplasia tissue sections. Expert
Systems with Applications, 193:116456.
Sirinukunwattana, K., Pluim, J. P., Chen, H., Qi, X., Heng,
P.-A., Guo, Y. B., Wang, L. Y., Matuszewski, B. J.,
Bruni, E., Sanchez, U., et al. (2017). Gland segmen-
tation in colon histology images: The glas challenge
contest. Medical image analysis, 35:489–502.
Suman, R., Mall, R., Sukumaran, S., and Satpathy, M.
(2010). Extracting state models for blackbox software
components. Journal of Object Technology - JOT,
9:79–103.
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S.,
Anguelov, D., Erhan, D., Vanhoucke, V., and Rabi-
novich, A. (2015). Going deeper with convolutions.
In Proceedings of the IEEE conference on computer
vision and pattern recognition, pages 1–9.
Yosinski, J., Clune, J., Nguyen, A., Fuchs, T., and Lipson,
H. (2015). Understanding neural networks through
deep visualization.
Zerdoumi, S., Sabri, A. Q. M., Kamsin, A., Hashem, I.
A. T., Gani, A., Hakak, S., Al-Garadi, M. A., and
Chang, V. (2018). Image pattern recognition in big
data: taxonomy and open challenges: survey. Multi-
media Tools and Applications, 77(8):10091–10121.
Association of Grad-CAM, LIME and Multidimensional Fractal Techniques for the Classification of H&E Images
447
