
Synergizing Data Imputation and Electronic Health Records for
Advancing Prostate Cancer Research: Challenges, and Practical
Applications
Abderrahim O. Batouche
1,2,3,∗ a
, Eugen Czeizler
2,3,∗ b
, Miika Koskinen
4 c
, Tuomas Mirtti
2,5 d
and Antti S. Rannikko
2,6 e
1
Doctoral Programme in Computer Science, University of Helsinki, Helsinki, Finland
2
Research Program in Systems Oncology, University of Helsinki, Helsinki, Finland
3
ICAN-Digital Precision Cancer Medicine Flagship, Helsinki, Finland
4
HUS Helsinki University Hospital, Helsinki, Finland
5
Department of Pathology, University of Helsinki and Helsinki University Hospital, Helsinki, Finland
6
Department of Urology, University of Helsinki and Helsinki University Hospital, Helsinki, Finland
fi fi
Keywords:
Data Mining, Electronic Health Records, Missing Data, Prostate Cancer.
Abstract:
The presence of detailed clinical information in electronic health record (EHR) systems presents promising
prospects for enhancing patient care through automated retrieval techniques. Nevertheless, it is widely ac-
knowledged that accessing data within EHRs is hindered by various methodological challenges. Specifically,
the clinical notes stored in EHRs are composed in a narrative form, making them prone to ambiguous for-
mulations and highly unstructured data presentations, while structured reports commonly suffer from missing
and/or erroneous data entries. This inherent complexity poses significant challenges when attempting auto-
mated large-scale medical knowledge extraction tasks, necessitating the application of advanced tools, such
as natural language processing (NLP), as well as data audit techniques. This work aims to address these
obstacles by creating and validating a novel pipeline designed to extract relevant data pertaining to prostate
cancer patients. The objective is to exploit the inherent redundancies available within the integrated structured
and unstructured data entries within EHRs in order to generate comprehensive and reliable medical databases,
ready to be used in advanced research studies. Additionally, the study explores potential opportunities arising
from these data, offering valuable prospects for advancing research in prostate cancer.
1 INTRODUCTION
Prostate cancer (PCa) is a prevalent disease known
for its indolent nature, often characterised by slow de-
velopment and protracted progression over time (Na-
tional Cancer Institute, 2023; The American Cancer
Society medical and editorial content team, 2019). As
such, one specific challenge in performing medical
research on PCa is dealing with incomplete medical
records and missing data, e.g., as a result of city re-
a
https://orcid.org/0000-0003-4181-9891
b
https://orcid.org/0000-0002-1607-1554
c
https://orcid.org/0000-0002-7267-5811
d
https://orcid.org/0000-0003-0455-9891
e
https://orcid.org/0000-0002-4261-3484
∗
These authors contributed equally to this work.
location or disease follow-up across different health
providers. This, in turn, can hinder the results of on-
going research studies analysing the effectiveness of
diagnoses and various treatment planning approaches
(Holmes et al., 2021). Ultimately, this can affect clin-
ical decision-making and the patient’s well-being.
To overcome the limitations of incomplete and/or
erroneous data, Electronic Health Records (EHRs)
mining has emerged as a crucial approach in medi-
cal research as well as within clinical practice(Yadav
et al., 2018). EHRs mining leverages advanced data
analytic and artificial intelligence (AI) approaches to
extract valuable insights from vast amounts of pa-
tient data(Ajmal et al., 2023; Javaid et al., 2022). By
identifying patterns, trends, and risk factors associ-
ated with prostate cancer, EHRs mining facilitates the
early detection of advanced diseases and the personal-
Batouche, A., Czeizler, E., Koskinen, M., Mirtti, T. and Rannikko, A.
Synergizing Data Imputation and Electronic Health Records for Advancing Prostate Cancer Research: Challenges, and Practical Applications.
DOI: 10.5220/0012350300003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 2, pages 77-86
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
77

isation of treatment strategies (Knighton et al., 2016;
Seneviratne et al., 2018; Henkel et al., 2022). How-
ever, challenges such as missing data and data secu-
rity must be addressed to ensure patient information
remains complete, confidential, and secure. Addi-
tionally, the lack of interoperability between differ-
ent EHR systems poses hurdles in data sharing and
aggregation, limiting the full potential of mining for
both prostate cancer research, as well as for general
improvement of patient care (De La Torre-D
´
ıez et al.,
2013). Overcoming these issues and promoting stan-
dardised data collection practices and protocols will
be pivotal in advancing the field of PCa treatment
through EHRs mining(Herp et al., 2023), as well as
the overall medical research in general.
In our work, we have designed and developed
a data preprocessing pipeline that can leverage rou-
tinely collected information from our EHRs (HUS
Datalake (Bruck, 2023; Pylk
¨
as, 2023; Misukka,
2022)) to efficiently and accurately retrieve and con-
solidate clinicians’ work on PCa treatment analysis.
Using Microsoft Azure machine learning studio and
batches from HUS datalake that are available at the
HUS Acamedic environment (a secure scalable data
analytics platform developed for medical research
(kuorttinen, 2023)), we developed an EHR mining
pipeline using Python libraries to read, process, and
provide curated data for further research applications.
One of the key clinical inputs exhibiting missing
entries within the EHR of a significant number of
PCa patients is the occurrence of curative treatment,
i.e., radical prostatectomy (RP) or radiation treatment
(RT). Since the imputation of such missing data is in-
evitable, we had to use a different approach to uncover
these lost data entries. Using routinely collected val-
ues of the prostate-specific antigen (PSA) lab mea-
surements, we were able to successfully identify and
even classify curative PCa treatments. To our knowl-
edge, this is the first attempt to approach the inference
of EHR missing treatment records through PSA time
series data.
Our approach enabled us to enhance our EHR by
incorporating approximately 2.8 thousand new cura-
tive treatment events, marking a notable 27% growth
compared to the treatment events available before-
hand. The explanation for this relatively large in-
crease is multi-folded. Some patients might have been
treated outside the (Helsinki and Uusima) district unit
whose database our study is based upon. Others might
have been treated within private practice units, which
again are not covered by our database. Finally, we can
assume that a proportion of these missing treatment
events are due to human error in correctly recording
them within the EHR.
Another key clinical information (as well as key
surrogate measurement within medical research anal-
ysis) which is most of the times not directly recorded
within EHRs, either in structured or non-structured
format, is the time instant when PCa patients are clas-
sified as having a biochemical recurrence (BCR). Af-
ter primary cancer treatment, BCR is achieved when
the PSA level in the blood surpasses a certain thresh-
old, thus indicating that the disease may be returning
or progressing. Thus, BCR status is an important in-
dicator both clinically, as it signs that further monitor-
ing or treatment may be needed to manage the condi-
tion (Stephenson et al., 2006; Artibani et al., 2018),
and from a (medical) data analysis perspective, as it
is a surrogate for PCa mortality (Zhao et al., 2022;
Artibani et al., 2018). By following the PSA mea-
surements as well as all EHR-available PCa treatment
records we were able to effectively determine (and re-
port) the status and timing of BCR for all PCa pa-
tients.
2 METHODS
2.1 Data Source
Our pipeline starts by identifying patients of interest
within a large academic EHR system (Figure 1). We
used the Finnprostate dataset, which is a large patient
registry study combining Finnish national healthcare
data with local hospital data (n=700,000) of men
suspected of having PCa (PSA measured) or diag-
nosed with PCa. From Finnprostate, we gathered a
HUS (Hospital District of Helsinki and Uusimaa) sub-
cohort of men (n=326,796) having comprehensive pa-
tient information regarding out-patient clinic and hos-
pital visits as well as data regarding laboratory tests,
medication prescriptions, radiological, pathological,
and surgical reports, as well as comorbidities covering
the years 1993 to 2019. The above data is embedded
within the regional HUS Acamedic datalake.
Medical research commonly encounters missing
data. Despite this prevalence, it is nowadays gener-
ally accepted to perform various data analysis tasks
on partially incomplete records, as long as the miss-
ing values are not substantial, and the analysis meth-
ods themselves can cope with specific uncertainties.
Moreover, the use of advanced imputation techniques
such as maximum likelihood (Wald, 1949), multi-
ple imputation(Schafer, 1999), or Bayesian methods
(Kong et al., 1994) have a good track record in ad-
dressing many of the missing data entries. However,
certain complex missing data records, such as the mo-
ment and type of a deployed treatment, or the first di-
HEALTHINF 2024 - 17th International Conference on Health Informatics
78

Figure 1: Data preprocessing pipeline for Prostate Cancer
research data.
agnostic biopsy of a tumour and its aggressiveness,
are very hard to be addressed by any of the available
computational imputation methods.
In our data processing work (yellow box, Figure
1), imputation was reinforced with customised algo-
rithms that rely on clinical guidelines, experts’ inter-
pretations, as well as the intrinsic information redun-
dancy available within EHR, in order to retrieve the
missing data. All created algorithms are described in
Table 1.
2.2 Missing Curative Treatment
Detection
The Treatment Detection Algorithm (DTX algo)
plays a pivotal role in enhancing our data quality by
identifying and incorporating missing curative treat-
ment records (in Algorithm 4). The algorithm takes
all patient’s data as input and returns a list of missing
curative treatments.
The Significant PSA Drop Algorithm (SIGDROP)
constitutes the initial phase of DTX, meticulously
tracking a patient’s PSA values subsequent to their
diagnostic biopsy (Algorithm 1). The algorithm takes
PSA measurements of patient i, and returns, if any:
• drop date: The date of the PSA drop, which is the
highest (maximum) point from where a significant
PSA drop starts; is subsequently considered as a
treatment date.
• nadir date: The date of the PSA nadir, which is
the lowest (minimum) point to where the signifi-
cant drop reached.
• PSA
min
: The minimum values (at the time
nadir date); this value is used to classify the drop
into radical prostatectomy or radiation therapy.
The algorithm’s operation commences with the
pursuit of the maximum PSA value (PSA
max
, lines
3-4), followed by an endeavour to identify the min-
imum value within the ensuing δ ≤ 12-month period
(lines 5-32). Upon successful identification of a de-
creasing value, at lines 15-16, the algorithm calcu-
lates α, which is the drop percentage that undergoes
rigorous testing to ascertain its adherence to predeter-
mined significance conditions (line 17). This process
is indispensable in establishing the genuineness of the
observed drop and confirming its clinical significance.
Having validated the drop as significant, and (line
6) with no EHR-recorded curative treatment between
the date of drop (d
max
) and the date of the nadir (d
min
),
DTX proceeds to collate all such identified drops, sys-
tematically categorising them into two distinct treat-
ment modalities: radiation therapy (RT) and radical
prostatectomy (RP) (Algorithm 2 line 7-10). This
classification not only facilitates comprehensive treat-
ment record augmentation but also provides valuable
missing insights into the patient’s therapeutic journey.
2.3 Biochemical Recurrence Detection
Biochemical recurrence (BCR) serves as a crucial in-
dicator for PCa mortality. However, its availability in
EHRs is not always guaranteed. In such cases, vari-
ous methods can be employed to retrieve and impute
this information. Our Detect Biochemical Recurrence
(DBCR algo) Algorithm is specifically designed to
analyse data from treated patients, identifying poten-
tial relapses and categorising patients as either having
experienced a BCR or not (Algorithm 7). To achieve
this outcome, DBCR utilises four (04) distinct func-
tions, each tailored to a specific task.
Clinical guidelines governing PSA relapse are
stringent and clearly defined (Van Den Broeck et al.,
2020), and these guidelines are meticulously inte-
grated into the PRP and PRT functions (Algorithms
5-6).
• PSA-based relapse after radical prostatectomy
PRP(p
i
): this function uses the European As-
sociation of Urology (EAU) guidelines (Van
Den Broeck et al., 2020) to detect whether a
Synergizing Data Imputation and Electronic Health Records for Advancing Prostate Cancer Research: Challenges, and Practical
Applications
79

Table 1: Summary of Algorithms.
Algorithm Name Input Output Complexity Short Description
SIGDROP PSA
i
drop date, nadir date, PSA
min
O(M) Detects significant PSA drop and related dates.
DTX PATIENT S LIST L O(M ∗N) Detects missing treatments based on PSA data.
CRT p
i
d
m1
O(1) Detects Clinical Relapse after RT.
CRP p
i
d
m1
O(1) Detects Clinical Relapse after RP.
PRP p
i
d
m2
O(N) Detects PSA Relapse after RP.
PRT p
i
d
m2
O(N) Detects PSA Relapse after RT.
DBCR T REAT ED PAT IENTS L
bcr
O(M ∗N) Main algorithm for BCR detection.
- RP=Radical prostatectomy, RT=Radiation therapy, BCR=Biochemical recurrence.
- In M*N, M is the number of PSA measurements and N is the number of patients.
Algorithm 1: SIGDROP - Significant PSA drop detec-
tion.
Input: PSA
i
Output: drop date, nadir date, PSA
min
1: M ← size(PSA
i
)
2: if M ≥ 0 then
3: PSA
max
← PSA
i
[1]
4: date PSA
max
← getDate(PSA
max
)
5: for j = 1 to M − 1 do
6: e ← PSA
i
[ j] − PSA
i
[ j + 1]
7: δ ← date PSA
next
− date PSA
max
8: if e ≤ 0 then
9: date PSA
next
← getDate(PSA
i
[ j + 1])
10: if (PSA
max
< PSA
i
[ j + 1]) or δ > 12m then
11: PSA
max
← PSA
i
[ j + 1]
12: date PSA
max
← getDate(PSA
max
)
13: end if
14: else
15: β ← PSA
max
− PSA
i
[ j + 1]
16: α ←
β
PSA
max
17: if (α ≥ 0.75 and β ≥ 3) or (α ≥ 0.5 and β ≥ 4) then
18: PSA
min
← PSA
i
[ j + 1]
19: else
20: if δ > 12m then
21: PSA
max
← PSA
i
[ j + 1]
22: date PSA
max
← getDate(PSA
max
)
23: else
24: γ ← date
PSA[ j + 2] − date PSA
max
25: if j + 2 ≤ M and γ > 12 then
26: PSA
max
← PSA
i
[ j + 1]
27: date PSA
max
← getDate(PSA
max
)
28: end if
29: end if
30: end if
31: end if
32: end for
33: end if
34: if PSA
min
exists then
35: drop date ← get date(PSA
max
)
36: nadir date ← get date(PSA
min
)
37: return drop date,nadir date,PSA
min
38: end if
39: return NULL
PSA-based relapse occurred after radical prostate-
ctomy. If an ultrasensitive PSA (Shen et al., 2005)
measurement psa
j
was taken for patient p
i
then
we take this into consideration to define the max-
imum threshold (lines 3-7).
• PSA-based relapse after radiation therapy
PRT (p
i
): this function also uses the EAU guide-
lines (Van Den Broeck et al., 2020) to detect
whether a PSA-based relapse occurred after
radiation therapy. The algorithm searches for the
first increase of 2 PSA units from a nadir value.
Going beyond this, our novel BCR detection
method is not solely reliant on PSA relapse; instead, it
incorporates expert knowledge and translates it into a
new tool for detecting BCR based on secondary treat-
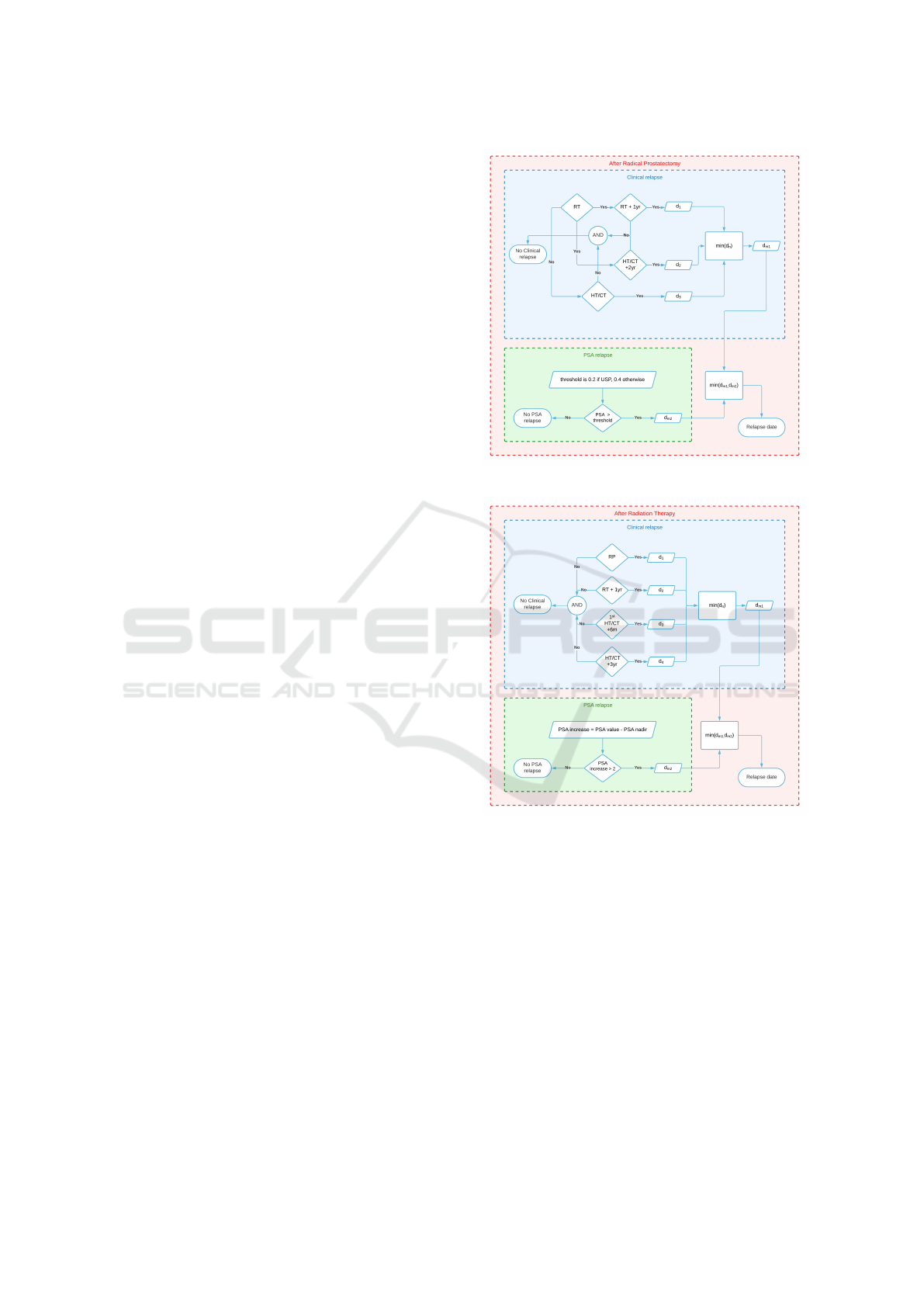
ments (Figures 2 and 3). The CRP and CRT functions
(Algorithms 2-3) have been developed to identify pos-
sible relapses that may have been missed (after an RP
or an RT primary treatment, respectively) either due
to the absence of PSA tests or because the curating
doctor decided on a secondary treatment before the
PSA value has crossed the EAU-guideline threshold.
The exact approaches used to define clinical relapse
after RP and RT primary treatments are described in
Figure 2 and Figure 3, respectively.
Algorithm 2: CRP - Clinical Relapse after RP.
Input: p
i
Output: d
m1
1: L ← [ ]
2: if lastRT Date(p
i
) > f irstRPDate(p
i
) then
3: if lastRT Date(p
i
) − f irstRPDate(p
i
) > 1yr then
4: L ← L + f irstRT DateA f terOneYear(p
i
)
5: end if
6: if hasHTCT (p
i
) and
lastHTCT Date(p
i
) > f irstRPDate(p
i
) then
7: if lastHTCT Date(p
i
) − f irstRPDate(p
i
) ≥ 2yr then
8: L ← L + f irstHTCT DateA f terOneYear(p
i
)
9: end if
10: end if
11: else
12: if hasHTCT (p
i
) and
lastHTCT Date(p
i
) > f irstRPDate(p
i
) then
13: L ← L + f irstHTCT DateA f terRp(p
i
)
14: end if
15: end if
16: d
m1
← getMin(L)
17: return d
m1
HEALTHINF 2024 - 17th International Conference on Health Informatics
80
Algorithm 3: CRT - Clinical Relapse after RT.
Input: p
i
Output: d
m1
1: L ← [ ]
2: if hasRP(p
i
) and
lastRPDate(p
i
) > f irstRT Date(p
i
) then
3: L ← L + f irstRpDateA f terRt(p
i
)
4: end if
5: if hasSecondRT (p
i
) and
secondRT Date(p
i
) − f irstRT Date(p
i
) > 1yr then
6: L ← L + secondRT Date(p
i
)
7: end if
8: if hasHTCT (p
i
) and
f irstHT CT Date(p
i
) − f irstRT Date(p
i
) ≥ 6m then
9: L ← L + f irstHT CT Date(p
i
)
10: end if
11: if hasHTCT (p
i
) and
f irstHT CT Date(p
i
) − f irstRT Date(p
i
) > 3yr then
12: L ← L + f irstHT CT Date(p
i
)
13: end if
14: d
m1
← getMin(L)
15: return d
m1
The DBCR Algorithm then uses all the outputs of
the above functions, namely the dates (d
1
, d
2
.d
3
, d
4
)
of possible BCR occurrences, and selects the earliest
date (if it exists) as the date of biochemical recurrence
for patient p
i
(Algorithm 7 lines 7-10).
2.4 Evaluation
Retrieving missing data is of utmost importance in
the pre-processing of EHR data for critical and sensi-
tive applications. Additionally, assessing the quality
of imputed data holds significant value as it provides
insights into the effectiveness of the methods and al-
gorithms employed. In our study, data evaluation in-
volves a two-tier validation process.
The first level (a.k.a. ’step-1’ evaluation) employs
automated tests, where we verify the accuracy of our
algorithms by taking records without missing treat-
ment data, applying the imputation algorithm, and
subsequently scrutinising the outcomes.
The second level (a.k.a. the ’step-2’ evaluation)
entails expert validation, wherein a random selection
of imputed data is manually inspected by domain ex-
perts, ensuring its correctness.
Due to the absence of biochemical recurrence data
in our EHR, we assessed the effectiveness of the
DBCR algorithms by manually evaluating patient out-
comes and employing descriptive statistics.
Figure 2: BCR definition after radical prostatectomy.
Figure 3: BCR definition after radiation therapy.
3 RESULTS
3.1 Curated Database
The initial phase of this work was to explore the HUS
datalake (Bruck, 2023; Pylk
¨
as, 2023) and extract the
most accurate and comprehensive prostate cancer data
suitable for subsequent medical research applications.
As a result, we successfully created a structured and
curated database that contains crucial patient informa-
tion, as defined in Table 3.
Synergizing Data Imputation and Electronic Health Records for Advancing Prostate Cancer Research: Challenges, and Practical
Applications
81

Table 2: Evaluation of DTX algorithm performance.
- Available CTx Estimated CTx Correct estimated CTx New estimated CTx
- DB DTX DTX ∩ DB True-Class False-Class DTX \ DB
PID 7563 9725 6962 (92%) 6294 (90%) 668 (10%) 2763 (+27%)
PID-RP 2495 2722 2233 (90%) 1929 (86%) 304 (14%) 0489 (+16%)
PID-RT 5068 7003 4729 (93%) 4365 (92%) 364 (08%) 2274 (+31%)
Table 3: The curated data tables.
Data Number of rows (%) Number of Patients (%)
T1: Pathology 23,393 12,277
GG1 6618 (28) 3652 (30)
GG2 6383 (27) 3313 (27)
GG3 4747 (20) 2503 (20)
GG4 2310 (10) 1195 (10)
GG5 3335 (14) 1614 (13)
T2: Treatment 40,369 9800
RP 2743 (7) 2742
RT 18,254 (45) 7248
HT 15,804 (39) 4088
CT 3568 (9) 514
T3: PSA 1,424,440 238,399
T4: MRI 20,103 15,807
T5: Medications 13,837,600 290,055
GG1–GG5 = Gleason grade group 1–5 (associated to each pathological entry)
RP=Radical prostatectomy, RT=Radiation therapy,
HT=Hormonal therapy, CT=Chemotherapy.
3.2 Treatments Data
Following the data curation and structuring, we have
implemented the DTX algorithm in order to detect
and impute the missing curative treatment data. As
a result, our database now incorporates n=2763 new
PCa-related treatment records, representing a 27% in-
crease compared to the original data found in the HUS
datalake. The number of patients with RP has in-
creased by 16% (n=489), while the number of those
with RT has increased by 31% (n=2274).
In Table 2 we present the results of ’step-1’ DTX
performance evaluation, i.e., estimated vs. known
(EHR-available) treatment records. We record an im-
putation performance of 92% (n=6962) correct esti-
mated curative treatments, i.e., treatments estimated
using the DTX algorithm that are also found in the ex-
isting database. Out of these, 90% (n=6294) are cor-
rectly classified as RP or RT, whereas 10% (n=669)
are wrongly classified. RP classification was 86%
correct, whereas RT classification reached 92%.
The ’step-2’ evaluation of the DTX algorithm was
performed vs. manual validation by domain experts,
where the experts were using the entire collection of
unstructured reports associated with the test subjects
in order to uncover their treatment history. The ’step-
2’ evaluation started by sampling 40 random patients,
i.e., 20 random RP + 20 random RT, that were de-
tected by the algorithm as having curative treatments
(CTx), however this treatment did not appear within
the EHR (DTX \ DB in Table 2). The results of this
manual validation are summarised in Table 4. Only
one patient from the RP group was unverifiable (no
data = treatment cannot be confirmed), while five RT
patients had the same situation. In addition, 95% of
RP patients were confirmed to have a curative PCa
treatment, and 60% of RT patients were confirmed.
In total 79% of the sampled patients (whose treat-
ments were not recorded within EHR) were confirmed
to have PCa curative treatment.
Table 4: Manual validation for DTX algorithm perfor-
mance.
- Sample Unverifiable Verifiable True CTx All True CTx
PID-RP 20 01 19 (95%) 18 (95%)
27 (79%)
PID-RT 20 05 15 (75%) 09 (60%)
3.3 BCR Data
Our DBCR algorithm successfully identified 2851 pa-
tients (Figures 4 and 5) who developed a biochemical
recurrence after a PCa curative treatment. These pa-
tients represent 27% of the treated patients.
Among the identified BCR patients, 70%
(n=2007) were detected using the PRP and PRT
algorithms, which are in accordance with the EAU
guidelines (Van Den Broeck et al., 2020).
However, around 30% (n=844) were identified us-
ing our new algorithms, CRP and CRT, formulated
based on the expertise of our clinicians’ team and
other contributors to this work. Notably, without ap-
plying these new algorithms, these cases might have
otherwise gone unnoticed.
Figure 4: BCR detected data: Bar plots representing the
distribution of the time from curative treatment to relapse.
HEALTHINF 2024 - 17th International Conference on Health Informatics
82
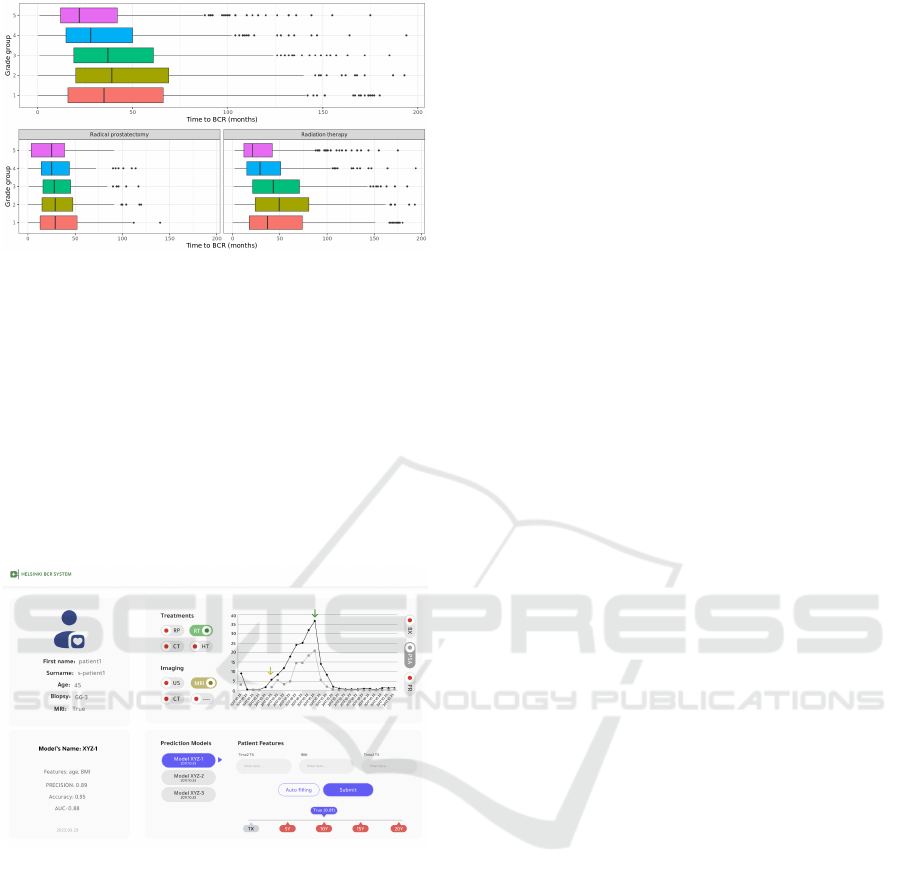
Figure 5: BCR detected data: Box plots of the time from
curative treatment to relapse distribution, by Gleason grade
groups, and by type of curative treatments.
3.4 Applications
After cleaning the data and improving its quality, we
have successfully carried out multiple applications.
The primary application involved developing a data
visualisation tool, enabling clinicians and researchers
to visualise the trajectory of PCa patients, includ-
ing their PSA values, treatments, pathological results,
medical prescriptions, and others (Figure 6).
Figure 6: Helsinki BCR system enables access to prostate
cancer patients’ trajectory and different BCR prediction
models.
Additionally, we have investigated the potential
grade inflation in PCa Gleason grade groups during
the MRI era; the research focused on patients in Glea-
son grade groups 1 and 2. This work hypothesised
that some patients in grade group 1 in the pre-MRI era
are nowadays, in the MRI era, classified and treated
as grade group 2 patients. With enough evidence, the
work proved the hypothesis which will open serious
discussions to reassess current risk stratification tools
and clinical decision-making. Updating guidelines on
cancer grading and treatments is crucial to be aligned
with the precision of modern MRI technology.
Furthermore, we are utilising the curated EHR
data to train machine learning models to predict
biochemical recurrence within the following 3-to-10
years from initial curative treatment. Knowing that
prostate cancer is a slow-developing cancer, BCR is
one of the most important and accurate surrogates
for prostate cancer mortality. Therefore, predicting
BCR would have a significant impact on treatment
decisions and treatment planning. Our (preliminary)
trained models achieved good performance (Accu-
racy=0.93, AUC=0.93, Precision=0.88) on an internal
validation. The models are trained on n=5262 patients
who have had PCa curative treatment.
4 DISCUSSION
Ensuring high data quality is essential when building
effective AI models and conducting significant statis-
tical analyses (Gudivada et al., 2017). This impor-
tance is particularly heightened in clinical research
and applications where decisions may directly impact
patients’ lives. Electronic Health Records (EHR),
such as the one available at HUS Acamedic, play
a critical role in this process, making it imperative
to develop robust exploration methods to harness the
available data.
In our work, we explored, curated, and aug-
mented bio-medical data from within Finnish health-
care records, with a specific focus on prostate cancer
patients. By establishing a new mining framework
and developing novel analysis algorithms, we suc-
cessfully consolidated our data, enabling us to con-
duct meaningful and impactful medical research.
One of our approaches was to use the time se-
ries data on patients’ PSA levels, a subset of medi-
cal data which is typically well collected and curated
within EHR, in order to infer the existence, and the
type, of EHR-missing curative treatment events. To
our knowledge, this is the first time PSA time series
data were used in this way, although, in (Bettencourt-
Silva et al., 2015), the authors employed a similar ap-
proach to generate a completeness score for the over-
all data quality of the cohort. Based on this approach,
we were able to consolidate our EHR by adding ap-
prox. 2.8k new curative treatment events, represent-
ing a 27% increase from the EHR-available treatment
events.
Another important outcome of our mining frame-
work was documenting the status and timing of our
PCa patients’ BCR. Differently than in previous EHR
mining frameworks for PCa medical data, see e.g.
(Park et al., 2021b; Park et al., 2021a), we define
BCR-status based on both PSA-level measurements
(after primary curative treatment, i.e., radiation ther-
apy –RT– or radical prostatectomy –RP–) as well
Synergizing Data Imputation and Electronic Health Records for Advancing Prostate Cancer Research: Challenges, and Practical
Applications
83

as based on secondary curative and adjuvant thera-
pies, i.e., PCa related hormonal- and/or chemother-
apy. This approach takes into consideration the clini-
cal reality that sometimes, curating doctors decide on
secondary therapies before the PSA level crosses the
threshold established by current EAU guidelines as
the BCR level. Using this approach, we accurately
captured an additional 844 BCR events (representing
a 42% increase from PSA-only detected BCR events),
which otherwise would either not have been found at
all or would have been given a significant later time-
stamp.
One important observation from our EHR data cu-
ration and analysis work is that there exists a large
amount of redundancy in these data sources. This is
particularly observable within the free text input writ-
ten by doctors during their medical checkups and/or
lab, pathological, or imaging reports. On the other
hand, due to a multitude of factors, including human
error, focusing on only one particular type of data
source at a time, such as lab results, pathological re-
ports, or even surgery records, one encounters a sig-
nificant amount of missing data entries.
Therefore, leveraging the data redundancy feature
in EHR not only makes it possible and highly ad-
vantageous to recover these missing data entries but
also validates and assesses the outcomes of our algo-
rithms. This is why, a ”data investigation” approach,
such as the one described in this manuscript, is more
relevant than classical ”data imputation” methods. In-
deed, these latter approaches provide only average-
like behaviours and also are completely inefficient in
detecting missing events, such as a radiation treatment
event altogether missing from within the EHR.
Strongly connected to the above reasoning, one
could not overlook the potential impact the use
of Large Language Models (LLM) could have in
detecting and augmenting the existing EHR data
(Thirunavukarasu et al., 2023). Such models could be
employed to extract (from the free text provided by
doctors) relevant information such as missing events,
e.g. treatments performed in different clinics, cities,
or even countries, or information that is usually not
structurally recorded within EHR, e.g., family history,
use of alcohol and tobacco products, general health
status of the patient, etc. During the current EHR
data analysis no LLM was employed; however, the
approach is currently actively analysed for future us-
age within our models.
5 CONCLUSION
This work demonstrates the challenges of mining
Finnish electronic health records for prostate cancer
(PCa) research, as well as the opportunities it of-
fers in gaining valuable insights. Our methodology,
when applied to the HUS datalake, enabled the de-
tection of missing treatments and biochemical recur-
rences (BCR), which led to a range of clinically rel-
evant findings, including patients’ timeline histories,
the Gleason grade group inflation finding, and the
BCR classification models. The results of our frame-
work highlight the potential of EHR data mining to
advance PCa research and guide personalised patient
care.
ACKNOWLEDGEMENTS
This work was supported by grants from the Can-
cer Society Finland, the Academy of Finland, Jane
and Aatos Erkko Foundation, and State funding for
university-level health research. It is a joint effort of
doctoral students, senior researchers, and clinicians at
the University of Helsinki and the University Hospital
of Helsinki.
REFERENCES
Ajmal, S., Ahmed, A. A. I., and Jalota, C. (2023). Natural
Language Processing in Improving Information Re-
trieval and Knowledge Discovery in Healthcare Con-
versational Agents. Journal of Artificial Intelligence
and Machine Learning in Management, 7(1):34–47.
Artibani, W., Porcaro, A. B., De Marco, V., Cerruto, M. A.,
and Siracusano, S. (2018). Management of Biochem-
ical Recurrence after Primary Curative Treatment for
Prostate Cancer: A Review. Urologia Internationalis,
100(3):251–262.
Bettencourt-Silva, J. H., Clark, J., Cooper, C. S., Mills, R.,
Rayward-Smith, V. J., and de la Iglesia, B. (2015).
Building data-driven pathways from routinely col-
lected hospital data: A case study on prostate cancer.
JMIR Med Inform, 3(3):e26.
Bruck, O. (2023). Welcome to the hospital dis-
trict of helsinki and uusimaa (hus) hematological
subdatalake data catalogue. Available online at:
https://www.oscarbruck.fi/datalake/. Accessed 2023-
08-22.
De La Torre-D
´
ıez, I., Gonz
´
alez, S., and L
´
opez-Coronado,
M. (2013). EHR Systems in the Spanish Public Health
National System: The Lack of Interoperability be-
tween Primary and Specialty Care. Journal of Medical
Systems, 37(1):9914.
HEALTHINF 2024 - 17th International Conference on Health Informatics
84

Gudivada, V., Apon, A., and Ding, J. (2017). Data quality
considerations for big data and machine learning: Go-
ing beyond data cleaning and transformations. Inter-
national Journal on Advances in Software, 10(1):1–
20.
Henkel, M., Horn, T., Leboutte, F., Trotsenko, P., Dugas,
S. G., Sutter, S. U., Ficht, G., Engesser, C., Matthias,
M., Stalder, A., Ebbing, J., Cornford, P., Seifert,
H., Stieltjes, B., and Wetterauer, C. (2022). Ini-
tial experience with AI Pathway Companion: Eval-
uation of dashboard-enhanced clinical decision mak-
ing in prostate cancer screening. PLOS ONE,
17(7):e0271183.
Herp, J., Braun, J.-M., Cantuaria, M. L., Tashk, A., Ped-
ersen, T. B., Poulsen, M. H. A., Krogh, M., Nadimi,
E. S., and Sheikh, S. P. (2023). Modeling of electronic
health records for time-variant event learning beyond
bio-markers—a case study in prostate cancer. IEEE
Access, 11:50295–50309.
Holmes, J. H., Beinlich, J., and Boland, M. R. (2021). Why
Is the Electronic Health Record So Challenging for
Research and Clinical Care? Methods of information
in medicine, 60(1-02):32–48.
Javaid, M., Haleem, A., Singh, R. P., Suman, R., and Rab,
S. (2022). Significance of machine learning in health-
care: Features, pillars and applications. International
Journal of Intelligent Networks, 3:58–73.
Knighton, A. J., Belnap, T., Brunisholz, K., Huynh, K., and
Bishoff, J. T. (2016). Using Electronic Health Record
Data to Identify Prostate Cancer Patients That May
Qualify for Active Surveillance. EGEMS (Washing-
ton, DC), 4(3):1220.
Kong, A., Liu, J. S., and Wong, W. H. (1994). Se-
quential imputations and bayesian missing data prob-
lems. Journal of the American Statistical Association,
89(425):278–288.
kuorttinen, E. (2023). HUS Acamedic - secure
operating environment. Available online at:
https://www.hus.fi/en/research-and-education/hus-
acamedic-secure-operating-environment. Accessed
2023-08-22.
Misukka, M. (2022). Standardizing electronic health
records in order to advance secondary use of hospital
data lakes - A case study on HUS data lake. Master’s
thesis, Aalto University. School of Science.
National Cancer Institute (2023). The natural his-
tory of prostate cancer. Available online at:
https://www.cancer.gov/types/prostate. Accessed
2023-08-03.
Park, J., Rho, M. J., Moon, H. W., Kim, J., Lee, C., Kim,
D., Kim, C.-S., Jeon, S. S., Kang, M., and Lee, J. Y.
(2021a). Dr. answer ai for prostate cancer: Predict-
ing biochemical recurrence following radical prostate-
ctomy. Technology in Cancer Research & Treatment,
20.
Park, J., Rho, M. J., Moon, H. W., Park, Y. H., Kim, C.-S.,
Jeon, S. S., Kang, M., and Lee, J. Y. (2021b). Prostate
cancer trajectory-map: clinical decision support sys-
tem for prognosis management of radical prostatec-
tomy. Prostate International, 9(1):25–30.
Pylk
¨
as, J. (2023). HUS facilitates clinical data ex-
ploitation through data lake. Available on-
line at: https://www.tietoevry.com/en/success-
stories/2019/hus-facilitates-clinical-data-exploitation-
through-an-integrated-hus-datalake-solution/. Ac-
cessed 2023-08-22.
Schafer, J. L. (1999). Multiple imputation: a primer. Statis-
tical Methods in Medical Research, 8(1):3–15. PMID:
10347857.
Seneviratne, M. G., Banda, J. M., Brooks, J. D., Shah,
N. H., and Hernandez-Boussard, T. M. (2018). Identi-
fying Cases of Metastatic Prostate Cancer Using Ma-
chine Learning on Electronic Health Records. AMIA
... Annual Symposium proceedings. AMIA Symposium,
2018:1498–1504.
Shen, S., Lepor, H., Yaffee, R., and Taneja, S. S.
(2005). ULTRASENSITIVE SERUM PROSTATE
SPECIFIC ANTIGEN NADIR ACCURATELY PRE-
DICTS THE RISK OF EARLY RELAPSE AFTER
RADICAL PROSTATECTOMY. Journal of Urology,
173(3):777–780.
Stephenson, A. J., Kattan, M. W., Eastham, J. A., Dotan,
Z. A., Bianco, F. J., Lilja, H., and Scardino, P. T.
(2006). Defining biochemical recurrence of prostate
cancer after radical prostatectomy: A proposal for a
standardized definition. Journal of Clinical Oncology,
24(24):3973–3978. PMID: 16921049.
The American Cancer Society medical and editorial con-
tent team (2019). Prostate cancer. Available
online at: https://www.cancer.org/cancer/prostate-
cancer/about/what-is-prostate-cancer.html. Accessed
2023-08-03.
Thirunavukarasu, A. J., Ting, D. S. J., Elangovan, K.,
Gutierrez, L., Tan, T. F., and Ting, D. S. W. (2023).
Large language models in medicine. Nature Medicine,
29(8):1930–1940.
Van Den Broeck, T., Van Den Bergh, R. C., Briers, E.,
Cornford, P., Cumberbatch, M., Tilki, D., De San-
tis, M., Fanti, S., Fossati, N., Gillessen, S., Grum-
met, J. P., Henry, A. M., Lardas, M., Liew, M., Ma-
son, M., Moris, L., Schoots, I. G., Van Der Kwast,
T., Van Der Poel, H., Wiegel, T., Willemse, P.-P. M.,
Rouvi
`
ere, O., Lam, T. B., and Mottet, N. (2020). Bio-
chemical Recurrence in Prostate Cancer: The Euro-
pean Association of Urology Prostate Cancer Guide-
lines Panel Recommendations. European Urology Fo-
cus, 6(2):231–234.
Wald, A. (1949). Note on the consistency of the maxi-
mum likelihood estimate. The Annals of Mathemat-
ical Statistics, 20(4):595–601.
Yadav, P., Steinbach, M., Kumar, V., and Simon, G. (2018).
Mining Electronic Health Records (EHRs): A Survey.
ACM Computing Surveys, 50(6):1–40.
Zhao, Y., Tao, Z., Li, L., Zheng, J., and Chen, X. (2022).
Predicting biochemical-recurrence-free survival using
a three-metabolic-gene risk score model in prostate
cancer patients. BMC Cancer, 22(1):239.
Synergizing Data Imputation and Electronic Health Records for Advancing Prostate Cancer Research: Challenges, and Practical
Applications
85

APPENDIX
Algorithm 4: DTX - Missing treatments detection.
Input: PAT IENT S LIST
Output: L
1: L ← [ ]
2: for all p
i
in PAT IENT S LIST do
3: PSA
i
← getPsa(p
i
)
4: T x
i
← getTreatments(p
i
)
5: (d
max
, d
min
, PSA
min
) ← SIGDROP(PSA
i
)
6: if TxExists(d
max
, d
min
, PSA
min
, T x
i
) = False then
7: if PSA
min
< 0.1 then
8: tx type ← ’RP’
9: else
10: tx type ← ’RT’
11: end if
12: L ← L + (p
i
,tx type, drop date)
13: end if
14: end for
15: return L
Algorithm 5: PRP - PSA Relapse after RP.
Input: p
i
Output: d
m2
1: PSA ← getPsaA f terRp(p
i
)
2: for psa
j
in PSA do
3: if usp(psa
j
) = TRUE then
4: th ← 0.2
5: else
6: th ← 0.4
7: end if
8: if psa
j
> th then
9: d
m2
← getDate(psa
j
)
10: return d
m2
11: end if
12: end for
13: return NULL
Algorithm 6: PRT - PSA Relapse after RT.
Input: p
i
Output: d
m2
1: PSA ← getPsaA f terRt(p
i
)
2: nadir ← getMax(PSA)
3: for psa
j
in PSA do
4: if nadir > psa
j
then
5: nadir ← psa
j
6: end if
7: inc ← psa
j
− nadir
8: if inc > 2 then
9: d
m2
← getDate(psa
j
)
10: return d
m2
11: end if
12: end for
13: return NULL
Algorithm 7: DBCR.
Input: TREATED PATIENT S
Output: L
bcr
1: L
bcr
← [ ]
2: for all p
i
in TREAT ED PAT IENT S do
3: d
1
← PRP(p
i
)
4: d
2
← CRP(p
i
)
5: d
3
← PRT (p
i
)
6: d
4
← CRT (p
i
)
7: if allAreNU LL(d
1
, d
2
, d
3
, d
4
) = FALSE then
8: bcr date ← getMin(d
1
, d
2
, d
3
, d
4
)
9: new bcr ← (p
i
, bcr date)
10: L
bcr
← L
bcr
+ new bcr
11: end if
12: end for
13: return L
bcr
HEALTHINF 2024 - 17th International Conference on Health Informatics
86
