
Detecting Overgrown Plant Species Occluding Other Species in
Complex Vegetation in Agricultural Fields Based on Temporal
Changes in RGB Images and Deep Learning
Haruka Ide
1
, Hiroyuki Ogata
2
, Takuya Otani
3
, Atsuo Takanishi
1
and Jun Ohya
1
1
Department of Modern Mechanical Engineering, Waseda University, 3-4-1, Ookubo, Shinjuku, Tokyo, Japan
2
Faculty of Science and Technology, Seikei University, 3-3-1, Kichijoji-kitamachi, Musashino-shi, Tokyo, Japan
3
Waseda Research Institute for Science and Engineering, Waseda University, 3-4-1, Ookubo, Shinjuku, Tokyo, Japan
Keywords: Synecoculture, Agriculture, Deep Learning, Robot Vision.
Abstract: Synecoculture cultivates useful plants while expanding biodiversity in farmland, but the complexity of its
management requires the establishment of new automated systems for management. In particular, pruning
overgrown dominant species that lead to reduced diversity is an important task. This paper proposes a method
for detecting overgrown plant species occluding other species from the camera fixed in a Synecoculture farm.
The camera acquires time series images once a week soon after seeding. Then, a deep learning based semantic
segmentation is applied to each of the weekly images. The plant species map, which consist of multiple layers
corresponding to the segmented species, is created by storing the number of the existence of that plant species
over weeks at each pixel in that layer. Finally, we combine the semantic segmentation results with the earlier
plant species map so that occluding overgrown species and occluded species are detected. As a result of
conducting experiments using six sets of time series images acquired over six weeks, (1) UNet-Resnet101 is
most accurate for semantic segmentation, (2) Using both segmentation and plant species map achieves
significantly higher segmentation accuracies than without plant species map, (3) Overgrown, occluding
species and occluded species are successfully detected.
1 INTRODUCTION
In the field of agriculture, with modernization, a
cropping method characterized by the cultivation of
specific plants as monocultures and the use of
chemical fertilizers and pesticides has been adopted
to enhance food productivity (Tudi et al., 2021).
However, such conventional farming practices render
plants vulnerable to pests, diseases, and weeds.
Furthermore, the continuous and increasing use of
chemicals not only disrupts the soil ecosystem but
also reduces the biodiversity of agricultural land and
its surrounding environment (Conway and Barbie,
1988; Geiger et al., 2010; Savci, 2012). Particularly,
reducing biodiversity is severe in conventional
farming, prompting the need for achieving more
sustainable farming methods (Norris, 2008).
In response to these issues, a new method called
“Synecoculture” has been proposed. Synecoculture is
an agricultural method in which various plant species
are grown in mixed, densely planted environments in
a single farm to promote self-organization of the
ecosystem, thereby increasing the biodiversity of the
farm and producing useful crops by enhancing
ecosystem functions. On farms where Synecoculture
is practiced, the rich biodiversity results in intensive
competition for survival among plant species. It is
believed that plants' inherent self-organization ability
allows the plants to grow without using pesticides or
chemical fertilizers. Therefore, Synecoculture is
considered to be a more sustainable agricultural
practice than conventional agriculture because
Synecoculture can increase the productive capacity of
multiple crops as the diversity of plant populations
expands.
Synecoculture is also expected to convert deserts
and wild areas into green spaces in the future. In such
cases, Synecoculture plantations could have very
large areas, and automation of their management is
essential. However, the vegetation in Synecoculture
plantations is so complex as mentioned earlier that it
is difficult to automate the management using
conventional agricultural machineries.
266
Ide, H., Ogata, H., Otani, T., Takanishi, A. and Ohya, J.
Detecting Overgrown Plant Species Occluding Other Species in Complex Vegetation in Agricultural Fields Based on Temporal Changes in RGB Images and Deep Learning.
DOI: 10.5220/0012352500003654
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 13th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2024), pages 266-273
ISBN: 978-989-758-684-2; ISSN: 2184-4313
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

The third and fourth coauthors of this paper focus
on the development of an automation robot designed
for the management of Synecoculture (Tanaka et al.,
2022). The 1st, 2nd and 5th coauthors are specifically
investigating the visual capabilities of this robot. Our
objective is to automate crucial tasks such as pruning
overgrown dominant plants, seeding, and harvesting
crops. Especially in Synecoculture farms, it is
important to increase plant diversity and the soil
should not be exposed. Therefore, it is not always
right to prune all the dominant species that are
commonly referred to as weeds. On the other hand, if
these dominant species are not pruned at all, only
these species will thrive, and they will overgrow and
inhibit the growth of other plants.
In order to prune dominant species that are
overgrown, occluding other species, and reduce
diversity, we have been working on a method using
image processing technologies that segment
dominant plants in the image acquired by the camera
observing Synecoculture farms and estimate pruning
points in a densely overgrown plantation. However,
due to its occlusive environments, it is difficult to
accurately segment dominant species and estimate
pruning points in agricultural areas with densely
mixed vegetation (Ide et al., 2022). It tunred out to us
that it is difficult to identify vegetation from an image
after plants have thrived.
The method proposed by this paper utlizes the fact
that there is little occlusion between plants during
short duration after seeding and the segmentation
accuracy for each plant is high. Our proposed method
acquires time series images using a fixed camera
according to some constant interval starting from a
time instance soon after the seeding. Each image of
the time series images are segmented, and plant
species map count the number of appearance of each
plant species at each pixel in the segmented images
over the time. By using the plant species map and
segmented time series images, even if useful species
are covered with other overgrown species,
information such as occluding areas, which should be
pruned, can be obtained.
This paper demonstrates that the utilization of
time-series data enables a more accurate
segmentation of the vegetation in Synecoculture
environments. Consequently, we have developed the
capability to identify plants that cause occlusion,
which is helpful to maintain the Synecoculture farm
automatically.
2 RELATED WORKS
Among agricultural robots already in practical use,
Raja et al. detected and pruned weeds that inhibited
the growth of useful species without pruning them
(Raja et al., 2020). They marked tomato and lettuce
stems, removing 83% of weeds without pruning.
However, due to the intricate and densely mixed
vegetation in this study, controlling specific plant
species through marking is deemed impossible.
In the field of machine learning, many studies
have been conducted to estimate plant species. In
recent years, deep learning has made it possible to
recognize a wide variety of plant species. Mortensen
et al. reported that the semantic segmentation model
achieved 79% accuracy, showing the effectiveness of
data augmentation and fine-tuning in plant species
estimation (Mortensen et al., 2016). Picon et al. used
Dual-PSPNet to classify diverse plant species,
achieving higher accuracy than the original PSPNet
model (Picon et al., 2022).
However, in all studies, recognition accuracy was
lowered for plants under the presence of occlusion,
which indicates that the presence of occlusion is a
major issue in classifying plant species using images.
To tackle the occlusion problem, Yu et al. improved
recognition accuracy over existing learning models
by developing original learning models suitable for
the presence of occlusion (Yu et al., 2022). However,
in Synecoculture farms, the sizes of plants vary due
to individual plant growth, which results in more
complex occlusions.
Therefore, it is necessary to enable highly
accurate plant species recognition in Synecoculture
environments, which could yield severe occlusions.
More specifically, when pruning, it is important to be
able to identify not only the plant species that are
visible from the camera, but also the plants that are
covered underneath.
3 PROPOSED METHOD
In Synecoculture farms, despite occlusions, it is
crucial to grasp the vegetation and identify occluded
useful species and occluding dominant species.
Our method leverages the initial stages of
Synecoculture farms, where occlusion is infrequent
right after seeding and germination. In these early
phases, small and isolated individual plants allow
accurate segmentation with existing models. As
shown in Figure 1, Our proposed method consists of
acquiring time series images, semantic segmentation,
Detecting Overgrown Plant Species Occluding Other Species in Complex Vegetation in Agricultural Fields Based on Temporal Changes in
RGB Images and Deep Learning
267

generating the plant species map and detecting
overgrown plant species occluding other species.
Figure 1: Over-view of our proposed method.
3.1 Semantic Segmentation
3.1.1 Model Architecture
UNet and Deeplab V3+ have been used as the main
learning models in many studies using semantic
sesgmentation with deep learning on plants (Li et al.,
2022; Wang et al., 2021; Zou et al., 2021; Kolhar and
Jagtap, 2023). In these studies, UNet and Deeplab
V3+ tend to perform better than other basic learning
models such as PSPNet and SegNet. Therefore, as a
result of comparing UNet and Deeplab V3+ in
Section 4.1.2, this paper uses Resnet101-UNet for the
subsequent processes such as for creating plant
species map.
Resnet50 and Resnet101 are used as the backbone
of the encoder part of the system. Recently, other
studies have shown that they are effective as the
backbone of semantic segmentation models
(Bendiabdallah et al., 2021; Nasiri et al., 2022;
Sharifzadeh et al., 2020).
Many studies have shown that fine-tuning of a
pre-trained model with a large dataset using a small
number of their own images is effective; therefore, we
use a backbone that was previously trained by
ImageNet and trained it in the form of fine-tuning.
In this paper, Section 4.1 investigate learning
models suitable for plant species detection in a
Synecoculture environment by changing the
combination of these encoders and decoders.
3.1.2 Implementation of Semantic
Segmentation
In this process, we obtain the semantic segmentation
result from each time series image. Before predicting
the species at each pixel, the original 1280 x 720
pixels image was cropped into 475 square pixels
before training models and prediction to reduce the
burden of computers. To prevent a decrease in
accuracy at the edges of cropped images, we used the
following method for image cropping. First, we
applied 400 pixels of zero padding to the outside of a
1280 x 720 pixels image and cropped the image to
475 x 475 pixels from the edge. When cropping the
image to 475 square pixels from the edge, we set the
strides to 100 pixels to generate overlaps between the
crop images. Each crop image outputs a semantic
segmentation result with a pre-trained model. Since
the overlapping regions of multi crop images produce
multiple predictions for a single pixel, the most
frequently predicted class was used as the prediction
class for that pixel. By outputting semantic
segmentation results as described above, a more
accurate 1280 x 720 pixels prediction result can be
obtained.
3.2 Creating Plant Species Map
To improve the accuracy of plant species estimation
under severe occlusions, the plant species map for
each species is generated using the plant species
estimation results from time series images. Figure 2
overviews how to create the plant species map.
First, the field is divided into several (six in this
paper) areas. A fixed bird’s eye view camera observes
each area as explained in Section 4.2 and acquires
RGB images at some predefined constant interval.
Next, for each of the acquired time series images,
we estimate the plant species at each pixel using a
semantic segmentation model that has already been
trained using training images as explained in Section
3.1.2. The output pixels by pixel-wise plant species
semantic segmentation results are then layered for
each plant species, as shown in Figure 2, where each
pixel of each plant species map stores a value
indicating whether or not that plant species is present:
specifically, 1 if the corresponding plant species is
present, and 0 otherwise.
Finally, the value of each corresponding pixel in
all the plant species layers are added over all of the
time series images so that the final plant species map
is obtained. This allows the presence of a plant to be
inferred from the plant map even under difficult-to-
recognize situations such as severe occlusions. That
is, even if a particular plant species is covered by
other plant species from some time instance, the
occluded plant species map is recorded as non-zero
pixels values, which means that the plant species map
is robust against occlusions.
ICPRAM 2024 - 13th International Conference on Pattern Recognition Applications and Methods
268
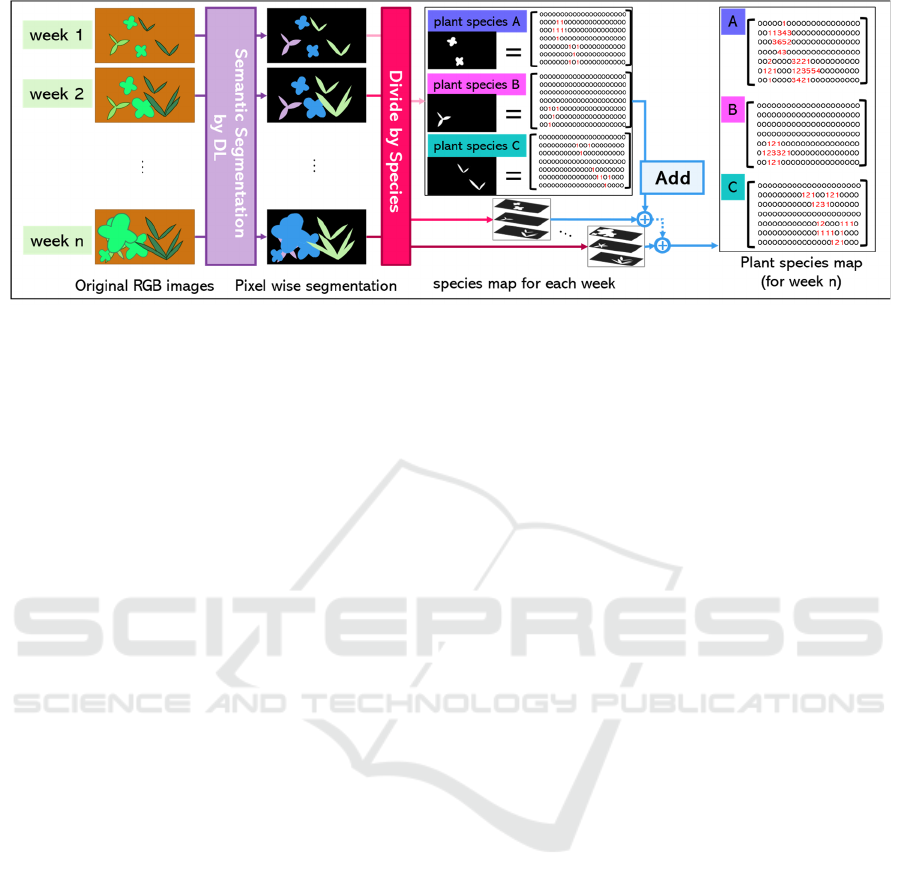
Figure 2: The method of creating plant species map.
3.3 Occlusion Area Detection
Even if a plant species is not detected in the
segmentation result of the image acquired at some
time instance, if that species is detected in the
segmentation result of an earlier image, we can know
the existence of that species from the plant species
map. Therefore, if a plant is not detected in the
segmentation result of the image at some time
instance and is confirmed that plant’s existence in the
plant species map, then that species can be judged to
be occluded by other species from an earlier time
instance.
The specific procedure for judging whether some
plant species overgrows from a time instance,
occluding other species, is as follows.
As a general case, we explain the case in which
the n-th week’s overgrowth is to be detected, where 2
≤ n ≤ 6. First, in the (n - 1)-th week’s plant species
map, whether the useful species (in this paper, green
pepper) exists at each pixel is checked. Next, in the
n-th week’s segmentation result, whether the useful
species exists is checked at the pixels whose image
coordinates are same as those of the (n - 1)-th week’s
plant species map’s pixels at which the useful species
exists. Finally, if all of the following three conditions
(1) to (3) are satisfied at a pixel, our proposed method
judges that a dominant species (apple mint or green
foxtail) overgrows at the pixel at the n-th week,
occluding the useful species.
(1) At a pixel in the (n - 1)-th week’s plant species
map, the useful species exists.
(2) At the n-th week’s segmentation result’s pixel
that corresponds to the pixel described in (1), the
useful species does not exit.
(3) At the n-th week’s segmentation result’s pixel
described in (2), a dominant species exists.
The above-mentioned processes are repeated for 2≤ n
≤ 6.
As our future work, the obtained positional
information on the overgrown, occluding non-useful
species could be utilized for determining the positions
to be pruned by our robotic system.
4 EXPERIMENTS AND RESULTS
4.1 Semantic Segmentation Model
As explained in Section 3.1 we compared the
common semantic segmentation models to chose
appropriate deep learning model for our data.
4.1.1 Training of Models
The image data used for training the models were
acquired once a week between May 2022 and June
2023 in farms in Akiruno, Tokyo, and Oiso,
Kanagawa Prefecture, where Synecoculture is
actually implemented. The data were acquired using
an Intel RealSense D435 RGBD camera, which
captured RGB images from a height of approximately
1.5 meters above the ground, looking vertically
downward at the farms.
A total of 137 1280 x 720 pixels images were
acquired, 127 were used to train the models and the
remaining 10 were used to test the models. The
acquired images were annotated so that "green
foxtail," "apple mint," "green pepper," "other plants,"
and "unvegetated area" were correctly labelled as
pixel wise segmentation. The train images were
cropped to 475 x 475 pixels before training.
Train images were further divided into train and
validation at a ratio of 7:3 for the training. In addition,
to compensate for the lack of datasets, data
Detecting Overgrown Plant Species Occluding Other Species in Complex Vegetation in Agricultural Fields Based on Temporal Changes in
RGB Images and Deep Learning
269

augmentation was applied to train images only. First,
we applied the gamma correction, which is
considered effective for plant detection (Saikawa et
al., 2019). In addition, we applied the Random
Shadow, Random Sun Flare, and Random Contrast
functions. Random Scale, Random Rotation, Color
Jitter, and Random Horizontal Flip, which are
commonly used, were also applied.
The GPU used for training was a 24GB NVIDIA
GeForce RTX 3090 with a learning rate of 0.001,
Adam as the optimization function, and a batch size
of 16. During the training process, mean IoU which is
explained in Section 4.1.2 is calculated, and the one
with the highest mean IoU value of validation during
each 200 epochs was used.
4.1.2 Model Performance Assessment
10 images of Synecoculture farms were used to test
the models, as described in section 4.1.1. To obtain
semantic segmentation results for each model, the
process described in section 3.1.2 was performed.
After the segmentation results for Deeplab V3+ or
UNet combined with Resnet50 or Resnet101 as
backbone were obtained, their accuracy was
calculated. Table 1 compares the accuracy of the
segmentation models on test images.
According to Table 1, overall UNet performs
better than DeeplabV3+. In particular, all the indices
of UNet with Resnet101 as the backbone are the
highest, while UNet with Renet50 almost the second
highest.
Table 1: Comparison of segmentation models.
Model
Recall
%
Precision
%
F1
%
mIoU
%
Resnet50-
UNet
72.55 78.32 74.82 61.01
Resnet101-
UNet
77.63 79.19 78.02 64.58
Resnet50-
DeeplabV3+
73.38 76.24 74.48 60.40
Resnet50-
DeeplabV3+
73.49 76.09 74.18 59.93
4.2 Acquisition of Time Series Data
As explained in Section 3.2, time series images for
the plant species map were acquired.
For acquiring time series images, 1280 x 720
pixels RGB images were acquired once a week over
the six weeks from May 2023 using an Intel
RealSense D435 RGBD camera pointing vertically
downward from a bird's eye view fixture installed at
the top of the 110mm-high frame. The camera was
installed in the same position as that of the
agricultural robot under development. The farm is
divided into six areas, at each of which the camera
was placed, so that (= 6 times 6) time series images
of the Synecoculture environment were acquired for
the six areas over a total of six weeks. In addition, we
pruned the overgrown apple mint and green foxtail in
7-th week and acquired post-pruning images in the
same way as the method explained earlier. We also
applied black masks to equipment in the images that
had no relevance to the plants.
4.3 Segmentation Results in Time
Series Images
We then segmented the six images for each week
using the Resnet101-UNet trained model, applying
the method described in Section 3.1.2. Figure 3 shows
examples of the segmentation results obtained by the
Resnet101-UNet trained model in the four weeks out
of the six weeks. As shown in Figure 3, the green
pepper, which was completely visible in early weeks,
is covered by apple mint and green foxtail, which tend
to dominate after week 5, and only a part of the leaves
of the green pepper is visible due to the occlusion
caused by the overgrown apple mint and green
foxtail. To solve this problem, as described in Section
3.2, the plant species map is proposed by this paper.
Figure 3: Segmentation results for each week.
4.4 Creating Plant Species Map
Figure 4 compares segmentation results for each
single image with the plant species map for all over
the six weeks, where the color in the right three
columns indicates as follows: dark blue and bright
orange if the count at a pixel is small and large,
ICPRAM 2024 - 13th International Conference on Pattern Recognition Applications and Methods
270
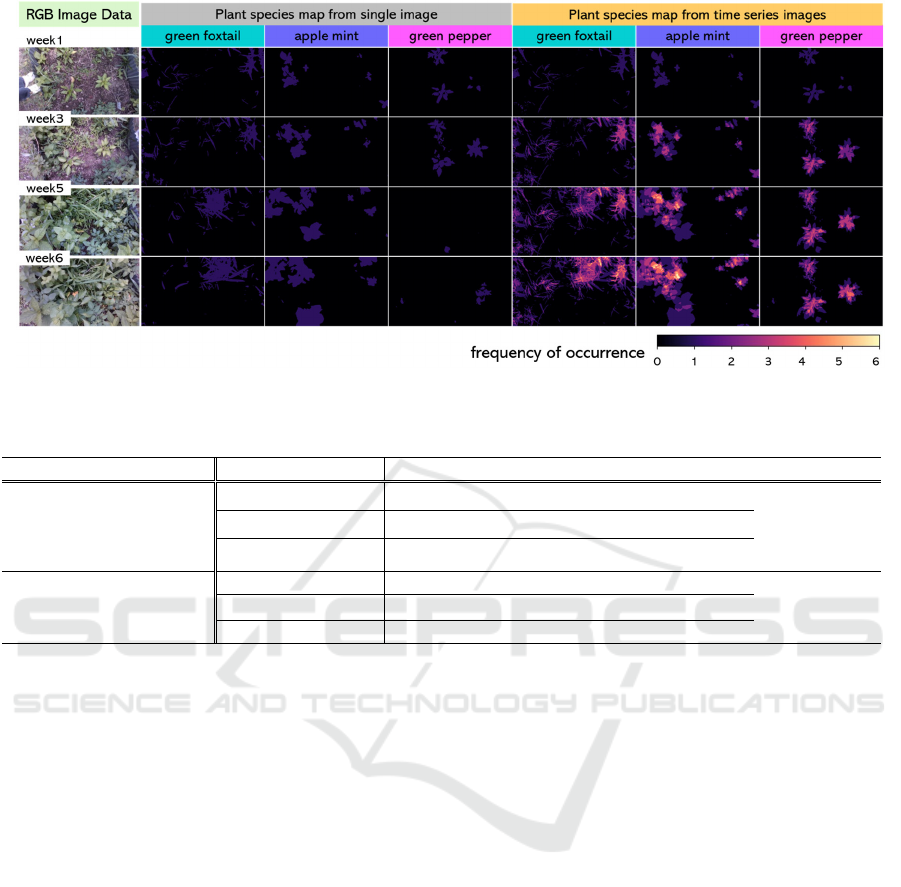
Figure 4: Visualization of plant species map.
Table 2: Comparison of segmentation accuracies with/without plant species map.
“with” / “without” Class (plant species) Precision % Recall % F1 % mean F1 %
“without” (Only using
week 6’s segmentation
result, without using plant
species map)
Green foxtail 83.49 46.18 59.47
46.68
Apple mint 97.51 56.17 71.28
Green pepper 88.43 4.90 9.28
“with” (plant species map
from our proposed method)
Green foxtail 72.24 88.71 79.63
80.32
Apple mint 95.05 93.25 94.14
Green pepper 83.09 56.41 67.20
respectively, while 0 or 1 in case of the segmentation
for single images in the middle three columns. As
shown in Figure 4, when a same plant species keeps
present and visible at the same place over weeks, the
value of the corresponding pixels in that plant species
map is increased, displaying brighter. In addition, we
can confirm that even after a plant species is occluded
by other species, we can locate the occluded species
in the map.
Next, we tested the validity of the plant species
map. Ground truth plant species map which indicates
the existence of each plant species for each pixel are
created from annotation data (ground truth of
semantic segmentation) for each time series image.
Table 2 shows the segmentation accuracies of the 6-
th week’s plant species map (“with”) and semantic
segmentation results which is predicted from only one
image of 6-th week without the plant species map
(“without”). According to Table 2, the plant species
map provides more accurate segmentations than
segmentation without using plant species map. In
particular, the Recall for all three classes (plant
species) are significantly lower in the case of the
"without" results, which using the 6-th week’s image
only, without using plant species map, indicating that
many plants are missed. The plant species map, being
additive, yields a lower Precision due to false
positives. However, the higher F1 score, representing
overall accuracy, suggests its effectiveness in
identifying complex vegetation.
4.5 Overgrown Plants Detection
Figure 5 shows examples of the result of using the
plant species map to detect areas occluded by
dominant species. In Figure 5, apple mint and green
foxtail occlude green pepper. From a single-week
segmentation result, locating occluded plant species
under overgrown conditions is challenging. However,
by performing the algorithm, explained in Section 3.3,
for the plant species map and segmentation result,
even plants that are completely occluded can be
detected.
Thus, we have confirmed that the plant species
map provides useful information for pruning. Our
future step is to develop a specific pruning algorithm
using the information obtained from the plant species
map.
Detecting Overgrown Plant Species Occluding Other Species in Complex Vegetation in Agricultural Fields Based on Temporal Changes in
RGB Images and Deep Learning
271

Figure 5: Results of overgrown plants detection.
5 DISSCUSION
To confirm the effectiveness of the proposed method
for detecting overgrown, occluding plant species
(Section 3.3), in 7-th week, we manually pruned the
dominant plants occluding the useful species (green
pepper) so that the useful species is visible. Figure 6
compares the location of the green peppers obtained
by Section 3.3’s method, which uses the plant species
map in the 6-th week, with the green peppers in the
segmentation result of the RGB image after pruning
apple mint and green foxtail in 7-th week as
mentioned earlier. Figure 6 shows that the position of
the green pepper obtained by the plant species map is
almost same as the actual position of the green
pepper. In particular, the locations of the roots of the
green pepper have not changed since the time of
sprouting; so, it can be said that maintaining the past
positional information by time series information is
effective for understanding vegetation in an
environment with severe occlusion.
Figure 6: Checking the position of the green peppers.
6 CONCLUSION AND FUTURE
WORK
This paper has proposed a method for detecting
overgrown plant species occluding other species from
bird's eye view camera images acquired in
Synecocluture farms. Conventional methods for
identifying vegetation from a single image using
semantic segmentation based on deep learning has
difficulty in identifying complex vegetations in
Synecoculture environments. To tackle this issue, our
proposed method consists of acquiring time series
RGB images, performing semantic segmentation for
each of the time series images, creating plant species
map, and detecting overgrown species occluding
other species.
The bird’s eye view camera acquires time series
images at a constant interval (in this paper, once a
week) soon after seeding. We then apply semantic
segmentation to each of the weekly images using a
deep learning model so that areas of the plant species
(in this paper, “apple mint”, “green foxtail” and
“green pepper”) are segmented. The plant species
map consists of multiple layers corresponding to the
segmented species, and at each pixel in each layer, the
number of the existence of that plant species over the
weeks is stored. Finally, we combine the semantic
segmentation results with the (one week) earlier plant
species map so that occluding overgrown species and
occluded species are detected.
Results of experiments using the 36 (= 6 images
(acquired at 6 places) times 6 weeks) time series
images are summarized as follows.
It turns out that UNet using Resnet101 as the
backbone achieves higher segmentation
accuracies than Resnet50 as the backbone or
Deeplab V3+. UNet-Resnet101 is decided to be
used for the semantic segmentation for each of
time series images.
As a result of comparing results of semantic
segmentation “with” and “without” the plant
species map, “with” achieves much higher
accuracies than “without”.
Using the segmentation result and plant species
map, areas of occluding, overgrown species and
occluded species are successfully detected.
In this paper, only green pepper was targeted as a
useful species and apple mint and green foxtail as
dominant species. However, since more plant species
are mixed and densely populated in Synecoculture
environments, our future work includes developing a
method that can deal with a larger number of densely
mixed plant species. In addition, based on this, a
method for determining the positions to be pruned by
our robotic system needs to be achieved.
ACKNOWLEDGEMENTS
This study was conducted with the support of the
Research Institute for Science and Engineering,
Waseda University; Future Robotics Organization,
Waseda University. This work was also supported by
Sustainergy Company (based in Japan), Sony
Computer Science Laboratories, Inc and New Energy
and Industrial Technology Development
ICPRAM 2024 - 13th International Conference on Pattern Recognition Applications and Methods
272

Organization (NEDO). We thank all these
organizations for the financial and technical support
provided. Synecoculture™ is a trademark of Sony
Group Corporation.
REFERENCES
Bendiabdallah, M, H., and Settouti, N. (2021). A
comparison of UNet backbone architectures for the
automatic white blood cells segmentation. WAS
Science Nature (WASSN) ISSN: 2766-7715, 4(1).
Chen, L. C., Zhu, Y., Papandreou, G., Schroff, F., and
Adam, H. (2018). Encoder-decoder with atrous
separable convolution for semantic image
segmentation. Proceedings of the European conference
on computer vision (ECCV):801-818.
Conway, G. R., and Barbie, E. B. (1988). After the green
revolution: sustainable and equitable agricultural
development. Futures, 20(6):651-670.
Geiger, F., Bengtsson, J., Berendse, F., Weisser, W. W.,
Emmerson, M., Morales, M. B., Ceryngier, P., Liira, J.,
Tscharntke, Teja., Winqvist, Camilla., Eggers, S.,
Bommarco, R., Pärt, T., Bretagnolle, V., Plantegenest,
M., Clement, L., Dennis, C., Palmer, C., Oñate, J.,
Guerrero, I., and Inchausti, Pablo. (2010). Persistent
negative effects of pesticides on biodiversity and
biological control potential on European farmland.
Basic and Applied Ecology, 11(2):97-105.
Ide H., Aotake S., Ogata H., Ohtani T., Takanishi A.,
Funabashi M. (2022). A method for detecting dominant
plants in fields from RGB images using Deep Learning.
The Institute of Image Electronics Engineers of Japan:
186-192 (in Japanese).
Kolhar, S., and Jagtap, J. (2023). Phenomics for Komatsuna
plant growth tracking using deep learning approach.
Expert Systems with Applications, 215, 119368.
Li, G., Cui, J., Han, W., Zhang, H., Huang, S., Chen, H.,
and Ao, J. (2022). Crop type mapping using Time series
Sentinel-2 imagery and UNet in early growth periods in
the Hetao irrigation district in China. Computers and
Electronics in Agriculture, 203, 107478.
Mortensen, A. K., Dyrmann, M., Karstoft, H., Jørgensen,
R. N., and Gislum, R. (2016). Semantic segmentation
of mixed crops using deep convolutional neural
network. CIGR-AgEng conference:26-29.
Nasiri, A., Omid, M., Taheri-Garavand, A., and Jafari, A.
(2022). Deep learning-based precision agriculture
through weed recognition in sugar beet fields.
Sustainable Computing. Informatics and Systems, 35,
100759.
Norris, K. (2008). Agriculture and biodiversity
conservation: opportunity knocks. Conservation letters,
1(1):2-11.
Picon, A., San-Emeterio, M. G., Bereciartua-Perez, A.,
Klukas, C., Eggers, T., and Navarra-Mestre, R. (2022).
Deep learning-based segmentation of multiple species
of weeds and corn crop using synthetic and real image
datasets. Computers and Electronics in Agriculture,
194, 106719.
Raja, R., Nguyen, T. T., Slaughter, D. C., and Fennimore,
S. A. (2020). Real-time robotic weed knife control
system for tomato and lettuce based on geometric
appearance of plant labels. Biosystems Engineering,
194:152-164.
Saikawa, T., Cap, Q. H., Kagiwada, S., Uga, H., and
Iyatomi, H. (2019, December). Aop: An anti-overfitting
pretreatment for practical image-based plant diagnosis.
2019 IEEE International Conference on Big Data (Big
Data):5177-5182.
Savci, S. (2012). Investigation of effect of chemical
fertilizers on environment. Apcbee Procedia, 1:287-
292.
Sharifzadeh, S., Tata, J., Sharifzadeh, H., and Tan, B.
(2020). Farm area segmentation in satellite images
using DeeplabV3+ neural networks. Data Management
Technologies and Applications. 8th International
Conference, DATA 2019, Prague, Czech Republic,
July 26–28, 2019, Revised Selected Papers 8:115-135.
Tanaka T., Masaya K., Terae K., Mizukami H., Murakami
M., Yoshida S., Aotake S., Funabashi M., Otani T., and
Takanishi A. (2022). Development of the Agricultural
Robot in SynecocultureTM Environment (1st Report,
Development of Moving Mechanism on the Farm and
Realization of Weeding and Harvesting), Journal of the
Robotics Society of Japan, 40(9):845-848 (in
Japanese).
Tudi, M., Daniel Ruan, H., Wang, L., Lyu, J., Sadler, R.,
Connell, D., Chu, C., and Phung, D. T. (2021).
Agriculture development, pesticide application and its
impact on the environment. International journal of
environmental research and public health, 18(3), 1112.
Wang, C., Du, P., Wu, H., Li, J., Zhao, C., and Zhu, H.
(2021). A cucumber leaf disease severity classification
method based on the fusion of DeeplabV3+ and UNet.
Computers and Electronics in Agriculture, 189,
106373.
Yu, H., Che, M., Yu, H., and Zhang, J. (2022).
Development of Weed Detection Method in Soybean
Fields Utilizing Improved DeeplabV3+ Platform.
Agronomy, 12(11), 2889.
Zou, K., Chen, X., Wang, Y., Zhang, C., and Zhang, F.
(2021). A modified UNet with a specific data
argumentation method for semantic segmentation of
weed images in the field. Computers and Electronics in
Agriculture, 187, 106242.
Detecting Overgrown Plant Species Occluding Other Species in Complex Vegetation in Agricultural Fields Based on Temporal Changes in
RGB Images and Deep Learning
273
