
Fundamental Limitations of Inverse Projections and Decision Maps
Yu Wang
a
and Alexandru Telea
b
Department of Information and Computing Science, Utrecht University, 3584 CS Utrecht, The Netherlands
Keywords:
Dimensionality Reduction, Inverse Projections, Decision Maps, Intrinsic Dimensionality.
Abstract:
Inverse projection techniques and decision maps are recent tools proposed to depict the behavior of a classifier
using 2D visualizations. However, which parts of the large, high-dimensional, space such techniques actually
visualize, is still unknown. A recent result hinted at the fact that such techniques only depict a two-dimensional
manifold from the entire data space. In this paper, we investigate the behavior of inverse projections and
decision maps in high dimensions with the help of intrinsic dimensionality estimation methods. We find that the
inverse projections are always surface-like no matter what decision map method is used and no matter how high
the data dimensionality is, i.e., the intrinsic dimensionality of inverse projections is always two. Thus, decision
boundaries displayed by decision maps are the intersection of the backprojected surface and the actual decision
surfaces. Our finding reveals a fundamental problem of all existing decision map techniques in constructing an
effective visualization of the decision space. Based on our findings, we propose solutions for future work in
decision maps to address this problem.
1 INTRODUCTION
Visualization of high-dimensional data is one of the
key challenges of information visualization (Munzner,
2014). For this goal, dimensionality reduction (DR)
methods, also called projections, are techniques of
choice. DR methods aim to map high-dimensional
data samples into a low-dimensional space while pre-
serving the so-called data structure. DR methods scale
very well both in the number of samples and num-
ber of dimensions and, as such, have gained a strong
foothold in the arena of visualization techniques for
high-dimensional data.
Inverse projection methods aim to construct the
opposite mapping to the one given by a DR tech-
nique – that is, to map points from a low-dimensional
to a high-dimensional space. Recently, inverse pro-
jections have been used to construct so-called deci-
sion maps (Rodrigues et al., 2019; Schulz et al., 2020;
Oliveira et al., 2022). These are two-dimensional im-
ages which aim to capture, in a dense way, the behavior
of a machine learning (ML) model after training.
While both projections and decision maps help
users understand the behavior of high-dimensional
data, they differ in a crucial respect: Decision maps
aim to depict the behavior of a ML model over the
a
https://orcid.org/0000-0001-6066-0279
b
https://orcid.org/0000-0002-1129-4628
entire (or at least a large part of the) high-dimensional
space its data comes from. If certain parts of this space
are not covered by a decision map, the users of the
map will have no idea of how the model behaves in
such areas. To trust a decision map, we thus need
to know (a) which parts of the data space it depicts;
and, if not the entire space is covered, (b) how we
can control the depicted spatial subset so as to obtain
actionable insights. In contrast, (inverse) projections
do not have this problem since they are applied to
a finite set of points to generate a corresponding set
of points. Simply put, (inverse) projections are used
to visualize sets; decision maps are used to visualize
functions (classification models).
A recent study (Wang et al., 2023) touched the
question (a) above for the particular case of a single
ML model used to classify a simple three-dimensional
dataset. Surprisingly, the results showed that decision
maps constructed for this case (by three different meth-
ods) only depict a surface embedded in the data space
(see Fig. 1). The rest of the three-dimensional data
space – that is, points not located on this surface – are
not depicted by the decision maps. We have no infor-
mation on how the studied ML model behaves for such
points, even though the model is likely to be used to
classify such samples.
The simple experiment in (Wang et al., 2023) im-
mediately raises several questions:
Wang, Y. and Telea, A.
Fundamental Limitations of Inverse Projections and Decision Maps.
DOI: 10.5220/0012352300003660
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 19th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2024) - Volume 1: GRAPP, HUCAPP
and IVAPP, pages 571-582
ISBN: 978-989-758-679-8; ISSN: 2184-4321
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
571
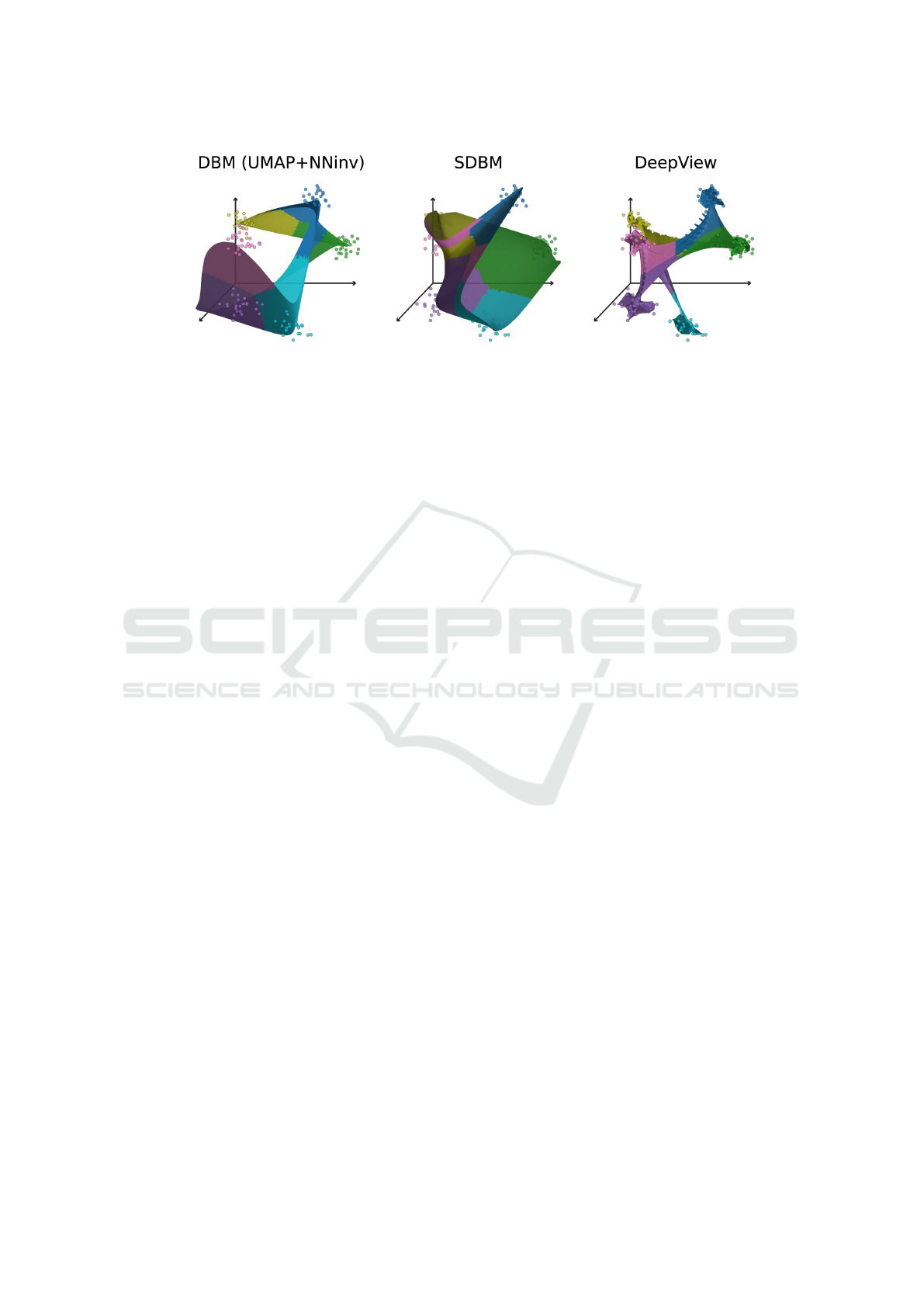
Figure 1: Surfaces created by backprojecting to the data space (3D) the decision maps constructed with three different
techniques of an already trained Logistic Regression classifier for a synthetic 6-blobs dataset, adapted from (Wang et al.,
2023). Each surface is colored to show how the classifier assigns one of the six classes to its points. 3D axes are added to the
background to make the shapes of the aforementioned surfaces easier to discern.
Q1.
Does the observation advanced in (Wang et al.,
2023) still hold for different ML models than the
one used in the above study?
Q2.
What exactly are the boundaries we see in Fig-
ure 1? How do these relate to the actual decision
boundaries in high dimensions?
Q3.
Which subsets of the input space do decision maps
cover when the dimensionality thereof is far larger
than three?
Q4.
Do different decision-map construction methods
provide different answers to the above questions?
In this paper, we report a series of experiments
that aim to answer these questions. We construct de-
cision maps for various datasets and classifiers us-
ing the three existing decision-map methods avail-
able in the literature (Rodrigues et al., 2019; Schulz
et al., 2020; Oliveira et al., 2022). Next, we pro-
pose a way to quantify their ability to densely sample
their data spaces based on intrinsic dimension estima-
tion (Bennett, 1969; Camastra, 2003; Campadelli et al.,
2015; Bac et al., 2021). We propose several novel visu-
alizations to analyze and compare the obtained results.
Our findings show that all decision map methods es-
sentially visualize only a surface embedded in high
dimensions, with the exception of areas located very
close to the training samples, no matter what the data
dimensionality is, and no matter which decision map
method is used.
The structure of this paper is as follows. Sec-
tion 2 introduces the background of this work. Sec-
tion 3 presents the visual evaluation for 3D data. Sec-
tion 4 demonstrates the qualitative evaluation for high-
dimensional data. Section 5 discusses our findings.
Finally, Section 6 concludes this paper and discusses
future work.
2 BACKGROUND
Notations. We start with the notations used next in
this work. Let x
= (x
1
,... ,x
n
)
,
x
i
∈ R
,
1 ≤ i ≤ n
, be
an
n
-dimensional data point or sample. Let
D = {
x
j
}
,
j = 1, 2,. .. ,N
, be a dataset of
N
samples, i.e., a table
with N rows (samples) and n columns (dimensions).
Classifiers. A classification model (or classifier) is a
function
f : R
n
→ C (1)
that maps a sample to a class label in a given label-set
C
. Classifiers
f
are typically constructed by aiming
to satisfy
f (
x
i
) = y
i
for a so-called training set
D
t
=
{(
x
i
,y
i
)}
with x
i
∈ D
,
y ∈ C
. A decision zone of
f
,
for class
y
, is the set of points
{
x
∈ R
n
| f (
x
) = y}
; the
boundaries between such decision zones are called the
decision (hyper)surfaces of f .
Projections. A projection, also known as dimension-
ality reduction operation, is a function
P : R
n
→ R
q
, (2)
that maps high dimensional data to a low dimensional
(
q ≪ n
) space. For visualization purposes, one most
typically uses
q = 2
. In this case, the projection of a
dataset
D
, which we denote as
P(D) = {P(
x
)|
x
∈ D}
is a 2D scatterplot. Tens of different projection tech-
niques exist with different abilities to capture data-
space similarities
R
n
by corresponding similarities in
the projection space
R
q
, computational speed, robust-
ness to noise, out-of-sample ability, and ease of use
and implementation. Extensive surveys cover all these
aspects (Espadoto et al., 2019a; Nonato and Aupetit,
2018; Huang et al., 2019; Sorzano et al., 2014; Engel
et al., 2012).
Inverse Projections. An inverse projection (or unpro-
jection (Espadoto et al., 2021a)) is a function
P
inv
: R
q
→ R
n
, (3)
IVAPP 2024 - 15th International Conference on Information Visualization Theory and Applications
572

which, roughly put, aims to reverse the effects of a
given projection
P
. In more detail, given a dataset
D ∈ R
n
and its projection scatterplot
P(D)
, an ideal
inverse projection would yield
P
inv
(P(
x
)) =
x for all
x
∈ D
, or more generally minimize, point-wise, the
difference between
D
and the so-called backprojec-
tion
D
′
= P
inv
(P(D))
of
D
. Note that
P
inv
is, in most
cases, not the mathematical inverse of
P
since projec-
tion functions
P
may not be injective, thus, they are not
invertible. This is so since
P
can be non-injective, i.e.,
it can map different data points to the same location
in
R
q
(think e.g., of PCA applied to map a point-set
that densely samples the surface of a 3D sphere to the
2D plane; there will be, for any projection plane PCA
computes, exactly two points projecting at the same
location). Also,
P
can be non-surjective. Indeed, pro-
jections that do not have the so-called out-of-sample
ability only map a given dataset
D ⊂ R
n
to
R
q
. How
data points outside
D
would map to
R
q
is not handled
by such techniques. This means that, by definition,
there will be points in
R
q
– more precisely, those not
covered by
P(D)
, which do not have a correspondent
in R
n
via P.
Inverse projections are used to extrapolate the in-
verse mapping constructed as outlined above to points
outside
P(D)
. This enables many applications such as
shape or image morphing (dos Santos Amorim et al.,
2012), data imputation (Espadoto et al., 2021a), and
constructing classifier decision maps (detailed further
below). In contrast to projections, for which many
algorithms exist, only a handful of inverse projection
techniques have been proposed. iLAMP (dos San-
tos Amorim et al., 2012) uses distance-based interpo-
lation with radial basis functions to reverse a specific
projection technique, LAMP (Joia et al., 2011), which
also uses the same interpolation. NNinv (Espadoto
et al., 2019b) constructs
P
inv
by deep learning to map
the points of any given 2D scatterplot
P(D)
, con-
structed by any projection technique
P
, to correspond-
ing points in
D
. SSNP (Espadoto et al., 2021b) uses
semi-supervised deep learning to construct both a di-
rect and inverse projection, thereby refining earlier
results based on unsupervised autoencoders for the
same goal (Hinton and Salakhutdinov, 2006).
Computing an inverse projection is a more compli-
cated – and actually ill-posed – task that computing a
direct projection, for several reasons. First, an inverse
projection has to revert the effects of a given direct
projection function
P
, which can be potentially quite
complicated. In contrast, constructing a direct projec-
tion does not need to consider any such earlier-given
mapping. Secondly, as mentioned above, direct pro-
jections often do not admit a mathematical inverse, so
all we can attempt to do is to compute an approximate
or pseudo-inverse. Thirdly, the key use-case for in-
verse projections is to infer data corresponding to 2D
locations where no sample point was projected by the
direct function
P
– and thereby generate data points
for which no ground-truth information is known (dos
Santos Amorim et al., 2012; Espadoto et al., 2021a).
The issue here is that, for such points, we have no for-
mal means of assessing when, and by how much, our
constructed
P
inv
is incorrect – thus no hard criterion
to optimize for. In this sense, inverse projections aim
to generalize the inverse mapping corresponding to a
given direct projection, much like ML algorithms aim
to extrapolate their working outside their training sets,
with the fundamental challenges that this extrapolation
implies. This is further discussed below.
Decision Maps. These techniques aim to construct
a dense visual representation of a given trained ML
model
f
. Given a 2D image
I = {
p
}
of 2D pixels p,
I
covers the range of
P(D)
, a corresponding backpro-
jection
I
inv
= {P
inv
(
p
)|
p
∈ I}
is constructed using an
inverse projection
P
inv
. Next, the pixels p are colored
to depict the labels
f (P
inv
(
p
))
inferred by the model
f
.
Same-color regions in
I
thus show the decision zones
of
f
; neighbor pixels of different colors show the deci-
sion boundaries of
f
. Decision maps can be used for a
wide range of tasks in explainable AI, such as under-
standing the shapes of the decision zones created by
a trained model (Espadoto et al., 2019b; Sohns et al.,
2023; Zhou et al., 2023), reasoning about the general-
ization ability of such models for unseen data (Schulz
et al., 2020), studying the agreements of different mod-
els (Espadoto et al., 2021a), and dynamic imputation
of data (Espadoto et al., 2021a).
Given that only a few inverse projection techniques
exist (see above), there are also only a few decision
map algorithms. DBM (Rodrigues et al., 2018) di-
rectly applies the above decision-map construction
method by using UMAP (McInnes et al., 2018) and
NNinv (Espadoto et al., 2019b) for the direct, respec-
tively inverse, projection techniques. Several exten-
sions thereof cover the use of additional direct and in-
verse projections and improve DBM’s noise-resistance
by filtering out poorly projected samples (Rodrigues
et al., 2019). SDBM (Oliveira et al., 2022) leverages
SSNP which, as already explained, provides both di-
rect and inverse projections. SDBM yields higher-
quality, smoother, decision maps than DBM but does
not allow one to freely choose
P
. Finally, Deep-
View (Schulz et al., 2020) leverages discriminative di-
mensionality reduction (Schulz et al., 2015) to enhance
UMAP (McInnes et al., 2018), which also provides an
inverse projection, to construct very high quality deci-
sion maps.
Fundamental Limitations of Inverse Projections and Decision Maps
573

Most work on decision maps only evaluate their
results qualitatively by visually comparing the results
of different such methods against each other for given
datasets and classifiers (Rodrigues et al., 2018; Ro-
drigues et al., 2019; Oliveira et al., 2022; Espadoto
et al., 2019c). Oliveira et al. (Oliveira et al., 2023) stud-
ied the stability of DBM and SDBM in presence of
small perturbations of the visualized model, conclud-
ing that these methods are quite robust to such changes.
More recently, Wang et al. (Wang et al., 2023) pro-
vided the most detailed evaluation of decision maps
we are aware of by combining classical ML perfor-
mance metrics (Schulz et al., 2020) with several novel
visual assessments. Additionally, as stated in Sec. 1,
Wang et al. (Wang et al., 2023) showed that DBM,
SDBM, and DeepView only depict a surface in the
case of visualizing a simple classifier trained on a syn-
thetic 3D dataset (Fig. 1). How this finding extends to
other classifiers and higher dimensions is however not
studied. Our paper aims to fill this gap.
To answer the questions in Sec. 1, we conduct
two studies. First, we extend the evaluation in (Wang
et al., 2023) to use additional classifiers and visualize
the actual decision boundaries, but still using a 3D
dataset (Sec. 3), thereby answering Q1-Q2. Next, we
investigate the behavior of inverse projections in higher
dimensions and for different datasets (Sec. 4), thereby
answering Q3. Our joint results answer Q4.
3 VISUAL EVALUATION
3.1 Method
We study the behavior of decision maps using six dif-
ferent classifiers: Logistic Regression (Cox, 1958),
Support Vector Machines (Cortes and Vapnik, 1995,
SVM), Random Forests (Breiman, 2001), Neural Net-
works, Decision Trees, and K-Nearest Neighbors
(KNN). All the models are implemented using Scikit-
Learn (Pedregosa et al., 2011) with default parameters,
with the exception of the Neural Network, which is
configured to have three hidden layers, each containing
256 units. We train the above classifiers using the well-
known Iris flower dataset (Fisher, 1988). For each clas-
sifier, we construct the backprojected decision-map
image
I
inv
(see Sec. 2), similar to those shown in Fig. 1.
To directly visualize these, we need to have
n = 3
di-
mensions. As such, we restrict the Iris dataset to its
last three features. For ease of visual interpretation of
the constructed images
I
inv
, we also restrict the dataset
to its last two classes (thus,
|C| = 2
,
N = 100
). Note
that these two classes are not fully linearly separable,
which makes our classification task more challenging
than the synthetic blob dataset used in Fig. 1.
3.2 Results
Figure 2 shows the actual decision zones and decision
boundaries and the backprojected decision maps for six
classifiers and three decision-map techniques (DBM,
SDBM, and DeepView). The actual 2D decision maps
are shown in Fig. 3. These results answer Q1-Q2 and
deepen the findings from (Wang et al., 2023) shown in
Fig. 1 by (1) more classifiers, (2) a more challenging
dataset (modified Iris), and (3) additional visualization
of the actual decision zones and surfaces.
The top row in Fig. 2 shows the backprojected de-
cision maps for the three decision-map techniques. In
all three cases, we see that the backprojected maps
are surfaces that go through the samples of the two
classes (yellow and purple). We however see also dif-
ferences between the three techniques. The backpro-
jected surfaces of DBM and SDBM are quite smooth
and, as such, cannot get very close to (all) the actual
data points. In contrast, DeepView creates a noisier
surface which ‘connects’ the data points. In other
words: DBM and SDBM depict the classifier’s be-
havior further from the training set (extrapolation),
whereas DeepView shows this behavior close to and
inside this set (interpolation). This insight can directly
help users to choose which technique they want to use
depending on where (in data space) they want to study
a given classifier. Another insight relates to the shapes
of the decision maps for different classifiers
f
. For
DBM and SDBM, the projections
P
of the data points
do not depend on
f
– see the scatterplots in Fig. 3. As
such, the shape of the backprojected surface is also
independent on
f
– see Fig. 2. In contrast, DeepView
uses discriminative dimensionality reduction (Schulz
et al., 2015), so its
P
depends on
f
. For different clas-
sifiers, this means different scatterplots (Fig. 3) and
thus different backprojected surfaces (Fig. 2). While
one can argue that this shows more information on
f
,
controlling how DeepView’s decision maps actually
sample the data space as a function of
f
is unclear.
As such, we believe that (S)DBM’s approach, where
this sampling only depends on the training set, is more
intuitive and stable.
We further study the differences between the back-
projected decision map and the actual behavior of the
classifier
f
as follows. We densely sample the 3D
data space on a voxel grid of size
150
3
(to limit com-
putational effort); compute, for each voxel, a color
encoding using
f
at that point; and draw this color-
coded volume half-transparently (Fig. 2, bottom 6
rows). The yellow, respectively purple, volumes are
IVAPP 2024 - 15th International Conference on Information Visualization Theory and Applications
574
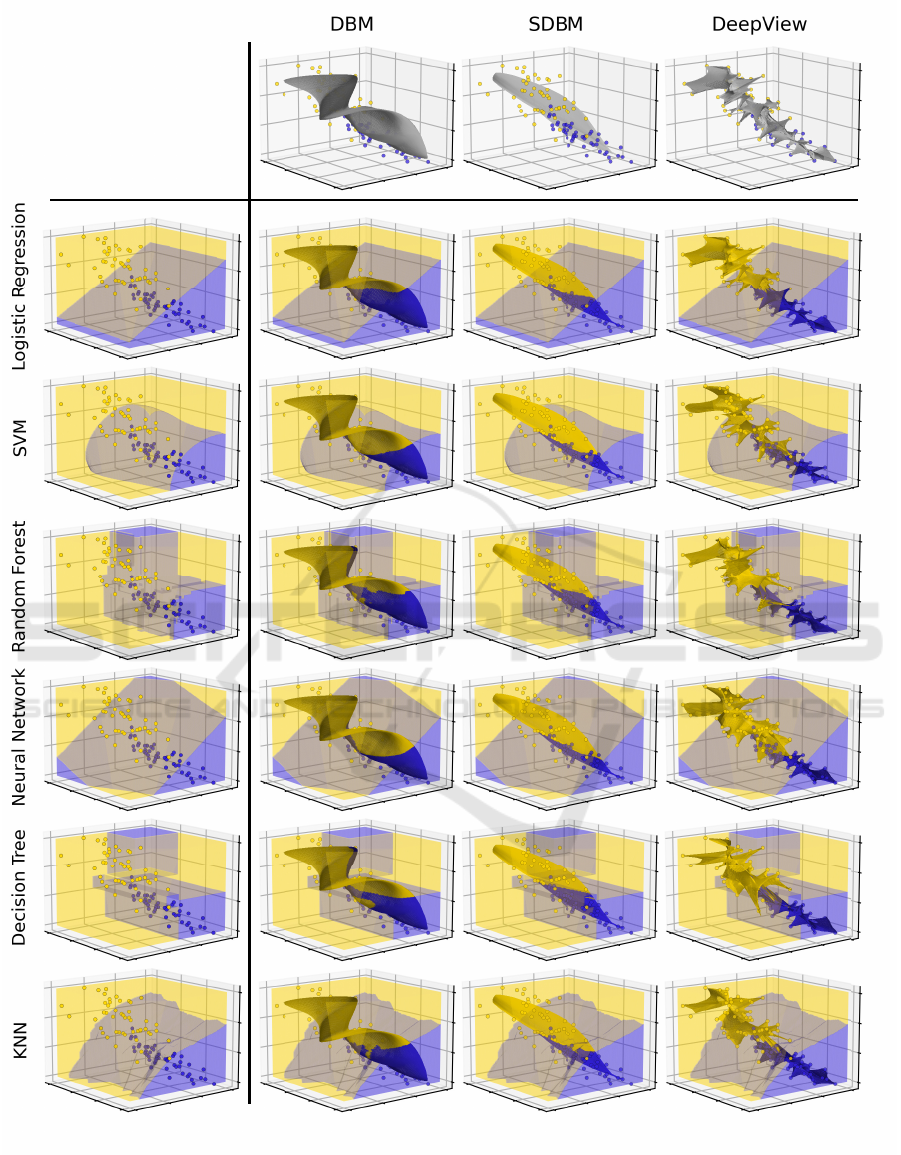
Figure 2: Decision maps of six classifiers for the modified Iris dataset (one row per classifier). The 3D decision zones, i.e., the
ground truth information, are indicated by yellow, respectively purple, with the decision surface separating them in pale brown.
The shaded surfaces embedded in the 3D space show the backprojection of the decision maps constructed by DBM, SDBM,
and DeepView.
Fundamental Limitations of Inverse Projections and Decision Maps
575
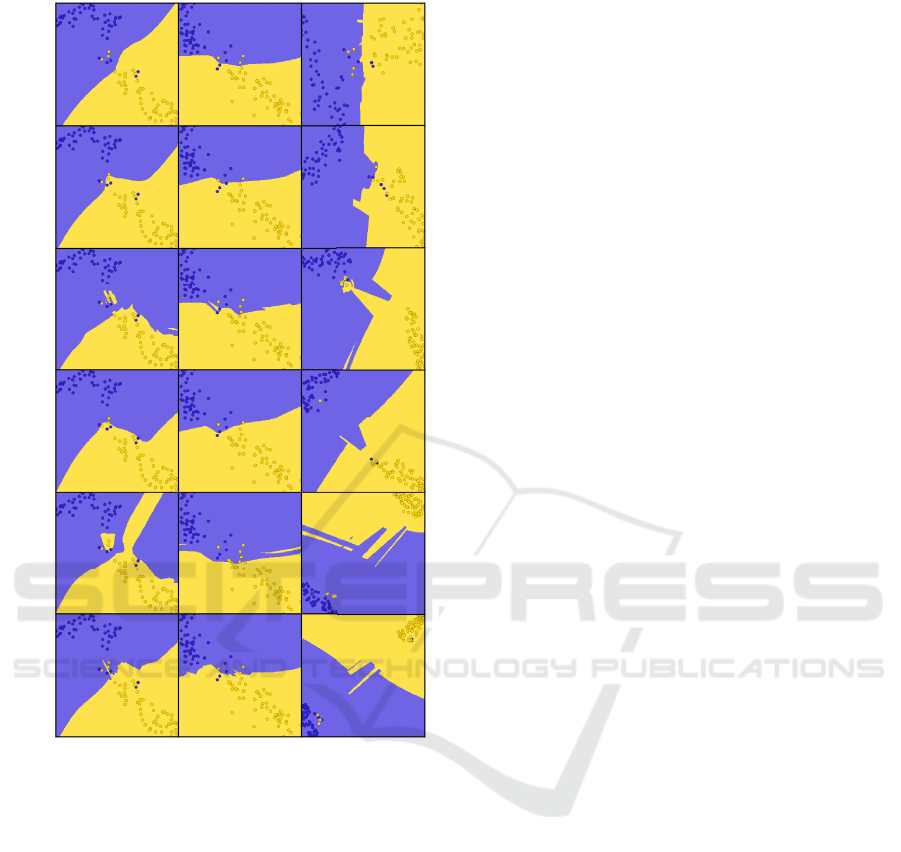
DBM SDBM DeepView
Logistic
regression
SVM
Random
forests
Neural
network
Decision
trees
KNN
Figure 3: The decision maps corresponding to Fig. 2.
thus the actual decision zones of
f
. Also, we draw
the actual boundary
S
that separates the two decision
zones (Fig. 2, bottom 6 rows, pale brown). This is the
actual decision boundary of
f
. We see that the back-
projected decision map
I
inv
(shaded surface in Fig. 2),
i.e., the part of the data space that a decision map visu-
alizes, is roughly orthogonal to, and intersecting, the
actual decision surface (pale brown in Fig. 2). That is,
the boundaries which we see in a decision map (curves
where yellow meets purple in Fig. 3) are the intersec-
tion
S ∩ I
inv
. This leads to two important insights: (1)
Decision maps capture global properties of the actual
decision boundaries quite well. For instance, we see
that the actual decision boundaries for SVM, Logistic
Regression, and Neural Networks are quite smooth
as compared to the other three classifiers (Fig. 2, left
column). The 2D decision maps also show this insight
(Fig. 3). (2) No decision map technique can actually
claim to visualize the entire decision boundaries of any
classifier. As a consequence, the way these techniques
sample the data space to construct their backprojected
surfaces is very relevant to the insights they produce.
For example, for Decision Trees, we see that the purple
decision zone is actually split into two disconnected
components (top and bottom purple cubes in Fig. 2,
leftmost column). However, only the DBM decision
map shows two separated purple zones (Fig. 3).
4 QUANTITATIVE EVALUATION
4.1 Method
For
n > 3
dimensional data, we cannot directly draw
the backprojected images
I
inv
. Recall now our ques-
tions Q3 (Sec. 1). To answer it, we measure how
far
I
inv
is, locally, from a two-dimensional manifold
embedded in
R
n
. For this, we use intrinsic dimension-
ality (ID) estimation (Bac et al., 2021) with a linear
ID estimation method, i.e., Principal Component Anal-
ysis (PCA), due to its intuitiveness, computational
efficiency, and popularity (Espadoto et al., 2019a; Bac
et al., 2021; Tian et al., 2021), as described next.
Local ID Estimation. Let
X
be a dataset embedded
in
R
n
. Let
S
i
be the
k
nearest neighbors in
X
of x
i
∈ X
.
Let
λ
λ
λ = (λ
1
,λ
2
,. .. ,λ
n
)
be the
n
eigenvalues of
S
i
’s
covariance matrix, sorted decreasingly, and normalized
to sum up to 1. The ID
d
i
of
S
i
is then the smallest
d
value so that the sum of the first
d
eigenvalues is larger
than a threshold θ (see Alg. 1).
Measurements. We perform two different ID mea-
surements as follows.
First, to study the quality of an inverse projection
P
inv
, for a given dataset
D
and its 2D projection
P(D)
,
we measure the average ID of the backprojection
D
′
=
P
inv
(P(D))
over all neighborhoods
S
i
(denoted
ID
D
′
)
and compare it with the ground-truth average ID of
D
(denoted
ID
D
).
ID
D
and
ID
D
′
are computed using
Alg. 1 with
D
and
D
′
as inputs, respectively. Ideally,
for a good inverse projection
P
inv
that reverses well
the effects of the direct projection
P
, we would obtain
ID
D
′
= ID
D
. Secondly, to study how well a decision
map covers the data space it aims to depict, we create
a uniform pixel grid
I
of size
500
2
and inverse project
I
via
P
inv
to obtain
I
inv
. We next measure the ID of the
backprojection
I
inv
at each pixel, denoted by
ID
p
.
ID
p
is computed using Alg. 1 with
I
inv
as input. We then
visualize
ID
p
at every location in
I
and also study its
average value over all pixels in I.
Figure 4 illustrates the process. From the data
points
D
, we compute a projection
P(D)
. Inversely
projecting these via
P
inv
yields the backprojection
D
′
.
IVAPP 2024 - 15th International Conference on Information Visualization Theory and Applications
576
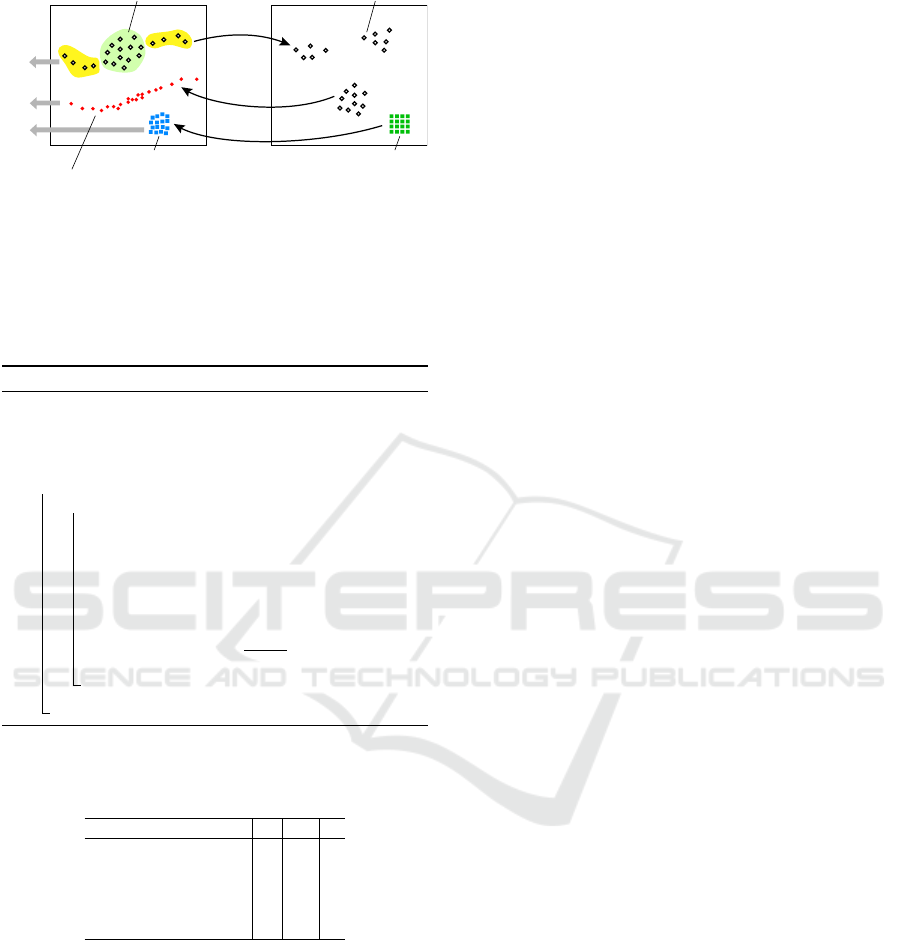
ℝ
n
ℝ
2
P
P
inv
dataset D projection P(D)
pixels Ibackprojection I
inv
backprojection D ’
P
inv
ID
D
ID
D’
ID
p
Figure 4: Computing the intrinsic dimensionality of data,
backprojection of data, and backprojection of pixels.
Inversely projecting a pixel set
I
yields the backpro-
jection
I
′
. In this example, the intrinsic dimensionality
ID
D
is the same to
ID
D
′
, but higher than
ID
D
′
, for the
yellow, respective green areas in D.
Algorithm 1: Intrinsic Dimensionality Estimation.
Data: X , set of data points in R
n
(can be D, D
′
, or I
inv
);
neighborhood size k = 120; threshold θ = 0.95
Result:
¯
d, the estimated ID of X (average among all local
neighborhoods)
1 begin
2 for x
i
∈ X do
3 Find the k nearest neighbors S of x
i
in X;
4 Compute the covariance matrix Cov of S;
5 Compute the eigenvalues λ
λ
λ = (λ
1
,λ
2
,. .., λ
n
) of Cov;
6 Sort λ
λ
λ in descending order;
7 Calculate ID d
i
of S as
d
i
= min
d
∑
d
j=1
λ
j
∑
n
i=1
λ
i
> θ
8 Calculate average ID
¯
d =
∑
i
d
i
/|X|;
Table 1: Datasets used for ID estimation. For each dataset,
we list the provenance, dimensionality (number of features)
n, number of samples N, and number of classes |C|.
Dataset n N |C|
Blobs 10D (synthetic) 10 5000 10
Blobs 30D (synthetic) 30 5000 10
Blobs 100D (synthetic) 100 5000 10
HAR (Anguita et al., 2012) 561 5000 6
MNIST (LeCun et al., 2010) 784 5000 10
Datasets. We use a mix of synthetic and real-world
datasets, all having
N = 5000
samples (Tab. 1). The
three synthetic datasets consist of
C = 10
isotropic
Gaussian blobs with ambient dimensionality
n
of 10,
30, and 100. The isotropy of the Gaussian blobs en-
sures that their ID is the same as their dimension count
n
. For real-world datasets, we use HAR (Anguita et al.,
2012) and MNIST (LeCun et al., 2010). The intrinsic
dimensionality of these datasets has been documented
in prior studies (El Moudden et al., 2016; Facco et al.,
2017; Aum
¨
uller and Ceccarello, 2019; Bahadur and
Paffenroth, 2019), enabling us to juxtapose our find-
ings with existing results (see further below). We
further use Logistic Regression as an example classi-
fier. Note that the choice of
f
does not affect the ID
estimation, as f is not involved in P
inv
’s construction.
Parameter Settings. We set
θ = 0.95
, thereby ac-
counting for 95% of the data variance in
S
i
, follow-
ing (Jolliffe, 2002; Fan et al., 2010; Tian et al., 2021).
The size
k
of the local neighborhood
S
i
is another pa-
rameter that needs careful setting. A too large
k
leads
to overestimating the local ID. A too small
k
leads to
noisy estimations. Note that
d + 1
independent vectors
are needed to span
d
dimensions, so
k
should be at
least equal to the actual ID of
S
i
(Verveer and Duin,
1995). For our studied datasets, we have ID ranging
from 13 to 33 for MNIST (Facco et al., 2017; Aum
¨
uller
and Ceccarello, 2019; Bahadur and Paffenroth, 2019);
the ID of HAR ranges from 15 to 61, depending on
the estimation method (El Moudden et al., 2016); our
other synthetic datasets have known ID values ranging
from 10 to 100 (see Tab. 1). As such, we globally set
k = 120 to cover all above cases.
4.2 Results
To answer Q3, we first need to see how well an in-
verse projection
P
inv
covers the data space
D
it aims to
depict. For this, we compare the estimated ID of the ac-
tual data (
ID
D
) with that of the round-trip constructed
by direct and inverse projections (
ID
D
′
). As explained
in Sec. 4.1, ideally
ID
D
′
= ID
D
. Table 2 shows our
results for the five studied datasets. First, we see that
the estimated
ID
D
aligns well with the ground-truth
values reported for most datasets. As such, we can
compare next the estimated
ID
D
to
ID
D
′
to judge how
good an inverse projection works. The last three rows
of Tab. 2 show a clear result: The inverse projection
always creates a far lower intrinsically-dimensional
dataset than the original one, with DeepView being the
closest (but still far away) from
ID
D
. This generalizes
our observations for 3D data discussed in Sec. 3.2:
Inverse projections used by (S)DBM create a roughly
two-dimensional, thus surface-like, sampling, of the
high-dimensional data space. In contrast, DeepView
shows a better ability to capture the ID of the data
– which corresponds to our observations showing its
backprojected surfaces having more complex shapes
that aim to connect the data points (Fig. 2).
To further answer Q3, we need to know how well
the pixels of a decision map cover the classifier space
it aims to depict. For this, we measure the ID of the
backprojected decision map image
ID
p
(see Sec. 4.1)
and compare it with the expected ID of the data. Fig-
ures 5
−
9 show these results for our five datasets. The
Fundamental Limitations of Inverse Projections and Decision Maps
577
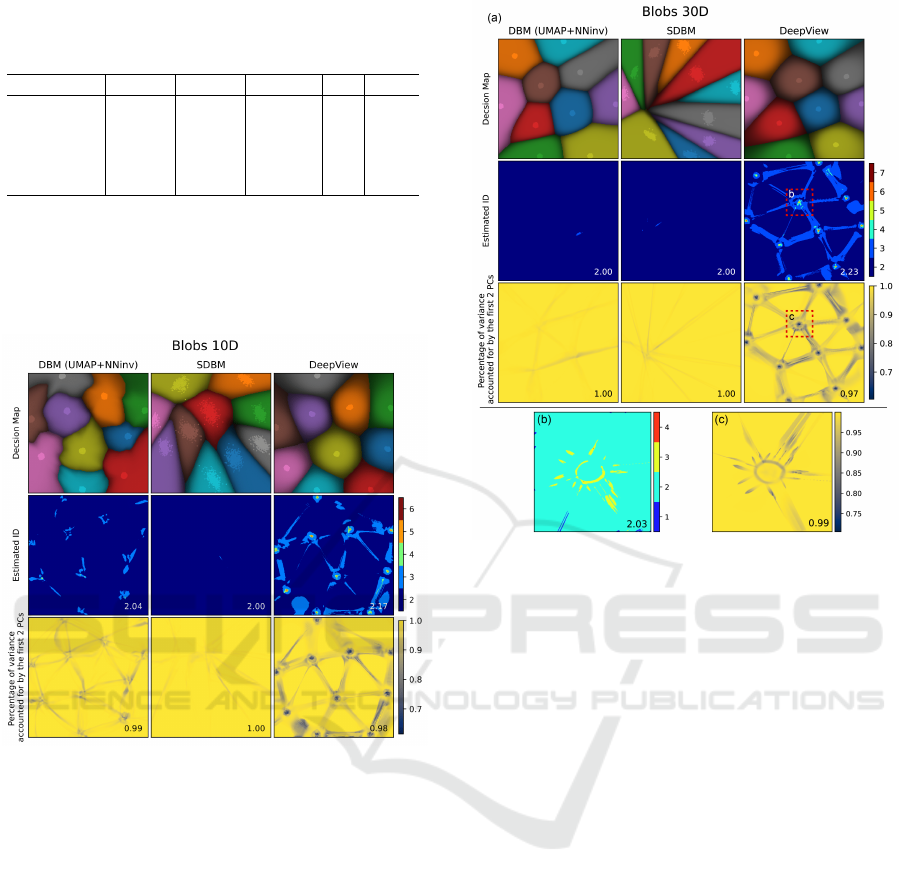
Table 2: Estimated intrinsic dimensionalities
ID
D
and
ID
D
′
for our studied datasets. We see that no inverse projection
method can capture the full ID of the data.
Blobs 10D Blobs 30D Blobs 100D HAR MNIST
Expected ID 10 30 100 15-61 13-33
ID
D
9.67 26.05 66.13 54.01 53.33
ID
D
′
DBM 2.37 2.00 2.00 2.94 3.85
ID
D
′
SDBM 2.07 2.00 2.00 2.02 1.96
ID
D
′
DeepView 3.74 3.58 3.67 6.85 6.02
top rows show the actual decision maps computed by
DBM, SDBM, and DeepView for the studied Logistic
Regression classifier. These images are only provided
for illustration purposes – the subsequent ID analysis
does not depend on the classifier choice.
Figure 5: Decision maps and ID estimation, Blob 10D.
The second rows in Figs. 5−9 show the estimated
ID
p
for each pixel, with the average
ID
p
value over
the entire map shown bottom-right in the figures. Strik-
ingly, the estimated ID for DBM and SDBM are ex-
actly 2 almost everywhere, which means that these
decision maps precisely correspond to surfaces embed-
ded in the high-dimensional space. This generalizes
our earlier observations (Sec. 3.2) to
n > 3
. DeepView
shows a more intricate pattern with higher
ID
p
values
close to the actual data points (peaking at
ID
p
= 9
for the HAR dataset); and lower values at map pix-
els far away from the data points (roughly equal to 2
but occasionally dipping to 1 in certain areas of the
MNIST and HAR decision maps). The average
ID
p
of
DeepView over all datasets is
2.20 ± 0.03
. To further
understand the higher
ID
p
values for DeepView, we
select areas having such high values (red squares in
Figs. 6 and 8, second rows). These areas are over-
sampled at a resolution of
500
2
pixels and
500 × 1000
pixels, respectively. The results (Figs. 6 and 8 (b))
Figure 6: Decision maps and ID estimation, Blob 30D
dataset. Bottom row: Zoom-ins of selected (red) regions.
show that
ID
p
actually is very close to 2 in such areas
as well. In other words, while DeepView constructs a
more complex-shaped, higher-curvature, surface than
(S)DBM, it still only maps data coming from a surface
in the high-dimensional space.
The third rows in Figs. 5
−
9 refine the above in-
sights by showing the percentage of data variance in
a neighborhood
S
captured by the eigenvectors cor-
responding to the two largest eigenvalues
λ
1
and
λ
2
(computed as in Sec. 4.1). We see that, in most map
regions, this value is close to 1, indicating again that
the decision maps correspond to mostly locally-planar
structures in high-dimensional space. Interestingly,
besides the darker areas corresponding to dense data
points in the projection for DeepView, we see a num-
ber of 1D filament-like darker structures that span the
image space, for all decision map techniques. These
indicate areas where the backprojected surface
I
inv
has high curvature. Also interestingly, these filaments
seem to connect the projected points in DeepView’s
map much like a Delaunay triangulation. This corre-
sponds, for the
n > 3
case, to what we observed earlier
for the
n = 3
case (Fig. 2) – that is, the DeepView
backprojected surface aims to tightly connect the data
samples.
IVAPP 2024 - 15th International Conference on Information Visualization Theory and Applications
578
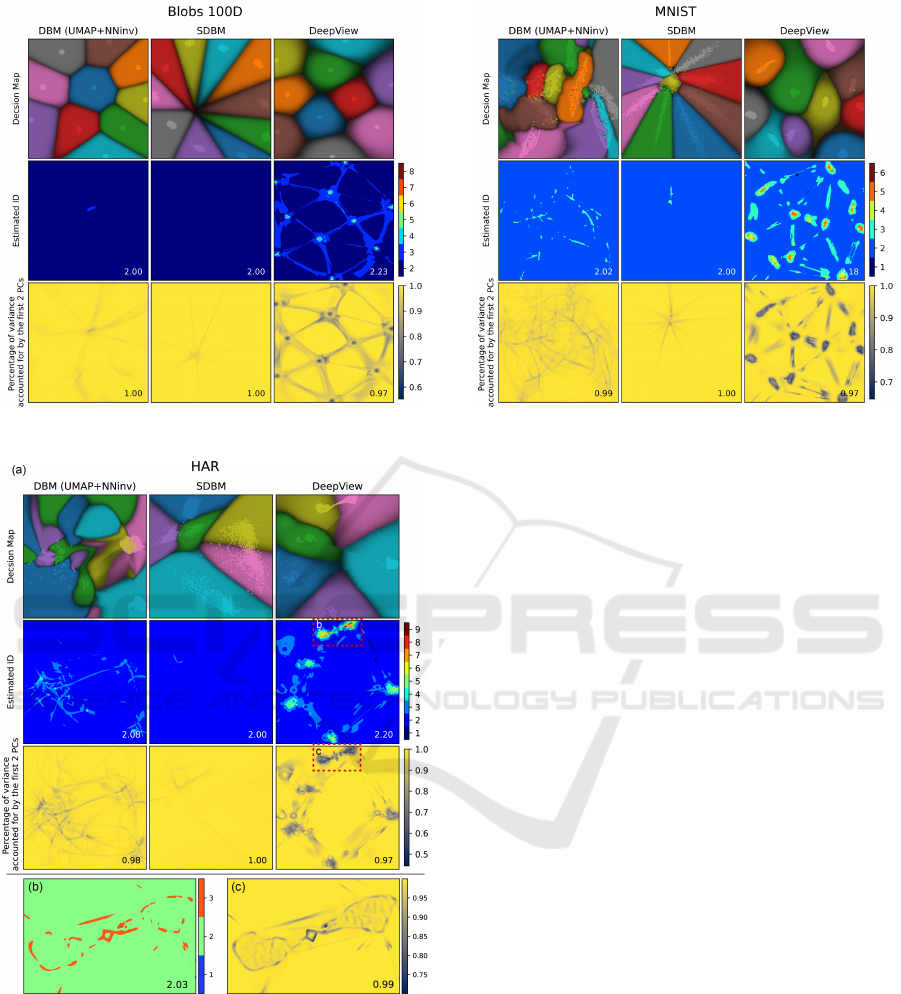
Figure 7: Decision maps and ID estimation, Blob 100D.
Figure 8: Decision maps and ID estimation, HAR. Bottom
row: Zoom-ins of selected (red) regions.
5 DISCUSSION
We next discuss our findings on the interpretation,
added value, and found limitations of decision maps,
and answer our original questions Q1-Q4.
Surface Behavior of Decision Maps: All three stud-
ied decision map techniques essentially depict surfaces
Figure 9: Decision maps and ID estimation, MNIST.
embedded in the high-dimensional data. This property
does not depend on the intrinsic or total dimensionality
of the studied dataset (Q1) or studied classifier (Q1).
Also, increasing the resolution of the decision map
does not change this aspect (see Figs. 6 and 8). As
such, current decision maps only depict a small part of
the behavior of a given classifier (Q3). The boundaries
that current decision maps show are actually only in-
tersections of these surfaces with the actual decision
boundaries in high-dimensional space (Q2). These are,
we believe, important and previously not highlighted,
limitations of current decision map techniques. We
argue that, without further explanatory tools that as-
sist the user in interpreting decision maps, the insights
given by such maps are limited and can be potentially
even misleading.
One could argue that this surface behavior is ev-
ident from the fact that inverse projections are de-
fined on the 2D image plane and, as such, they will
always construct a surface embedded in higher dimen-
sions. However, this is not necessarily so. Space-
filling curves, known since long (Peano, 1890), can
map lower-dimensional intervals to completely cover
higher-dimensional ones. Very recent equivalents exist
for space-filling surfaces (Paulsen, 2023). By compos-
ing such operations, we can imagine dense mappings
between intervals in spaces of any two dimensions
q
and
n
,
q < n
. While it is true that the existing inverse
projection functions we know of (DBM, SDBM, Deep-
View) do arguably not have this fractal-like behavior,
given that they are constructed by composing locally
differentiable mappings, it was not clear – before our
study – how far they are from piecewise-planar map-
pings. This is especially hard to gauge a priori since
Fundamental Limitations of Inverse Projections and Decision Maps
579

these methods use internally complex nonlinear mecha-
nisms such as deep learning. Our study – in particular,
the ID estimation – evaluated precisely this aspect,
with the aforementioned differences highlighted be-
tween these methods. Moreover, our study proposes
a methodology by which new DBM methods – which
may possibly use mappings like the Peano curves men-
tioned above, or any kinds of mappings – can be evalu-
ated to gauge how much of the high-dimensional data
space they actually cover.
Comparing Decision Map Methods. In terms of
the abovementioned surfaces, different decision map
techniques sample the high-dimensional space quite
differently (Q4). As such, they produce different maps
for the same classifier (which, obviously, has a single
fixed set of actual decision surfaces). Each such map
provides unique insights for the same classifier (see
e.g. Fig. 3). While how the decision maps sample the
high-dimensional space depends only on the training
points for (S)DBM, those of DeepView also depend
on the actual classifier. Overall, (S)DBM sample the
classifier along a smoother surface which is farther
away from the training set, whereas DeepView yields
a less smooth surface that aims to connect the training
samples. Both surface types have their advantages and
limitations: Smoother surfaces farther from training
samples are easier to interpret and show better how
a classifier extrapolates from its training set but are
harder to control in terms of where they are actually
constructed; tighter surfaces that connect the training
samples are easier to control in terms of location but
they only interpolate the classifier behavior close to
and between the training points. Importantly, none of
the studied techniques aims to sample a classifier close
to its actual decision boundaries – which, arguably, are
the most interesting areas to understand.
Limitations. Our findings have several generalization
challenges. First, we only used two real-world datasets.
More such datasets would be needed to strengthen
our observations. A challenge here is to find datasets
having well-documented estimations of the intrinsic
dimensionality. Separately, additional methods to es-
timate the intrinsic dimensionality can be used along
our current linear model, e.g., non-linear methods or
methods that automatically adapt the size
k
of the local
neighborhood to account for varying data densities.
We plan to consider such methods in future work.
6 CONCLUSIONS
We have presented an analysis of the limitations of cur-
rent inverse projection and decision map techniques
used to construct visualizations of the behavior of ma-
chine learning classification models. For this, we have
compared the decision zones and boundaries depicted
by three such techniques – DBM, SDBM, and Deep-
View – with the actual zones and boundaries that are
created by six classifiers on a three-dimensional real-
world dataset. We found out that the studied maps only
capture a two-dimensional surface embedded in the
data, with different map techniques offering different
trade-offs on how this surface ranges between interpo-
lation and extrapolation of the classifier behavior with
respect to its training set. We further extended our anal-
ysis to high-dimensional data by comparing the intrin-
sic dimensionality of the data with that of the inverse
projection and backprojection of the map to the data
space. We found that all studied map techniques still
only consider locally two-dimensional, thus surface-
like, subsets of the data space for visualization. Fur-
thermore, we showed evidence that the extrapolation
vs interpolation behavior of (S)DBM vs DeepView
generalize also to higher-dimensional data spaces. Our
work highlights fundamental limitations of all studied
decision map techniques in terms of how much of a
classifier’s behavior they capture, but also where and
how they choose to capture this behavior. These limi-
tations are essential to understand when interpreting a
decision map.
We see two possible ways to further overcome the
surface-like limitation of decision maps. First, differ-
ent inverse-projection techniques can be developed to
focus the sampling of the high-dimensional space on
areas where one wants to study a classifier’s behavior
in more detail, e.g., around actual decision boundaries,
rather than on the (less-interesting) areas containing
same-label samples. This is a very complex challenge:
How to design such a sampling method to capture
large, complex, areas of a high-dimensional space
with only a 2D map? An alternative possibility is to
acknowledge that it is impossible to capture all such
areas by a single map. As such, mechanisms can be of-
fered to allow users to interactively specify which data
regions they want to explore. In this direction, Sohns
et al. (Sohns et al., 2023) have proposed an interactive
tool for exploring high-dimensional decision bound-
aries. Yet, this tool only works on local neighborhoods
and uses a linear projection (PCA), which has been
shown to have poor cluster separation (Espadoto et al.,
2019c). Moreover, this technique works only on tabu-
lar data and faces scalability challenges. We believe
that such approaches can be extended by additional in-
teraction mechanisms that allow users to parameterize
the inverse projection to sample specific regions of the
data space by e.g. controlling the shape and position
of the backprojected surface with respect to the actual
decision boundaries and/or training-set samples.
IVAPP 2024 - 15th International Conference on Information Visualization Theory and Applications
580

REFERENCES
Anguita, D., Ghio, A., Oneto, L., Parra, X., and Reyes-Ortiz,
J. L. (2012). Human activity recognition on smart-
phones using a multiclass hardware-friendly support
vector machine. In Proc. Intl. Workshop on ambient
assisted living, pages 216–223. Springer.
Aum
¨
uller, M. and Ceccarello, M. (2019). The Role of Lo-
cal Intrinsic Dimensionality in Benchmarking Nearest
Neighbor Search. arXiv:1907.07387 [cs].
Bac, J., Mirkes, E. M., Gorban, A. N., Tyukin, I., and
Zinovyev, A. (2021). Scikit-Dimension: A Python
Package for Intrinsic Dimension Estimation. Entropy,
23(10):1368.
Bahadur, N. and Paffenroth, R. (2019). Dimension Esti-
mation Using Autoencoders. arXiv:1909.10702 [cs,
stat].
Bennett, R. (1969). The intrinsic dimensionality of signal
collections. IEEE Trans. Inform. Theory, 15(5):517–
525.
Breiman, L. (2001). Random Forests. Mach. Learn., 45(1):5–
32.
Camastra, F. (2003). Data dimensionality estimation meth-
ods: a survey. Pattern Recognit., 36(12):2945–2954.
Campadelli, P., Casiraghi, E., Ceruti, C., and Rozza, A.
(2015). Intrinsic dimension estimation: Relevant tech-
niques and a benchmark framework. Math. Probl. Eng.,
2015:1–21.
Cortes, C. and Vapnik, V. (1995). Support-vector networks.
Mach. Learn., 20(3):273–297.
Cox, D. R. (1958). Two further applications of a model for
binary regression. Biometrika, 45(3/4):562–565.
dos Santos Amorim, E. P., Brazil, E. V., Daniels, J., Joia,
P., Nonato, L. G., and Sousa, M. C. (2012). iLAMP:
Exploring high-dimensional spacing through backward
multidimensional projection. In Proc. IEEE VAST,
pages 53–62.
El Moudden, I., El Bernoussi, S., and Benyacoub, B. (2016).
Modeling human activity recognition by dimensional-
ity reduction approach. In Proc. IBIMA, pages 1800–
1805.
Engel, D., H
¨
uttenberger, L., and Hamann, B. (2012). A
survey of dimension reduction methods for high-
dimensional data analysis and visualization. In Proc.
IRTG workshop, volume 27, pages 135–149. Schloss
Dagstuhl–Leibniz-Zentrum fuer Informatik.
Espadoto, M., Appleby, G., Suh, A., Cashman, D., Li,
M., Scheidegger, C. E., Anderson, E. W., Chang, R.,
and Telea, A. C. (2021a). UnProjection: Leverag-
ing Inverse-Projections for Visual Analytics of High-
Dimensional Data. IEEE TVCG, pages 1–1.
Espadoto, M., Hirata, N., and Telea, A. (2021b). Self-
supervised Dimensionality Reduction with Neural Net-
works and Pseudo-labeling. In Proc. IVAPP, pages
27–37. SciTePress.
Espadoto, M., Martins, R., Kerren, A., Hirata, N., and Telea,
A. (2019a). Toward a quantitative survey of dimension
reduction techniques. IEEE TVCG, 27(3):2153–2173.
Espadoto, M., Rodrigues, F. C. M., Hirata, N. S. T., and
Hirata Jr, R. (2019b). Deep Learning Inverse Multidi-
mensional Projections. In Proc. EuroVA, page 5.
Espadoto, M., Rodrigues, F. C. M., and Telea, A. (2019c).
Visual analytics of multidimensional projections for
constructing classifier decision boundary maps. In
Proc. IVAPP. SCITEPRESS.
Facco, E., d’Errico, M., Rodriguez, A., and Laio, A. (2017).
Estimating the intrinsic dimension of datasets by a min-
imal neighborhood information. Sci Rep, 7(1):12140.
Fan, M., Gu, N., Qiao, H., and Zhang, B. (2010). Intrinsic
dimension estimation of data by principal component
analysis. arXiv:1002.2050 [cs].
Fisher, R. A. (1988). Iris Plants Database. UCI Machine
Learning Repository.
Hinton, G. E. and Salakhutdinov, R. R. (2006). Reducing the
dimensionality of data with neural networks. Science,
313(5786):504–507.
Huang, X., Wu, L., and Ye, Y. (2019). A review on dimen-
sionality reduction techniques. Int. J. Pattern Recognit.
Artif. Intell., 33(10):1950017.
Joia, P., Coimbra, D., Cuminato, J. A., Paulovich, F. V., and
Nonato, L. G. (2011). Local affine multidimensional
projection. IEEE TVCG, 17(12):2563–2571.
Jolliffe, I. T. (2002). Principal component analysis for spe-
cial types of data. Springer.
LeCun, Y., Cortes, C., and Burges, C.
(2010). MNIST handwritten digit database.
http://yann.lecun.com/exdb/mnist.
McInnes, L., Healy, J., and Melville, J. (2018). UMAP:
Uniform Manifold Approximation and Projection for
Dimension Reduction. arXiv:1802.03426 [cs, stat].
Munzner, T. (2014). Visualization analysis and design. CRC
press.
Nonato, L. and Aupetit, M. (2018). Multidimensional pro-
jection for visual analytics: Linking techniques with
distortions, tasks, and layout enrichment. IEEE TVCG,
25:2650–2673.
Oliveira, A. A. A. M., Espadoto, M., Hirata, R., and Telea,
A. C. (2023). Stability Analysis of Supervised Decision
Boundary Maps. SN COMPUT. SCI., 4(3):226.
Oliveira, A. A. A. M., Espadoto, M., Hirata Jr, R., and
Telea, A. C. (2022). SDBM: Supervised Decision
Boundary Maps for Machine Learning Classifiers. In
Proc. IVAPP, pages 77–87.
Paulsen, W. (2023). A Peano-based space-filling surface of
fractal dimension three. Chaos, Solitons & Fractals,
168.
Peano, G. (1890). Sur une courbe, qui remplit toute une aire
plane. Mathematische Annalen, 36(1):157–160.
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V.,
Thirion, B., Grisel, O., Blondel, M., Prettenhofer,
P., Weiss, R., and Dubourg, V. (2011). Scikit-learn:
Machine learning in Python. J. Mach. Learn. Res.,
12:2825–2830.
Rodrigues, F. C. M., Espadoto, M., Hirata, R., and Telea,
A. C. (2019). Constructing and Visualizing High-
Quality Classifier Decision Boundary Maps. Infor-
mation, 10(9):280.
Fundamental Limitations of Inverse Projections and Decision Maps
581

Rodrigues, F. C. M., Hirata, R., and Telea, A. C. (2018).
Image-based visualization of classifier decision bound-
aries. In Proc. SIBGRAPI, pages 353–360. IEEE.
Schulz, A., Gisbrecht, A., and Hammer, B. (2015). Using
discriminative dimensionality reduction to visualize
classifiers. Neural Process. Lett., 42:27–54.
Schulz, A., Hinder, F., and Hammer, B. (2020). DeepView:
Visualizing Classification Boundaries of Deep Neural
Networks as Scatter Plots Using Discriminative Dimen-
sionality Reduction. In Proc. IJCAI, pages 2305–2311.
Sohns, J.-T., Garth, C., and Leitte, H. (2023). Decision
Boundary Visualization for Counterfactual Reasoning.
Comput. Graph. Forum, 42(1):7–20.
Sorzano, C. O. S., Vargas, J., and Montano, A. P. (2014).
A survey of dimensionality reduction techniques.
arXiv:1403.2877 [stat.ML].
Tian, Z., Zhai, X., van Driel, D., van Steenpaal, G., Espadoto,
M., and Telea, A. (2021). Using multiple attribute-
based explanations of multidimensional projections
to explore high-dimensional data. Comput. Graph.,
98:93–104.
Verveer, P. J. and Duin, R. P. W. (1995). An evaluation
of intrinsic dimensionality estimators. IEEE PAMI,
17(1):81–86.
Wang, Y., Machado, A., and Telea, A. (2023). Quantitative
and Qualitative Comparison of Decision-Map Tech-
niques for Explaining Classification Models. Algo-
rithms, 16(9):438.
Zhou, T., Cai, Y., An, M., Zhou, F., Zhi, C., Sun, X., and
Tamer, M. (2023). Visual interpretation of machine
learning: Genetical classification of apatite from vari-
ous ore sources. Minerals, 13(4):491.
IVAPP 2024 - 15th International Conference on Information Visualization Theory and Applications
582
