
Unsupervised Domain Adaptation for Medical Images with an Improved
Combination of Losses
Ravi Kant Gupta, Shounak Das and Amit Sethi
Department of Electrical Engineering, Indian Institute of Technology Bombay, Mumbai, India
Keywords:
Adversarial, Deep Learning, Domain Adaptation, Medical Image.
Abstract:
This paper presents a novel approach for unsupervised domain adaptation that is tested on H&E stained his-
tology and retinal fundus images. Existing adversarial domain adaptation methods may not effectively align
different domains of multimodal distributions associated with classification problems. Since our objective is to
enhance domain alignment and reduce domain shifts between these domains by leveraging their unique char-
acteristics, we propose a tailored loss function to address the challenges specific to medical images. This loss
combination not only makes the model accurate and robust but also faster in terms of training convergence.
We specifically focus on leveraging texture-specific features, such as tissue structure and cell morphology, to
enhance adaptation performance in the histology domain. The proposed method – Domain Adaptive Learning
(DAL) – was extensively evaluated for accuracy, robustness, and generalization. We conducted experiments
on the FHIST and a retina dataset and the results show that DAL significantly surpasses the ViT-based and
CNN-based state-of-the-art methods by 1.41% and 6.56% respectively for FHIST dataset while also showing
improved results for the retina dataset.
1 INTRODUCTION
In traditional supervised learning, a model is trained
using labeled data from the same domain as the test
data. Obtaining labels for medical data is challeng-
ing due to the intricacies of medical expertise, mak-
ing it costly and time-consuming. The need for
specialized knowledge, meticulous review, and ethi-
cal considerations contribute to the difficulty in ac-
quiring accurate and reliable annotations for medi-
cal datasets. However, when the distribution of the
source and target domains differs significantly, the
model’s performance may suffer due to the domain
shift. This domain shift can be because of color
variation, data acquisition bias, distributional differ-
ences, domain-specific factors, covariate shift, stain-
ing techniques in medical images, etc. Unsupervised
domain adaptation (UDA) techniques aim to miti-
gate this domain shift by aligning the feature distri-
butions or learning domain-invariant representations
by using only unlabeled samples from the target do-
main. By learning domain-invariant representations,
adversarial-based UDA models can effectively reduce
the domain discrepancy and improve the generaliza-
tion performance on the target domain. This ap-
proach has shown promising results in various do-
mains, such as image classification, object detec-
tion, and semantic segmentation. However, while
adversarial-based UDA has achieved notable success,
challenges still exist. These include addressing the
sensitivity to hyper-parameter tuning, handling the
high-dimensional feature space, and effectively cap-
turing complex domain shifts.
To address the aforementioned challenge, we de-
velop a UDA approach that surpasses the state-
of-the-art performance for medical images. We
present our findings from developing convolution
neural networks (CNNs) for such tasks based on
FHIST dataset (Shakeri et al., 2022), which is com-
posed of several histology datasets, namely CRC-
TP (Javed et al., 2020), LC25000 (Borkowski et al.,
), BreakHis (Spanhol et al., 2016), and NCT-CRC-
HE-100K (Kather et al., 2018). We framed our ex-
periments on CRCTP and NCT with six classes (Be-
nign, Tumor, Muscle, Stroma, Debris, and Inflamma-
tory). The t-distributed stochastic neighbor embed-
ding (tSNE) (van der Maaten and Hinton, 2008) plot
in Figure 1 of source data distribution (circle shape)
and target data distribution (square shape) while the
color of classes differs with light and dark versions of
the same color for the FHIST dataset. The sample im-
ages of each domain with different classes are shown
Gupta, R., Das, S. and Sethi, A.
Unsupervised Domain Adaptation for Medical Images with an Improved Combination of Losses.
DOI: 10.5220/0012328100003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 1, pages 205-215
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
205

in Figure 2. To show our model’s robustness we also
tested it on a retinal dataset from Glaucoma Screening
(AIROGS) challenge of ISBI-2021 (De Vente et al.,
2023). This retinal dataset is collected from three dif-
ferent centres and consists of two classes (glaucoma
positive and glaucoma Negative). Glaucoma is one of
the main causes of irreversible blindness and impaired
vision in the world. It affects the optic nerve, which
connects the eye with the brain, and leads to progres-
sive visual field damage. The sample images of each
domain with different classes are shown in Figure 3.
Our research attempts to meet three key objec-
tives: firstly, to reduce the discordance between
source and target domains in medical images; sec-
ondly, to harness the distinctive attributes of FHIST
and Retinal, such as cellular morphology and tissue
structure, to elevate adaptation performance specif-
ically within the medical domain; and finally, to
transcend the limitations of current UDA techniques,
to achieve state-of-the-art accuracy, resilience, and
generalization capabilities compared to the previous
methods.
Our adoption of deep learning for unsupervised
domain adaptation in medical images is driven by its
potential to enhance model generalization, extract op-
timal features, enable versatile cross-domain applica-
tions, and achieve field-advancing progress. By tai-
loring the combination of loss functions which leads
to improved convergence and robustness, and with the
leverage of deep learning’s power, we aim to surpass
current methods, benefiting various applications. In-
spired by a conditional domain adversarial network
(CDAN) (Long et al., 2018a), the core idea is to
simultaneously train a feature extractor (typically a
deep neural network) and a domain classifier (dis-
criminator) to distinguish between source and target
domains. We have examined different CNN-based
feature extractor as ResNet-50 (He et al., 2016a),
ResNet-101 (He et al., 2016a), ResNet-152 (He et al.,
2016a), VIT (Dosovitskiy et al., 2020), and Con-
vMixer (Trockman and Kolter, 2022) to extract mean-
ingful features. The feature extractor aims to learn
domain-invariant representations, while the domain
classifier tries to classify the domain of the extracted
features correctly. During training, the feature extrac-
tor and domain classifier are optimized in an adver-
sarial manner. The feature extractor aims to fool the
domain classifier by generating indistinguishable fea-
tures across domains, while the domain classifier tries
to classify the domains correctly. To achieve this, we
propose a combination of loss function pseudo label
maximum mean discrepancy (PLMMD) along with
other losses such as maximum information loss (en-
tropy loss) (Krause et al., 2010), maximum mean dis-
Figure 1: Snapshot of t-SNE plot of source (CRC-TP) (Cir-
cle shape) and target (NCT) (Square shape), clearly shows
significant difference between source and target data distri-
bution.
crepancy (MMD) loss (Gretton et al., 2012), mini-
mum class confusion (MCC) loss (Jin et al., 2020),
etc. This combination of loss functions has the fol-
lowing specific advantages : Employing MCC loss
enhances classification models by minimizing class
confusion, particularly in scenarios with imbalanced
class distributions. With maximum information loss,
our model is encouraged to learn tightly clustered
target features with uniform distribution, such that
the discriminative information in the target domain
is retained, while MDD loss measures the difference
between the mean embeddings of two distributions,
helping to quantify the dissimilarity between domains
and facilitating domain adaptation techniques. The
loss PLMMD enhances unsupervised domain adapta-
tion by selectively emphasizing domain-invariant fea-
tures through weight assignments. The benefit of this
loss is, that training convergence is faster as compared
to other scenarios. With the help of this novel com-
bination of the loss function our method surpasses
not only the CNN-based model state-of-the-art but
also the transformer-based model for the medical im-
ages. To justify our claims for medical images, we use
the FHIST dataset (Shakeri et al., 2022) and Retina
dataset.
Our stated goals were achieved by proposing an
improved combination of loss functions tailored to
address the unique challenges of H&E stained his-
tology images in FHIST dataset and Retinal dataset.
The performance evaluation was focused on accu-
racy, robustness, and generalization, to surpass state-
of-the-art techniques in both domains. Furthermore,
the research explored potential cross-domain applica-
tions in medical image analysis and computer vision,
offering promising advancements in practical unsu-
pervised domain adaptation with the help of various
BIOIMAGING 2024 - 11th International Conference on Bioimaging
206
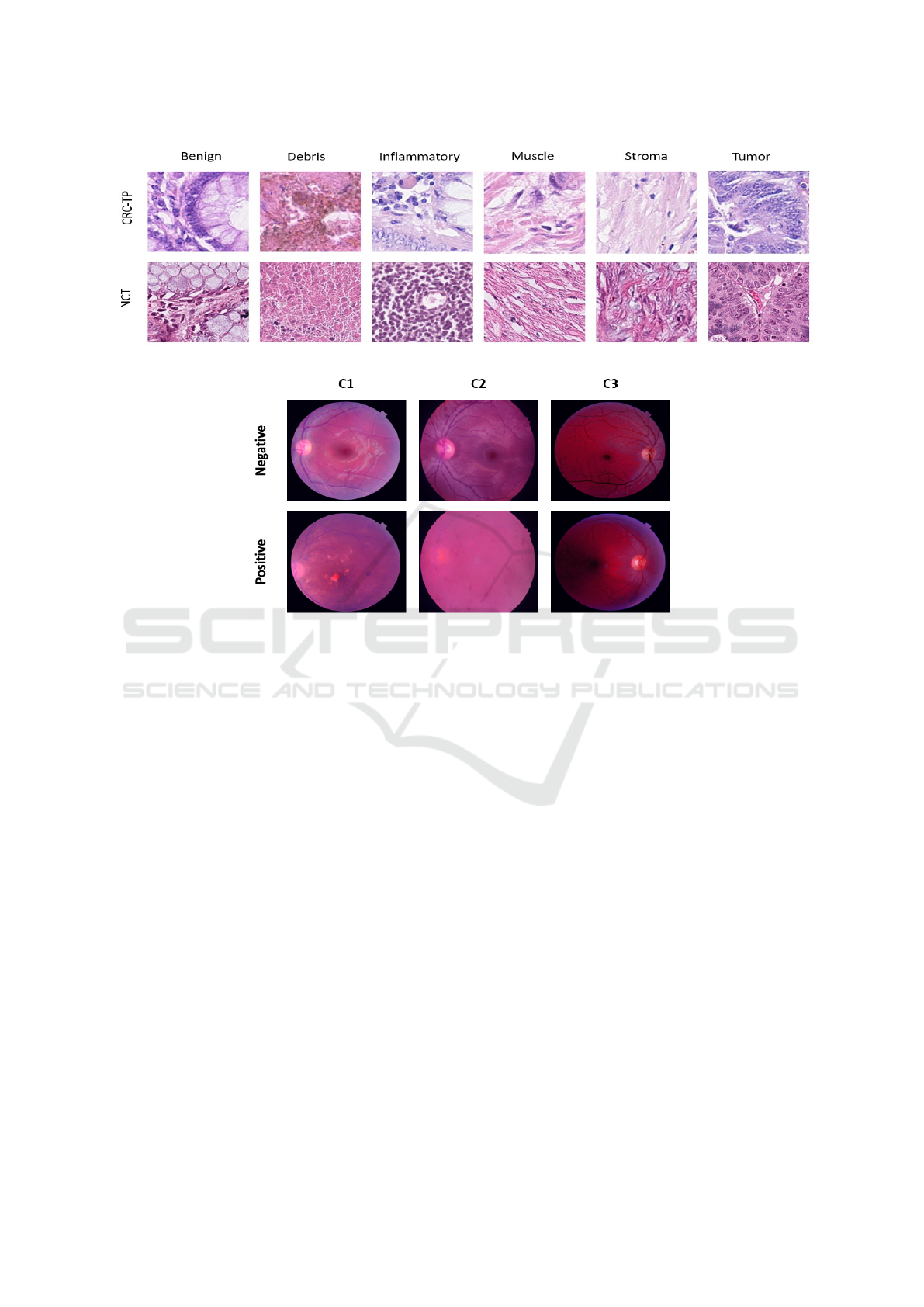
Figure 2: Snapshot of sample images of each class from CRC-TP (top row) and NCT (bottom row) of FHIST dataset.
Figure 3: Snapshot of sample images of each class from C1 (row) and NCT (bottom row) of Retinal dataset.
combinations of loss functions with different existing
models.
2 BACKGROUND AND RELATED
WORK
In unsupervised domain adaptation, we have a source
domain D
s
= {(x
s
i
,y
s
i
)}
n
s
i=1
of n
s
labeled examples
and a target domain D
s
= {(x
t
i
,y
t
i
)}
n
t
i=1
of n
t
unla-
beled examples. The source domain and target do-
main are sampled from joint distributions P(x
s
,y
s
)
and Q(x
t
,y
t
) respectively. Notably, the two distribu-
tions are initially not aligned, that is, P ̸= Q.
Domain adversarial neural network
(DANN) (Ganin et al., 2016) is a framework of
choice for UDA. It is a two-player game between
domain discriminator D, which is trained to distin-
guish the source domain from the target domain, and
the feature representation F trained to confuse the
domain discriminator D as well as classify the source
domain samples. The error function of the domain
discriminator corresponds well to the discrepancy be-
tween the feature distributions P( f ) and Q( f ) (Ganin
and Lempitsky, 2015), a key to bound the target risk
in the domain adaptation theory (Ben-David et al.,
2010).
Alignment-based domain adaptation is another
typical line of work that leverages a domain-
adversarial task to align the source and target do-
mains as a whole so that class labels can be trans-
ferred from the source domain to the unlabeled tar-
get one (Ganin et al., 2016; Pinheiro, 2018; Tzeng
et al., 2017; Zhang et al., 2018). Another typi-
cal line of work directly minimizes the domain shift
measured by various metrics, e.g., maximum mean
discrepancy (MMD) (Gretton et al., 2012). These
methods are based on domain-level domain align-
ment. To achieve class-level domain alignment, the
works of (Long et al., 2018b; Pei et al., 2018) uti-
lize the multiplicative interaction of feature represen-
tations and class predictions so that the domain dis-
criminator can be aware of the classification bound-
ary. Based on the integrated task and domain classi-
fier, (Tang and Jia, 2020) encourages a mutually in-
hibitory relation between category and domain pre-
dictions for any input instance. The works of (Chen
et al., 2019a; Xie et al., 2018) align the labeled source
centroid and pseudo-labeled target centroid of each
shared class in the feature space. Some work uses
individual task classifiers for the two domains to de-
Unsupervised Domain Adaptation for Medical Images with an Improved Combination of Losses
207
tect non-discriminative features and reversely learn
a discriminative feature extractor (Lee et al., 2019;
Saito et al., 2017a; Saito et al., 2017b). Certain other
works focus attention on transferable regions to de-
rive a domain-invariant classification model (Kurmi
et al., 2019; Wang et al., 2019; Wen et al., 2019).
To help achieve target-discriminative features, (Kang
et al., 2018; Sankaranarayanan et al., 2017) generate
synthetic images from the raw input data of the two
domains via GANs (Goodfellow et al., 2014). The re-
cent work of (Chen et al., 2019b) improves adversar-
ial feature adaptation, where the discriminative struc-
tures of target data may be deteriorated (Xu et al.,
2019). The work of (Zhao et al., 2019) adapts the
feature norms of the two domains to a large range of
values so that the learned features are not only task-
discriminative but also domain-invariant.
3 PROPOSED METHOD
The challenge of domain shift in a cross-domain clas-
sification task using unsupervised domain adaptation
leverages the knowledge from a labeled source do-
main to improve the performance of a classifier on
an unlabeled target domain. We propose a tailored
loss function that minimizes the domain discrepancy
and aligns feature distributions across domains. Our
datasets even differ in image sizes for the source do-
main and the target domain. Before training, the im-
ages were subjected to data augmentation such as hor-
izontal flip, vertical flip, and normalization to ensure
consistency. To facilitate domain adaptation, we in-
troduce a structure-preserving colour normalization
technique to normalize the stain appearance of images
across domains (Vahadane et al., 2016). The normal-
ization process aims to preserve the local structure
while removing domain-specific variations. There-
fore, the images of both dataset were colour normal-
ized (Vahadane et al., 2016).
From the color-normalized images, we extracted
features using ResNet-52 trained on ImageNet (He
et al., 2016b). Our proposed model architecture is
based on a deep neural network with convolutional
and fully connected layers, specifically tailored for
domain adaptation.
In this work, we design a method to train a deep
network N : x → y which reduces the shifts in the
data distributions across domains, such that the tar-
get risk r
t
= E
(x
t
,y
t
)∼Q
[N(x
t
) ̸= y
t
] can be bounded by
the source risk r
s
= E
(x
s
,y
s
)∼P
[N(x
s
) ̸= y
s
] plus the dis-
tribution discrepancy disc(P, Q) quantified by a novel
conditional domain discriminator. To minimize do-
main cross-domain discrepancy (Ganin et al., 2016) in
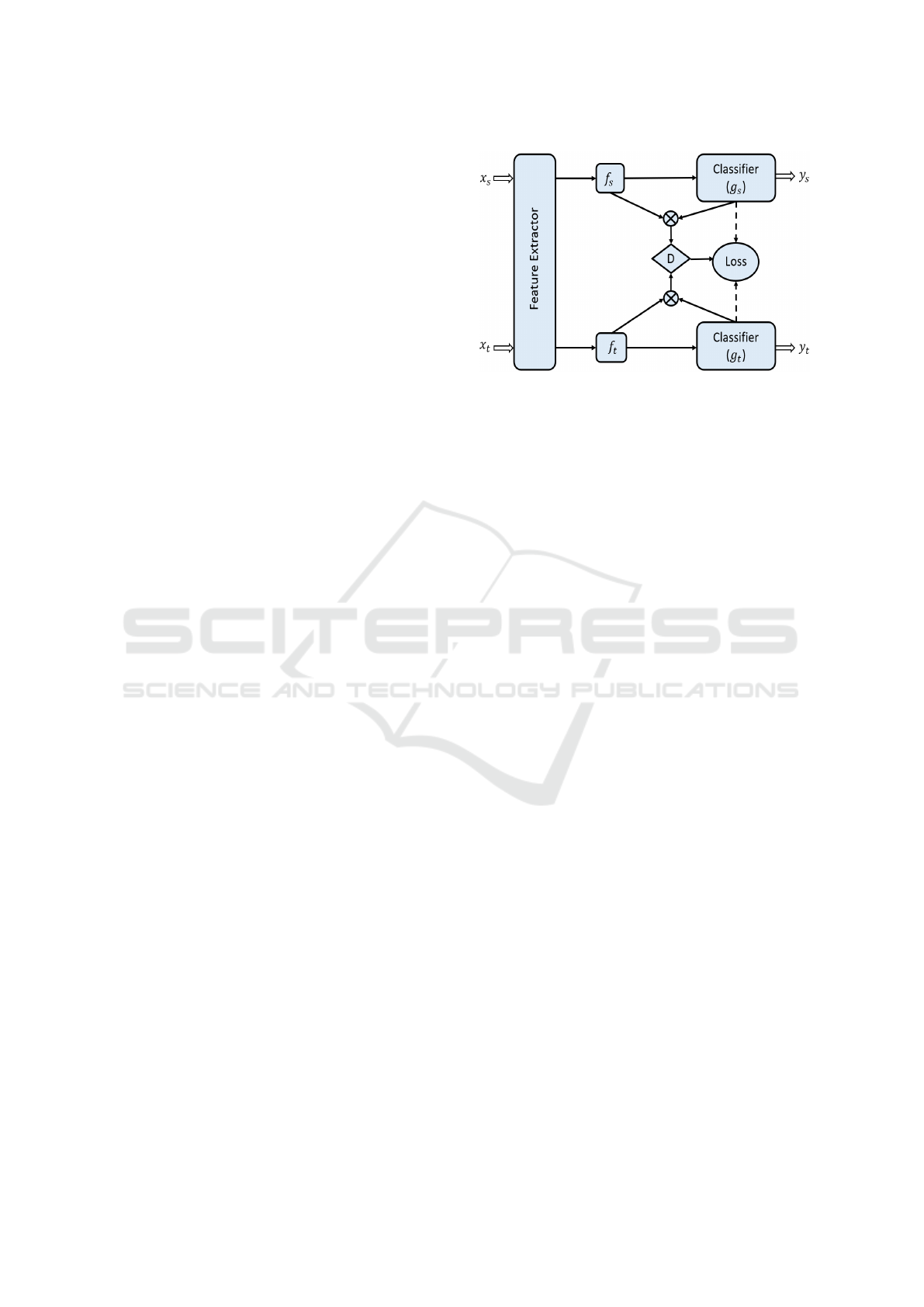
Figure 4: Architecture of the proposed networks, where
domain-specific feature representation f and classifier pre-
diction g embody the cross-domain gap to be reduced
jointly by the conditional domain discriminator D.
adversarial learning Generative Adversarial Networks
(GANs) (Goodfellow et al., 2014) play a vital role.
Features are represented by f = F(x) and classifier
prediction, g = N(x) generated from deep network N.
We improve existing adversarial domain adapta-
tion methods in two directions. First, when the joint
distributions of feature and class, i.e. P(x
s
,y
s
) and
Q(x
t
,y
t
), are non-identical across domains, adapting
only the feature representation f may be insufficient.
A quantitative study (Yosinski et al., 2014) shows that
deep representations eventually transition from gen-
eral to specific along deep networks, with transfer-
ability decreased remarkably in the domain-specific
feature layer f and classifier layer g. Second, due to
the nature of multi-class classification, the feature dis-
tribution is multimodal, and hence adapting feature
distribution may be challenging for adversarial net-
works.
By conditioning, domain variances in feature rep-
resentation f and classifier prediction g can be mod-
eled simultaneously. This joint conditioning allows us
to bridge the domain gap more effectively, enabling
the adapted model to capture and align the underly-
ing data distributions between the source and target
domains. Consequently, incorporating classifier pre-
diction as a conditioning factor in domain adaptation
holds great potential for achieving improved trans-
ferability and generating domain-invariant represen-
tations in challenging cross-domain scenarios.
We formulate Conditional Domain Adversarial
Network (CDAN) (Long et al., 2018a) as a mini-
max optimization problem with two competitive er-
ror terms: (a) E(N) on the source classifier N, which
is minimized to guarantee lower source risk; (b)
E(D,N) on the source classifier N and the domain
discriminator D across the source and target domains,
which is minimized over D but maximized over f =
BIOIMAGING 2024 - 11th International Conference on Bioimaging
208

F(x) and g = N(x):
L
clc
(x
s
i
,y
s
i
) = E
(x
s
i
,y
s
i
)∼D
s
L(N(x
s
i
),y
s
i
) (1)
L
dis
(x
s
,x
t
) = − E
x
s
i
∼D
s
log[D( f
s
i
,g
s
i
)]
− E
x
t
j
∼D
t
log[1 − D( f
t
j
,g
t
j
)],
(2)
where L is the cross-entropy loss, and h = ( f , g)
is the joint variable of feature representation f and
classifier prediction g. The minimax game of CDAN
is
min
N
L
clc
(x
s
i
,y
s
i
) − λL
dis
(x
s
,x
t
)
min
D
L
dis
(x
s
,x
t
),
(3)
where λ is a hyper-parameter between the two objec-
tives to trade off source risk and domain adversary.
We condition domain discriminator D on the clas-
sifier prediction g through joint variable h = ( f ,g) to
potentially tackle the two aforementioned challenges
of adversarial domain adaptation. A simple condi-
tioning of D is D( f ⊕ g), where we concatenate the
feature representation and classifier prediction in vec-
tor f ⊕g and feed it to conditional domain discrimina-
tor D. This conditioning strategy is widely adopted by
existing conditional GANs (Goodfellow et al., 2014).
However, with the concatenation strategy, f and g are
independent of each other, thus failing to fully cap-
ture multiplicative interactions between feature repre-
sentation and classifier prediction, which are crucial
to domain adaptation. As a result, the multimodal in-
formation conveyed in classifier prediction cannot be
fully exploited to match the multimodal distributions
of complex domains (Song et al., 2009). The multi-
linear map is defined as the outer product of multi-
ple random vectors. The multilinear map of infinite-
dimensional nonlinear feature maps has been suc-
cessfully applied to embed joint distribution or con-
ditional distribution into reproducing kernel Hilbert
spaces (Song et al., 2009; Song and Dai, 2013). Be-
sides the theoretical benefit of the multilinear map
x ⊗ y over the concatenation x ⊕ y (Song et al., 2009;
Song et al., 2013). Taking advantage of the multilin-
ear map, in this paper, we condition D on g with the
multilinear map. Superior to concatenation, the mul-
tilinear map x ⊗ y can fully capture the multimodal
structures behind complex data distributions. A dis-
advantage of the multilinear map is dimension explo-
sion.
We enable conditional adversarial domain adapta-
tion over domain-specific feature representation f and
classifier prediction g. We jointly minimize with re-
spect to (1) source classifier N and feature extractor F,
minimize (2) domain discriminator D, and maximize
(2) feature extractor F and source classifier N. This
yields the mini-max problem of Domain Adversarial
Networks:
min
G
E
(x
i
s
,y
i
s
)∼D
s
L(G(x
i
s
),y
i
s
)
+ λ
E
x
i
s
∼D
s
log[D(T (h
i
s
))]
+E
x
j
t
∼D
t
log[1 − D(T(h
j
t
))]
max
D
E
x
i
s
∼D
s
log[D(T (h
i
s
))] + E
x
j
t
∼D
t
log[1 − D(T(h
j
t
))],
(4)
where λ is a hyper-parameter between the source clas-
sifier and conditional domain discriminator, and note
that h = ( f , g) is the joint variable of domain-specific
feature representation f and classifier prediction g for
adversarial adaptation.
The general problem of adversarial domain adap-
tation of the proposed model for classification can be
formulated as follows:
L = min
N
L
clc
(x
s
i
,y
s
i
) − λL
dis
(x
s
,x
t
)
+βL
IM
+ γL
MCC
+ δL
MDD
+ ηL
W MMD
(5)
where λ, β, γ, δ and η are hyper parameters, L
MCC
is minimum class confusion loss, L
MDD
is maximum
mean discrepancy loss, L
W MDD
represents weighted
maximum mean discrepancy loss and L
IM
represents
information maximization loss. All individual losses
have their own specialty and this novel combina-
tion of loss significantly surpasses the performance
of CNN-based models as well as transformer-based
models. A detailed description of all the losses is
given below in the losses section.
3.1 Losses
3.1.1 Maximum Mean Discrepancy
Maximum mean discrepancy (MMD) is a kernel-
based statistical test used to determine whether given
two distributions are the same (Gretton et al., 2012).
Given an random variable X , a feature map φ maps X
to an another space F such that φ(X ) ∈ F. Assuming
F satisfies the necessary conditions, we can benefit
from the kernel trick to compute the inner product in
F:
X,Y such that k(X,Y ) = ⟨φ(X),φ(Y )⟩
F
, (6)
where k is gram matrix produced using the kernel
function.
MMD is the distance between feature means. That
means for a given probability measure P on X, fea-
ture means is an another feature map that takes φ(X)
and maps it to the means of every coordinate of φ(X):
µ
p
(φ(X)) = [E[φ(X
1
)],...., E[φ(X
m
)]]
T
(7)
Unsupervised Domain Adaptation for Medical Images with an Improved Combination of Losses
209

The inner product of feature means of X ∼ P and Y ∼
Q can be written in terms of kernel function such that:
⟨µ
p
(φ(X)),µ
q
(φ(Y ))⟩
F
= E
P,Q
[⟨φ(X),φ(Y )⟩
F
]
= E
P,Q
[k(X,Y )]
(8)
Given X, Y maximum mean discrepancy is the dis-
tance between feature means of X, Y :
MMD
2
(P,Q) = ||µ
P
− µ
Q
||
2
F
(9)
MMD
2
(P,Q) = ⟨µ
P
− µ
Q
,µ
P
− µ
Q
⟩
= ⟨µ
P
,µ
P
⟩ − 2⟨µ
P
,µ
Q
⟩ + ⟨µ
Q
,µ
Q
⟩
(10)
Using the equation (8), finally above expression be-
comes
L
MMD
= MMD
2
(P,Q)
= E
P
[k(X, X)] − 2E
P,Q
[k(X,Y )] + E
Q
[k(Y,Y )]
(11)
3.1.2 Pseudo Label Maximum Mean
Discrepancy
We calculated the PLMMD using a similar proce-
dure to calculating MMD loss in equation (11). How-
ever, our proposed loss differs in terms of weights as-
signed to each similarity term. Hence we can define
PLMMD loss as:
L
PLMMD
= w
XX
E
P
[k(X, X)] − 2w
XY
E
P,Q
[k(X,Y )]
+w
YY
E
Q
[k(Y,Y )],
(12)
where, w
XX
represent weight to get similarity within
the source domain, similarly, w
YY
are weights for sim-
ilarity within the target domain, and w
XY
are weights
to get similarity within source and target domain. For
calculating the weights, first, we generated pseudo
labels for the target using a source classifier. After
that, the source and target pseudo-label is normal-
ized to account for class imbalances. For each class
common to both datasets, dot products of normalized
vectors are computed to quantify instance relation-
ships. Calculated dot products are normalized by the
count of common classes, ensuring fairness. This re-
turns three weight arrays, representing relationships
between instances in the source dataset, target dataset,
and source-to-target pairs.
3.1.3 Minimum Class Confusion
The minimum class confusion loss L
MCC
(Jin et al.,
2020) seeks to minimize confusion terms between
classes j and j
′
, such that j ̸= j
′
where the indices
are exhaustive over the set of classes. On the target
domain, the class confusion term between two classes
j and j
′
is given by:
C
j j
′
=
ˆ
y
⊺
· j
ˆ
y
⊺
· j
′
A much more nuanced and meaningful formula-
tion of the class confusion would be:
C
j j
′
=
ˆ
y
⊺
· j
W
ˆ
y
⊺
· j
′
, (13)
where the matrix W is a diagonal matrix. The diago-
nal terms W
ii
are given as the softmax outputs of the
entropies in classifying a sample i.
ˆ
y
i j
is given as:
ˆ
y
i j
=
exp(Z
i j
/T )
∑
c
j
′
=1
exp(Z
i j
′
/T )
, (14)
where c is the number of classes, T is the tempera-
ture coefficient, and Z
i j
is the logistic output of the
classifier layer for the class j and the sample i.
After normalizing the class confusion terms, the
final MCC Loss function is given as:
L
MCC
=
1
c
c
∑
j=1
c
∑
j
′
̸= j
|C
j j
′
|, (15)
which is the sum of all the non-diagonal elements of
the class confusion matrix. The diagonal terms rep-
resent the ”certainty” in the classifier, while the non-
diagonal terms represent the ”uncertainty” in classi-
fication. The MCC loss can be added in conjunction
with other domain adaptation methods.
3.1.4 Information Maximization Loss
The Information Maximization loss is designed to
encourage neural networks to learn more informa-
tive representations by maximizing the mutual infor-
mation between the learned features and the input
data (Krause et al., 2010). This type of loss aims
to guide the model to capture relevant and distinctive
patterns in the data, which can be especially valu-
able in scenarios where unsupervised learning, do-
main adaptation, or feature learning are important.
The assumptions that p
t
= softmax(N( f (x
t
))) are ex-
pected to retain as much information about x
t
as pos-
sible, and decision boundary should not cross high-
density regions, but instead lie in low-density regions,
which is also known as cluster assumption. These
two assumptions can be met by maximizing mutual
information between the empirical distribution of the
target inputs and the induced target label distribution,
which can be formally defined as:
I(p
t
;x
t
) = H(
p
t
) −
1
n
t
n
t
∑
j=1
H(p
t j
)
= −
K
∑
k=1
p
tk
log(p
tk
) +
1
n
t
n
t
∑
j=1
K
∑
k=1
p
tk j
log(p
tk j
), (16)
where, p
t j
= softmax(G
c
(G
f
(x
t j
))), p
t
= E
x
t
[p
t
],
and K is the number of classes. Maximizing
BIOIMAGING 2024 - 11th International Conference on Bioimaging
210

Figure 5: Snapshots of 2D tSNE plots of the target (NCT) domain sample features before training (leftmost), after three
epochs (middle), and after six epochs (right).
−
1
n
t
∑
n
t
j=1
H(p
t j
) enforces the target predictions close
to one-hot encoding, therefore the cluster assumption
is guaranteed. To ensure global diversity, we also
maximize H(p
t
) to avoid every target data being as-
signed to the same class. With I(p
t
;x
t
), our model
is encouraged to learn tightly clustered target features
with uniform distribution, such that the discriminative
information in the target domain is retained.
4 EXPERIMENTATION AND
RESULTS
4.1 Dataset and Implementation
Dataset: To evaluate the proposed method, we
introduce the FHIST dataset, a proposed benchmark
for the few-shot classification of histological im-
ages (Shakeri et al., 2022) and Retinal Dataset from
ISBI-2021 Challenge(De Vente et al., 2023). FHIST
is composed of several histology datasets, namely
CRC-TP (Javed et al., 2020), LC25000 (Borkowski
et al., ), BreakHis (Spanhol et al., 2016), and NCT-
CRC-HE-100K (Kather et al., 2018). For each class,
there are close to 20,000 images in the CRC-TP
domain with an image size of 150X150 pixels and
around 10,000 images of size 224X224 pixels in
the NCT domain. We performed experiments with
CRC-TP as the source and NCT as the target and
vice versa. The tSNE plots shown in Figure 5 depict
the distribution of target (NCT) at different stages of
training. Different colors map different class types in
the tSNE plot. We have plotted five classes in tSNE
which are Benign, Tumor, Debris, Inflammatory,
and Muscle + Stroma with 200 sample points from
each five classes. We combined the last two classes
because of their physiological as well as feature
intertwining. The first plot(leftmost) shows the data
distribution of NCT(as target) at epoch 0, and the
second one shows the data distribution of NCT after
three epochs, and the last one (rightmost) shows
the target(NCT) data distribution after six epochs of
domain adaptation. These histology datasets consist
of different tissue types and different organs. We
consider each tissue type as a class label with one-hot
encoding in the classification task. We framed
our experiments on CRC-TP and NCT with six
classes (Benign, Tumor, Muscle, Stroma, Debris, and
Inflammatory). On the other hand, we demostrated
our algorithm on Retinal dataset, in which the images
were originally acquired for a diabetic retinopathy
screening program. We divided this dataset in three
different centres on the basis of center information
available in AIROGS challenge of ISBI-2021. This
dataset is having 2427 samples in Centre 1 (C1), 497
image samples in Centre 2 (C2) and 276 images in
Centre 3 (C3). All centres have images of different
sizes.
Implementation: All the experiments were con-
ducted on an NVIDIA A100 in PyTorch, using the
CNN-based neural network (ResNet-50) pre-trained
on ImageNet (He et al., 2016b) as the backbone
for our proposed model. The base learning rate is
0.00001 with a batch size of 32, and we train mod-
els by 20 epochs. The hyper-parameters were β=0.05
, γ=1.4 , δ=0.54 and η=0.54 for the experiments of
CRC-TP → NCT and NCT → CRC-TP as well as
for the experiments on the Retinal Dataset. We used
AdamW (Loshchilov and Hutter, 2019) with a mo-
mentum of 0.9, and a weight decay of 0.001 as the
optimizer. We adopt the standard protocol for un-
supervised domain adaptation (UDA) where all la-
beled source samples and unlabeled target samples
are utilized for training. To report our results for each
transfer task, we use center-crop images from the tar-
get domain and report the classification performance.
Unsupervised Domain Adaptation for Medical Images with an Improved Combination of Losses
211

Table 1: Accuracy (%) on the FHIST dataset (Shakeri et al., 2022) with two different UDA tasks and their average, where all
methods are fine-tuned on their respective backbone model.
Method Backbone Feature Extractor CRC-TP → NCT NCT → CRC-TP Average
ResNet-50 (He et al., 2016a) 40.7 32.9 36.8
DANN (Ganin et al., 2016) 73.5 66.6 70.0
CDAN (Long et al., 2018a) ResNet-50 66.2 61.4 63.8
GVB-GD (Cui et al., 2020) 73.9 66.7 70.3
CHATTY (Wagle et al., 2023) 81.6 67.9 74.7
TVT (Yang et al., 2023) ViT 86.4 73.2 79.8
Proposed Method ResNet-50 87.7 74.8 81.2
Table 2: Accuracy (%) on the Retinal dataset (De Vente et al., 2023) with four different UDA tasks.
Experiment Without Domain Adaptation Proposed Method
Trained on C1 and tested on C2 62.1 86.5
Trained on C1 and tested on C3 75.2 79.6
Trained on C1 + C2 and tested on C3 77.8 80.1
Trained on C1 + C3 and tested on C2 64.8 87.1
For a fair comparison with prior works, we also con-
duct experiments with the same backbone as ViT-
based (Dosovitskiy et al., 2020) as TVT (Yang et al.,
2023), ResNet-50 (He et al., 2016a), DANN (Ganin
et al., 2016), CDAN (Long et al., 2018a), GVB-
GD (Cui et al., 2020), CHATTY+MCC (Wagle et al.,
2023) on FHIST dataset.
4.2 Results
Our analysis in Table 1 depicts results with differ-
ent methods and feature extractors for the FHIST
dataset. The top five methods are CNN model us-
ing ResNet-50 as a feature extractor trained on Im-
ageNet dataset while TVT uses ViT based model
pre-trained on ImageNet-21k dataset. Our proposed
method is a CNN-based model that utilizes ResNet-50
as a backbone with a novel combination of loss func-
tions. Our model outperforms CNN-based models
such as ResNet-50, DANN, CDAN, GVB-GD, and
CHATTY+MCC, and surpasses the state of the Art
(SoTA) CNN results by 6.56%. At the same time, our
method also surpasses the transformer-based SoTA by
1.41%. We achieved an accuracy of 87.7% for CRC-
TP to NCT domain adaptation and 74.8% for NCT to
CRC-TP with an average accuracy of 81.26% for both
tasks, as mentioned in Table 1 with bold text. Table 2
depicts our model’s performance for Retinal dataset
by consider permutation combination of centres as
source and target respectively. Results in Table 2
shows comparison between with and without domain
adaptation. In our experiments, we explored the per-
formance of our model in domain adaptation scenar-
ios, particularly focusing on multiple source domains
and a single target domain. When we employed do-
main adaptation techniques and considered center C1
and C3 as sources, with C2 as the target, we achieved
an accuracy of 87.1%. In contrast, without domain
adaptation, the accuracy was notably lower at 64.8%.
Similarly, when we switched the roles of C1 and C2
as sources and evaluated against C3, we obtained ac-
curacy rates of 80.1% with domain adaptation and
77.8% without it. Furthermore, we conducted ex-
periments to emphasize the significance of domain
generalization in the presence of multiple source do-
mains. Specifically, when our model was trained on
C1 and tested on C2, it yielded accuracy of 86.5%
and 62.1% with and without domain adaptation, re-
spectively. Likewise, training on source C1 and tar-
geting C3 resulted in accuracy rates of 79.6% with
domain adaptation and 75.2% without domain adap-
tation. These findings underscore the importance of
domain adaptation and domain generalization tech-
niques in enhancing the robustness and adaptability of
our model across diverse source and target domains.
5 DISCUSSION AND
CONCLUSION
In this study, we have demonstrated that utilizing dif-
ferent combinations of loss functions with a CNN
such as ResNet-50 can lead to significant improve-
ments in unsupervised domain adaptation (UDA)
performance that can surpass the performance of
ViTs using other UDA methods. By leveraging the
strengths of various loss functions tailored to specific
domain characteristics, we have surpassed the state-
of-the-art (SOTA) performance for histology images.
We conducted ablation studies to understand the im-
pact of the different feature extractors such as Con-
BIOIMAGING 2024 - 11th International Conference on Bioimaging
212

vMixer (Trockman and Kolter, 2022) and ResNet-
101 (He et al., 2016a). However, the performance
in these cases was worse than our reported results.
To know the impact of individual loss and a com-
bination of losses, we performed extensive exper-
iments. Through comprehensive experiments, we
discovered that Minimum Class Confusion (MCC)
loss functions offer an enhancement to classifica-
tion models by mitigating class confusion, particu-
larly when faced with imbalanced class distributions.
In parallel, we observed that information maximiza-
tion losses aid the classifier in selecting the most cer-
tain samples for domain alignment. In our proposed
approach, the Pseudo Label Maximum Mean Dis-
crepancy (PLMMD) accelerates training convergence
(comparison with CHATTY model) and notably en-
hances domain alignment by incorporating weighted
considerations. Additionally, the Maximum Mean
Discrepancy (MMD) loss effectively narrows the gap
between the mean embeddings of the two distribu-
tions. By artfully combining these distinctive loss
functions, we not only surpass the current state-of-
the-art but also achieve a comprehensive solution that
advances the field of classification models in diverse
scenarios.
REFERENCES
Ben-David, S., Blitzer, J., Crammer, K., Kulesza, A.,
Pereira, F., and Vaughan, J. (2010). A theory of
learning from different domains. Machine Learning,
79:151–175.
Borkowski, A. A., Bui, M. M., Thomas, L. B., Wilson, C. P.,
DeLand, L. A., and Mastorides, S. M. Lc25000 lung
and colon histopathological image dataset.
Chen, C., Xie, W., Huang, W., Rong, Y., Ding, X., Huang,
Y., Xu, T., and Huang, J. (2019a). Progressive fea-
ture alignment for unsupervised domain adaptation.
In Proceedings of the IEEE/CVF conference on com-
puter vision and pattern recognition, pages 627–636.
Chen, X., Wang, S., Long, M., and Wang, J. (2019b). Trans-
ferability vs. discriminability: Batch spectral penal-
ization for adversarial domain adaptation. In Chaud-
huri, K. and Salakhutdinov, R., editors, Proceedings of
the 36th International Conference on Machine Learn-
ing, volume 97 of Proceedings of Machine Learning
Research, pages 1081–1090. PMLR.
Cui, S., Wang, S., Zhuo, J., Su, C., and Tian, Q. (2020).
Gradually vanishing bridge for adversarial domain
adaptation. pages 12452–12461.
De Vente, C., Vermeer, K., Jaccard, N., Wang, H., Sun, H.,
Khader, F., Truhn, D., Aimyshev, T., Zhanibekuly, Y.,
Le, T.-D., Galdran, A., Ballester, M., Carneiro, G.,
Devika, R., Hrishikesh, P., Puthussery, D., Liu, H.,
Yang, Z., Kondo, S., Kasai, S., Wang, E., Durvasula,
A., Heras, J., Zapata, M., Araujo, T., Aresta, G., Bo-
gunovic, H., Arikan, M., Lee, Y., Cho, H., Choi, Y.,
Qayyum, A., Razzak, I., Van Ginneken, B., Lemij, H.,
and Sanchez, C. (2023). Airogs: Artificial intelligence
for robust glaucoma screening challenge.
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn,
D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer,
M., Heigold, G., Gelly, S., et al. (2020). An image is
worth 16x16 words: Transformers for image recogni-
tion at scale. arXiv preprint arXiv:2010.11929.
Ganin, Y. and Lempitsky, V. (2015). Unsupervised do-
main adaptation by backpropagation. In International
conference on machine learning, pages 1180–1189.
PMLR.
Ganin, Y., Ustinova, E., Ajakan, H., Germain, P.,
Larochelle, H., Laviolette, F., March, M., and Lem-
pitsky, V. (2016). Domain-adversarial training of neu-
ral networks. Journal of machine learning research,
17(59):1–35.
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B.,
Warde-Farley, D., Ozair, S., Courville, A., and Ben-
gio, Y. (2014). Generative adversarial nets. In Ghahra-
mani, Z., Welling, M., Cortes, C., Lawrence, N., and
Weinberger, K., editors, Advances in Neural Infor-
mation Processing Systems, volume 27. Curran Asso-
ciates, Inc.
Gretton, A., Borgwardt, K. M., Rasch, M. J., Sch
¨
olkopf,
B., and Smola, A. (2012). A kernel two-sample test.
Journal of Machine Learning Research, 13(25):723–
773.
He, K., Zhang, X., Ren, S., and Sun, J. (2016a). Deep resid-
ual learning for image recognition. In Proceedings of
the IEEE conference on computer vision and pattern
recognition, pages 770–778.
He, K., Zhang, X., Ren, S., and Sun, J. (2016b). Deep resid-
ual learning for image recognition. In 2016 IEEE Con-
ference on Computer Vision and Pattern Recognition
(CVPR), pages 770–778.
Javed, S., Mahmood, A., Werghi, N., Benes, K., and Ra-
jpoot, N. (2020). Multiplex cellular communities in
multi-gigapixel colorectal cancer histology images for
tissue phenotyping. 29:9204–9219.
Jin, Y., Wang, X., Long, M., and Wang, J. (2020). Minimum
class confusion for versatile domain adaptation. In
Computer Vision–ECCV 2020: 16th European Con-
ference, Glasgow, UK, August 23–28, 2020, Proceed-
ings, Part XXI 16, pages 464–480. Springer.
Kang, G., Zheng, L., Yan, Y., and Yang, Y. (2018). Deep ad-
versarial attention alignment for unsupervised domain
adaptation: The benefit of target expectation maxi-
mization. In Computer Vision – ECCV 2018: 15th Eu-
ropean Conference, Munich, Germany, September 8-
14, 2018, Proceedings, Part XI, page 420–436, Berlin,
Heidelberg. Springer-Verlag.
Kather, J. N., Halama, N., and Marx, A. (2018). 100,000
histological images of human colorectal cancer and
healthy tissue.
Krause, A., Perona, P., and Gomes, R. (2010). Discrimina-
tive clustering by regularized information maximiza-
tion. In Lafferty, J., Williams, C., Shawe-Taylor, J.,
Zemel, R., and Culotta, A., editors, Advances in Neu-
Unsupervised Domain Adaptation for Medical Images with an Improved Combination of Losses
213

ral Information Processing Systems, volume 23. Cur-
ran Associates, Inc.
Kurmi, V. K., Kumar, S., and Namboodiri, V. P. (2019). At-
tending to discriminative certainty for domain adapta-
tion. CoRR, abs/1906.03502.
Lee, C., Batra, T., Baig, M. H., and Ulbricht, D. (2019).
Sliced wasserstein discrepancy for unsupervised do-
main adaptation. CoRR, abs/1903.04064.
Long, M., Cao, Z., Wang, J., and Jordan, M. I. (2018a).
Conditional adversarial domain adaptation. Advances
in neural information processing systems, 31.
Long, M., Cao, Z., Wang, J., and Jordan, M. I. (2018b).
Conditional adversarial domain adaptation. In Pro-
ceedings of the 32nd International Conference on
Neural Information Processing Systems, NIPS’18,
page 1647–1657, Red Hook, NY, USA. Curran As-
sociates Inc.
Loshchilov, I. and Hutter, F. (2019). Decoupled weight
decay regularization. In International Conference on
Learning Representations.
Pei, Z., Cao, Z., Long, M., and Wang, J. (2018). Multi-
adversarial domain adaptation. In Proceedings of the
Thirty-Second AAAI Conference on Artificial Intelli-
gence and Thirtieth Innovative Applications of Arti-
ficial Intelligence Conference and Eighth AAAI Sym-
posium on Educational Advances in Artificial Intelli-
gence, AAAI’18/IAAI’18/EAAI’18. AAAI Press.
Pinheiro, P. O. (2018). Unsupervised domain adaptation
with similarity learning. In Proceedings of the IEEE
conference on computer vision and pattern recogni-
tion, pages 8004–8013.
Saito, K., Ushiku, Y., Harada, T., and Saenko, K.
(2017a). Adversarial dropout regularization. CoRR,
abs/1711.01575.
Saito, K., Watanabe, K., Ushiku, Y., and Harada, T.
(2017b). Maximum classifier discrepancy for unsu-
pervised domain adaptation. CoRR, abs/1712.02560.
Sankaranarayanan, S., Balaji, Y., Castillo, C. D., and Chel-
lappa, R. (2017). Generate to adapt: Aligning do-
mains using generative adversarial networks. CoRR,
abs/1704.01705.
Shakeri, F., Boudiaf, M., Mohammadi, S., Sheth, I., Havaei,
M., Ayed, I. B., and Kahou, S. E. (2022). Fhist: a
benchmark for few-shot classification of histological
images. arXiv preprint arXiv:2206.00092.
Song, L. and Dai, B. (2013). Robust low rank kernel embed-
dings of multivariate distributions. In Burges, C., Bot-
tou, L., Welling, M., Ghahramani, Z., and Weinberger,
K., editors, Advances in Neural Information Process-
ing Systems, volume 26. Curran Associates, Inc.
Song, L., Fukumizu, K., and Gretton, A. (2013). Kernel em-
beddings of conditional distributions: A unified kernel
framework for nonparametric inference in graphical
models. IEEE Signal Processing Magazine, 30(4):98–
111.
Song, L., Huang, J., Smola, A., and Fukumizu, K. (2009).
Hilbert space embeddings of conditional distributions
with applications to dynamical systems. In Proceed-
ings of the 26th Annual International Conference on
Machine Learning, ICML ’09, page 961–968, New
York, NY, USA. Association for Computing Machin-
ery.
Spanhol, F. A., Oliveira, L. S., Petitjean, C., and Heutte, L.
(2016). A dataset for breast cancer histopathological
image classification. IEEE Transactions on Biomedi-
cal Engineering, 63(7):1455–1462.
Tang, H. and Jia, K. (2020). Discriminative adversarial do-
main adaptation. Proceedings of the AAAI Conference
on Artificial Intelligence, 34(04):5940–5947.
Trockman, A. and Kolter, J. Z. (2022). Patches are all you
need? arXiv preprint arXiv:2201.09792.
Tzeng, E., Hoffman, J., Saenko, K., and Darrell, T. (2017).
Adversarial discriminative domain adaptation. In Pro-
ceedings of the IEEE Conference on Computer Vision
and Pattern Recognition (CVPR).
Vahadane, A., Peng, T., Sethi, A., Albarqouni, S., Wang,
L., Baust, M., Steiger, K., Schlitter, A. M., Esposito,
I., and Navab, N. (2016). Structure-preserving color
normalization and sparse stain separation for histolog-
ical images. IEEE Transactions on Medical Imaging,
35(8):1962–1971.
van der Maaten, L. and Hinton, G. (2008). Visualizing data
using t-sne. Journal of Machine Learning Research,
9(86):2579–2605.
Wagle, M., Gupta, R. K., Sethi, A., et al. (2023). Chatty:
Coupled holistic adversarial transport terms with yield
for unsupervised domain adaptation. arXiv preprint
arXiv:2304.09623.
Wang, X., Li, L., Ye, W., Long, M., and Wang, J. (2019).
Transferable attention for domain adaptation. In Pro-
ceedings of the Thirty-Third AAAI Conference on Ar-
tificial Intelligence and Thirty-First Innovative Appli-
cations of Artificial Intelligence Conference and Ninth
AAAI Symposium on Educational Advances in Artifi-
cial Intelligence, AAAI’19/IAAI’19/EAAI’19. AAAI
Press.
Wen, J., Liu, R., Zheng, N., Zheng, Q., Gong, Z., and Yuan,
J. (2019). Exploiting local feature patterns for unsu-
pervised domain adaptation. Proceedings of the AAAI
Conference on Artificial Intelligence, 33(01):5401–
5408.
Xie, S., Zheng, Z., Chen, L., and Chen, C. (2018). Learn-
ing semantic representations for unsupervised domain
adaptation. In Dy, J. and Krause, A., editors, Pro-
ceedings of the 35th International Conference on Ma-
chine Learning, volume 80 of Proceedings of Machine
Learning Research, pages 5423–5432. PMLR.
Xu, R., Li, G., Yang, J., and Lin, L. (2019). Larger norm
more transferable: An adaptive feature norm approach
for unsupervised domain adaptation. In Proceedings
of the IEEE/CVF international conference on com-
puter vision, pages 1426–1435.
Yang, J., Liu, J., Xu, N., and Huang, J. (2023). Tvt: Trans-
ferable vision transformer for unsupervised domain
adaptation. In 2023 IEEE/CVF Winter Conference on
Applications of Computer Vision (WACV), pages 520–
530.
Yosinski, J., Clune, J., Bengio, Y., and Lipson, H. (2014).
How transferable are features in deep neural net-
works? CoRR, abs/1411.1792.
BIOIMAGING 2024 - 11th International Conference on Bioimaging
214

Zhang, W., Ouyang, W., Li, W., and Xu, D. (2018). Col-
laborative and adversarial network for unsupervised
domain adaptation. In Proceedings of the IEEE Con-
ference on Computer Vision and Pattern Recognition
(CVPR).
Zhao, H., Combes, R. T. D., Zhang, K., and Gordon, G.
(2019). On learning invariant representations for do-
main adaptation. In Chaudhuri, K. and Salakhutdi-
nov, R., editors, Proceedings of the 36th International
Conference on Machine Learning, volume 97 of Pro-
ceedings of Machine Learning Research, pages 7523–
7532. PMLR.
Unsupervised Domain Adaptation for Medical Images with an Improved Combination of Losses
215
