
CaRe-CNN: Cascading Refinement CNN for Myocardial Infarct
Segmentation with Microvascular Obstructions
Franz Thaler
1,2 a
, Matthias A. F. Gsell
1 b
, Gernot Plank
1 c
and Martin Urschler
3 d
1
Gottfried Schatz Research Center: Medical Physics and Biophysics, Medical University of Graz, Graz, Austria
2
Institute of Computer Graphics and Vision, Graz University of Technology, Graz, Austria
3
Institute for Medical Informatics, Statistics and Documentation, Medical University of Graz, Graz, Austria
Keywords:
Machine Learning, Image Segmentation, Myocardial Infarction.
Abstract:
Late gadolinium enhanced (LGE) magnetic resonance (MR) imaging is widely established to assess the vi-
ability of myocardial tissue of patients after acute myocardial infarction (MI). We propose the Cascading
Refinement CNN (CaRe-CNN), which is a fully 3D, end-to-end trained, 3-stage CNN cascade that exploits
the hierarchical structure of such labeled cardiac data. Throughout the three stages of the cascade, the label
definition changes and CaRe-CNN learns to gradually refine its intermediate predictions accordingly. Further-
more, to obtain more consistent qualitative predictions, we propose a series of post-processing steps that take
anatomical constraints into account. Our CaRe-CNN was submitted to the FIMH 2023 MYOSAIQ challenge,
where it ranked second out of 18 participating teams. CaRe-CNN showed great improvements most notably
when segmenting the difficult but clinically most relevant myocardial infarct tissue (MIT) as well as microvas-
cular obstructions (MVO). When computing the average scores over all labels, our method obtained the best
score in eight out of ten metrics. Thus, accurate cardiac segmentation after acute MI via our CaRe-CNN allows
generating patient-specific models of the heart serving as an important step towards personalized medicine.
1 INTRODUCTION
Cardiovascular diseases are the leading cause of death
worldwide among which myocardial infarction (MI)
is one of the most prevalent diseases
1
. MI is caused by
a decrease or complete cessation of blood flow in the
coronary arteries which reduces perfusion in the sup-
plied myocardial tissue, leading to a metabolic under-
supply that impairs cardiac function and, ultimately,
may result in myocardial necrosis. The accurate as-
sessment of tissue damage after acute MI is highly
relevant as the extension of myocardial necrosis is an
important risk factor for developing heart failure. On
one hand, viable myocardial tissue with a potential
for functional recovery on restoration of normal blood
supply by revascularization might recover (Wrob-
lewski et al., 1990; Perin et al., 2002), which may
improve the functional capacity and survival (Van der
Wall et al., 1996; Kim and Manning, 2004). On the
other hand, precise delineation of infarcted myocar-
a
https://orcid.org/0000-0002-6589-6560
b
https://orcid.org/0000-0001-7742-8193
c
https://orcid.org/0000-0002-7380-6908
d
https://orcid.org/0000-0001-5792-3971
1
https://www.who.int/health-topics/cardiovascular-
diseases, last accessed on October 8, 2023
dial tissue is crucial to determine the risk of further
adverse cardiovascular events like ventricular tachy-
cardia which may lead to sudden death (Rosenthal
et al., 1985; Hellermann et al., 2002). For example,
the presence of microvascular obstructions, charac-
terized by a damaged microvasculature resulting in a
’no-reflow’ phenomenon preventing blood flow from
penetrating beyond the myocardial capillary bed, is
linked to adverse ventricular remodeling and an in-
creased risk of future cardiovascular events (Hami-
rani et al., 2014; Rios-Navarro et al., 2019). Thus,
the accurate assessment of post-MI tissue damage is
of pivotal importance. In clinical practice magnetic
resonance (MR) imaging is used to quantify areas of
impaired myocardial function e.g. by estimating the
end-diastolic wall thickness of the left ventricle, or
by evaluating the contractile reserve, i.e. the myocar-
dial stress-to-rest ratio (Kim et al., 1999; Schinkel
et al., 2007). One of the most accurate methods is
late gadolinium enhanced (LGE) MR imaging, where
the contrast agent accumulates in impaired tissue ar-
eas, thus allowing to visualize the transmural extent of
tissues affected by MI (Selvanayagam et al., 2004).
However, analyzing LGE MR images to char-
acterize tissue viability in an accurate and efficient
manner remains a significant challenge. Nowadays,
Thaler, F., Gsell, M., Plank, G. and Urschler, M.
CaRe-CNN: Cascading Refinement CNN for Myocardial Infarct Segmentation with Microvascular Obstructions.
DOI: 10.5220/0012324800003660
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 19th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2024) - Volume 3: VISAPP, pages
53-64
ISBN: 978-989-758-679-8; ISSN: 2184-4321
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
53
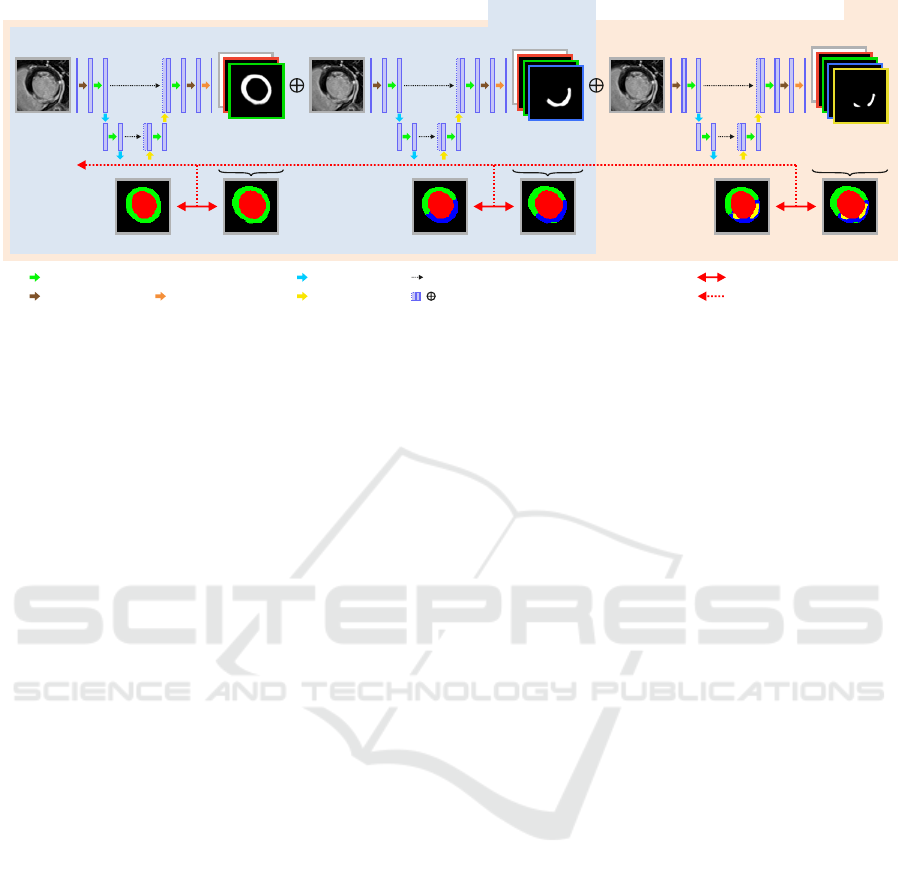
Stage 1 Stage 2 Stage 3
Image x Image x Image x
Label y
3
Label y
2
Label y
1
Label Pred. y
1
^
Pred. p
1
^
Pred. p
2
^
Label Pred. y
2
^
Label Pred. y
3
^
Pred. p
3
^
D8
M1, M12
convolution-dropout-convolution
pooling
upsampling
2x convolution
final convolution
skip-connection
concatenation
/
Generalized Dice Loss
backpropagation
Figure 1: Overview of the proposed CaRe-CNN architecture for segmenting cardiac LGE MR images after MI. CaRe-CNN
is a 3-stage CNN cascade that exploits the hierarchical label definition of the data and refines intermediate predictions in
consecutive stages. The whole architecture is trained end-to-end and all data is processed in 3D. As MVO can only be present
for data of the D8 subgroup, we consider Stage 2 predictions as final predictions for data of the M1 and M12 subgroups.
deep learning-based Convolutional Neural Networks
(CNNs) are widely adopted to medical image analy-
sis tasks like the detection of diseases in medical im-
ages (Esteva et al., 2017; Feng et al., 2022), or im-
age segmentation of the brain (Akkus et al., 2017),
the vertebrae (Payer et al., 2020), or the heart (Chen
et al., 2020). From cardiac LGE MR data, healthy and
necrotic myocardial tissue can be assessed by CNN-
based medical image segmentation, where each voxel
of an LGE MR image is assigned the respective label.
Accurate cardiac segmentation of patients after MI
can provide a foundation for generating anatomically
accurate patient-specific models of the heart, which,
in turn, can be used e.g., to create cardiac digital twin
models of human electrophysiology (Gillette et al.,
2021) to identify potential patient-specific causes for
arrhythmia improving personalized therapy planing
(Campos et al., 2022).
Due to the challenging nature of fully automated
infarct segmentation, some approaches in the liter-
ature rely on manual segmentations of the full my-
ocardium such that a distinction between healthy and
infarcted tissue only needs to be learned within that
region (Zabihollahy et al., 2018; Moccia et al., 2019).
Instead of using LGE MR data, (Xu et al., 2018)
uses cine MR data without contrast agents and a
Long Short-Term Memory-based Recurrent Neural
Network (Graves et al., 2013) to predict myocardial
infarct tissue from motion. In contrast to that, (Fahmy
et al., 2018) automatically segment both, healthy and
infarcted tissue from LGE MR images by employing
a 2D CNN based on the U-Net (Ronneberger et al.,
2015) architecture. In another fully-automated seg-
mentation approach, (Chen et al., 2022) employed
two consecutive 2D U-Net-like CNNs as a cascade,
where the first network learns to segment the full
myocardium, while the second is trained to refine
the prediction to obtain the infarct region. The au-
thors show that the consecutive setup achieves bet-
ter Dice and Jaccard scores, but worse volume esti-
mation compared to a parallel setup of two CNNs.
The semi-supervised myocardial infarction segmen-
tation approach in (Xu et al., 2022) proposes to use
attention mechanisms to obtain the coarse location of
the myocardial infarction before refining the predic-
tion step-by-step. In order to allow training from un-
labeled data, they use an adversarial learning model
that provides a training objective even when ground
truth labels are not available. The EMIDEC chal-
lenge held in conjunction with the International Con-
ference on Medical Image Computing and Computer-
Assisted Intervention (MICCAI) in 2020 aimed to au-
tomatically segment myocardial infarct regions from
LGE MR images in their segmentation track (Lalande
et al., 2022). Different one- and two-stage approaches
mostly based on U-Net-like architectures were sub-
mitted by the challenge participants. The highest
scores in the segmentation track were achieved by
(Zhang, 2021) who employed a coarse to fine two-
stage approach, where initial predictions are obtained
from a 2D U-Net variant before all 2D predictions are
stacked to a 3D volume. The stacked prediction in
combination with the LGE MR image is then refined
by a 3D U-Net variant to obtain the final prediction.
In this work, we propose the Cascading Refine-
ment CNN (CaRe-CNN), which – differently to re-
lated work – is a fully 3D, end-to-end trained 3-stage
CNN cascade that exploits the hierarchical structure
of cardiac LGE MR images after MI and sequen-
tially refines the predicted segmentations. Further,
we propose a series of post-processing steps that take
anatomical constraints into account to obtain more
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
54
consistent qualitative predictions. Our CaRe-CNN
was submitted to the Myocardial Segmentation with
Automated Infarct Quantification (MYOSAIQ) chal-
lenge which was held in conjunction with the Interna-
tional Conference on Functional Imaging and Mod-
eling of the Heart (FIMH) 2023. We evaluate our
method by comparing to state-of-the-art methods sub-
mitted to the MYOSAIQ challenge where our CaRe-
CNN ranked second out of 18 participating teams.
2 METHOD
In this work we propose CaRe-CNN, a cascading re-
finement CNN to semantically segment different car-
diac structures after MI from LGE MR images in 3D.
An overview of CaRe-CNN is provided in Fig. 1.
2.1 Notation and Definitions
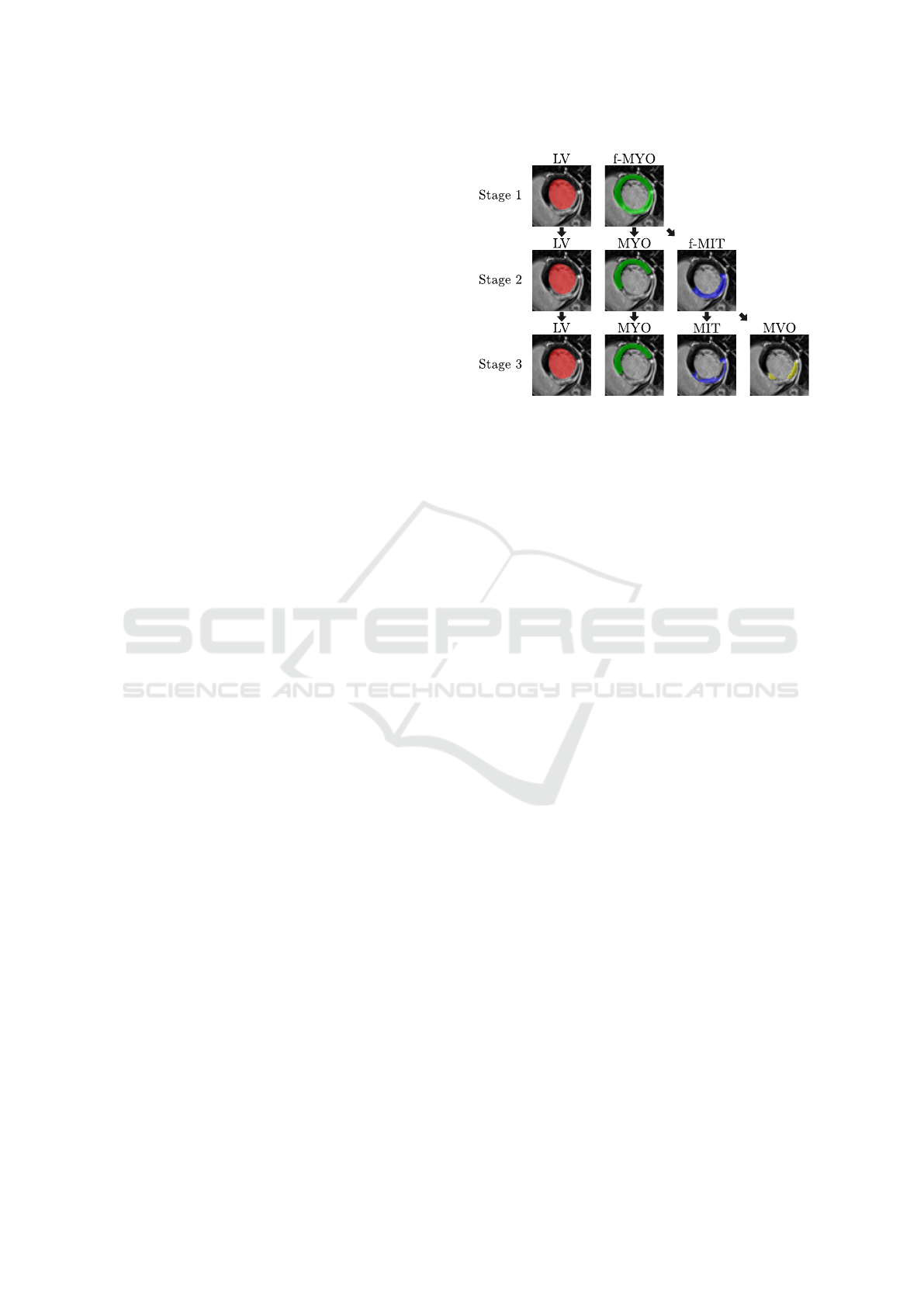
Throughout this work, we will refer to the labels
as left ventricle cavity (LV), healthy myocardium
(MYO), myocardial infarct tissue (MIT) and mi-
crovascular obstruction (MVO). For further disam-
biguation of intermediate results at the different
stages of our method, we additionally define the full
myocardium (f-MYO) as MYO ∪ MIT ∪ MVO and
the full myocardial infarct tissue (f-MIT) as MIT ∪
MVO. A visualization of the label definitions at dif-
ferent stages is provided in Fig. 2. While all scans in
the dataset are LGE MR images after MI, the dataset
can be split into three subgroups (D8, M1, M12) de-
pending on how much time has passed since the MI,
see Section 3.1. Importantly, MVO is exclusive to the
D8 subgroup and the subgroup information is well-
known for every image in the training and test set.
2.2 Cascading Refinement CNN
Our CaRe-CNN architecture exploits the hierarchi-
cal structure of the semantic labels and is set up as
a cascade of three consecutive 3D U-Net-like archi-
tectures (Ronneberger et al., 2015) which are trained
end-to-end. Throughout this work, we will refer
to each of these consecutive parts of the processing
pipeline as stages numbered from 1 to 3. By design,
any subsequent stage of CaRe-CNN receives the pre-
diction of the preceding stage as additional input, such
that the prediction is gradually refined, see Fig. 1.
After randomly choosing and preprocessing a 3D
image x with ground truth y from the training set, the
image x is provided as input to CaRe-CNN. Stage 1
of CaRe-CNN aims to distinguish between the LV,
the f-MYO and the background based on the image
Figure 2: Visualization of the hierarchical label definitions
per stage as used by CaRe-CNN. While LV remains un-
changed, f-MYO can be separated into MYO and f-MIT of
which the latter can be separated into MIT and MVO.
information. By denoting the Stage 1 model as M
1
(·)
with trainable parameters θ
1
, the output prediction
ˆ
p
1
of Stage 1 for image x can be expressed as:
ˆ
p
1
= M
1
(x;θ
1
). (1)
Please note that the output prediction
ˆ
p refers to the
model output without activation function. In Stage 2,
CaRe-CNN learns to predict the LV, the healthy
MYO, the f-MIT and the background by refining the
Stage 1 prediction. To allow consecutive refinement
of
ˆ
p
1
in Stage 2, we provide
ˆ
p
1
concatenated with the
original image x in the channel dimension as input to
the Stage 2 model. This way,
ˆ
p
1
can be refined based
on the original image information which is crucial for
our cascading CNN as the label definition of the in-
dividual stages is not the same. The Stage 2 model
M
2
(·) with trainable parameters θ
2
is defined as:
ˆ
p
2
= M
2
(
ˆ
p
1
⊕ x;θ
2
), (2)
where ⊕ refers to a concatenation in the channel di-
mension and
ˆ
p
2
refers to the output prediction of
Stage 2, again without any activation function. Lastly,
Stage 3 aims to distinguish all labels, i.e., the LV,
MYO, MIT, MVO as well as the background. To con-
tinue our CNN cascade, we concatenate the prediction
ˆ
p
2
and the image x in the channel dimension to pro-
vide both as input to the Stage 3 model M
3
(·) of our
cascading CNN. Formally, the output prediction
ˆ
p
3
of
Stage 3 can be expressed as:
ˆ
p
3
= M
3
(
ˆ
p
2
⊕ x;θ
3
), (3)
where θ
3
refers to the trainable parameters of Stage 3
and ⊕ defines the concatenation operator.
2.3 Training Objective
In our training pipeline the segmentation loss is com-
puted for each stage individually and backpropagation
CaRe-CNN: Cascading Refinement CNN for Myocardial Infarct Segmentation with Microvascular Obstructions
55
through all stages is allowed to update model weights
in an end-to-end manner for the whole cascade. As
the label definition varies from stage to stage, we
adapt the ground truth labels such that they follow the
label definition of the respective stage as defined in
Fig. 2. For every stage, we compute the generalized
Dice loss between the ground truth y and the label
prediction
ˆ
y = softmax(
ˆ
p) of that stage. Formally, the
generalized Dice loss L
GD
(·) is expressed as:
L
GD
(y,
ˆ
y) = 1 − 2
∑
K
k=1
w
k
·
∑
M
m=1
ˆ
y
m
· y
m
∑
K
k=1
w
k
·
∑
M
m=1
ˆ
y
2
m
+ y
m
, (4)
where K represents the number of all labels and M is
the number of voxels. The label weight w
k
for label k
is computed as the ratio of voxels M
k
with label k in
the ground truth compared to the number of all voxels,
i.e. w
k
=
M
k
M
. The square term
ˆ
y
2
m
is used to account
for class imbalance.
During training only images that actually contain
the MVO label are forwarded through Stage 3 as im-
ages with missing labels might lead to unstable train-
ing which can greatly impact the performance at that
stage. In order to provide a loss at every stage for ev-
ery iteration while also allowing all training images to
be selected at some point, we always randomly pick
two training images per iteration: One image with and
one without the MVO label. The overall training ob-
jective of CaRe-CNN for all stages and a single image
can then be expressed as:
L(y,
ˆ
y) = λ
1
L
GD
(y
1
,
ˆ
y
1
;θ
1
)
| {z }
update M
1
+λ
2
L
GD
(y
2
,
ˆ
y
2
;θ
1
,θ
2
)
| {z }
update M
1
and M
2
+ δ
MVO
· λ
3
L
GD
(y
3
,
ˆ
y
3
;θ
1
,θ
2
,θ
3
)
| {z }
update M
1
, M
2
and M
3
,
(5)
where the stage weights λ
1
, λ
2
and λ
3
serve as
weights between the individual loss terms and are set
to 1. The term δ
MVO
is set to 1 if ground truth y con-
tains MVO anywhere and is 0 otherwise. Finally, we
provide the mean loss over the batch to the optimizer.
2.4 Inference
As the subgroup (D8, M1, M12) for every image in
the test set is known as well, we utilize the subgroup
information for test set data to determine the final pre-
diction as encouraged by the MYOSAIQ challenge
organizers. Specifically, we consider the label pre-
diction
ˆ
y
3
of Stage 3 as the final label prediction
ˆ
y
f
only for D8 data, while we use the Stage 2 label pre-
diction
ˆ
y
2
as the final label prediction
ˆ
y
f
for M1 and
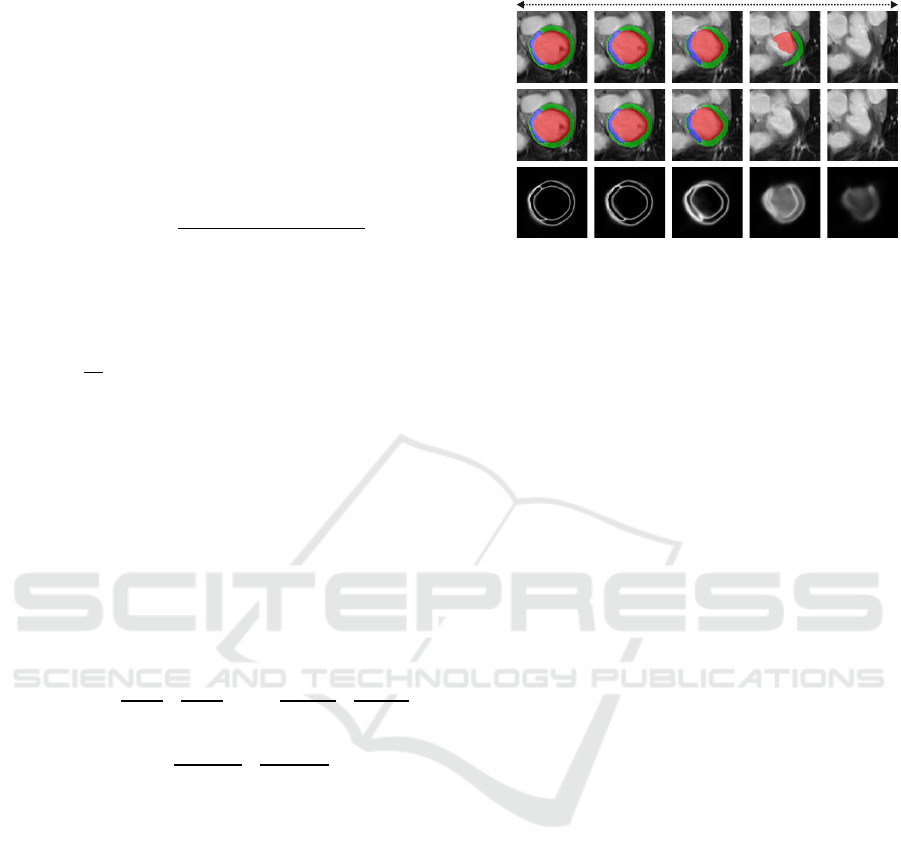
Apex Base
Pred.
Pred.
w PP
Unc.
Figure 3: Predictions of CaRe-CNN (row 1) are in some
cases incomplete for the top-most slice towards the base
of the left ventricle (col. 4). The model’s uncertainty is
computed as the entropy of the softmax prediction (row 3),
where bright values indicate a higher uncertainty. The
highest uncertainty occurs in the incompletely labeled slice
(col. 4). This motivates our post-processing (PP) where, in
this case, the incomplete prediction is removed (row 2).
M12 data. The final label prediction
ˆ
y
f
is defined as:
ˆ
y
f
=
(
ˆ
y
2
if x ∈ {M1, M12}
ˆ
y
3
if x ∈ {D8}.
(6)
To further improve the final prediction of our method,
we independently trained N = 10 CaRe-CNNs with
random weight initialization and random data aug-
mentation. These N models were used as an ensem-
ble for which the final label prediction is obtained by
averaging the final label predictions of the individual
models. The average inference time per image for the
whole ensemble with post-processing takes roughly
8 seconds using an NVIDIA GeForce RTX 3090.
2.5 Post-Processing
As can be observed in Fig. 3 (bottom row), after train-
ing on the data our CaRe-CNN remains ’uncertain’
about how far the heart should be segmented towards
the base which may result in a top-most slice that is
incompletely labeled. Even though such incomplete
model predictions in themselves are not incorrect,
we decided to implement a series of post-processing
steps to obtain more consistent predictions that take
anatomical constraints into account.
As a first step of our post-processing pipeline, we
employ a disconnected component removal strategy,
where any components that are disconnected from
the largest component in 3D as well as in-plane in
2D are removed. In 3D, a connected component
analysis is performed where all foreground labels are
treated as one label and a 3D 6-connected kernel is
applied. Any independent region that is disconnected
from the largest connected component is removed.
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
56
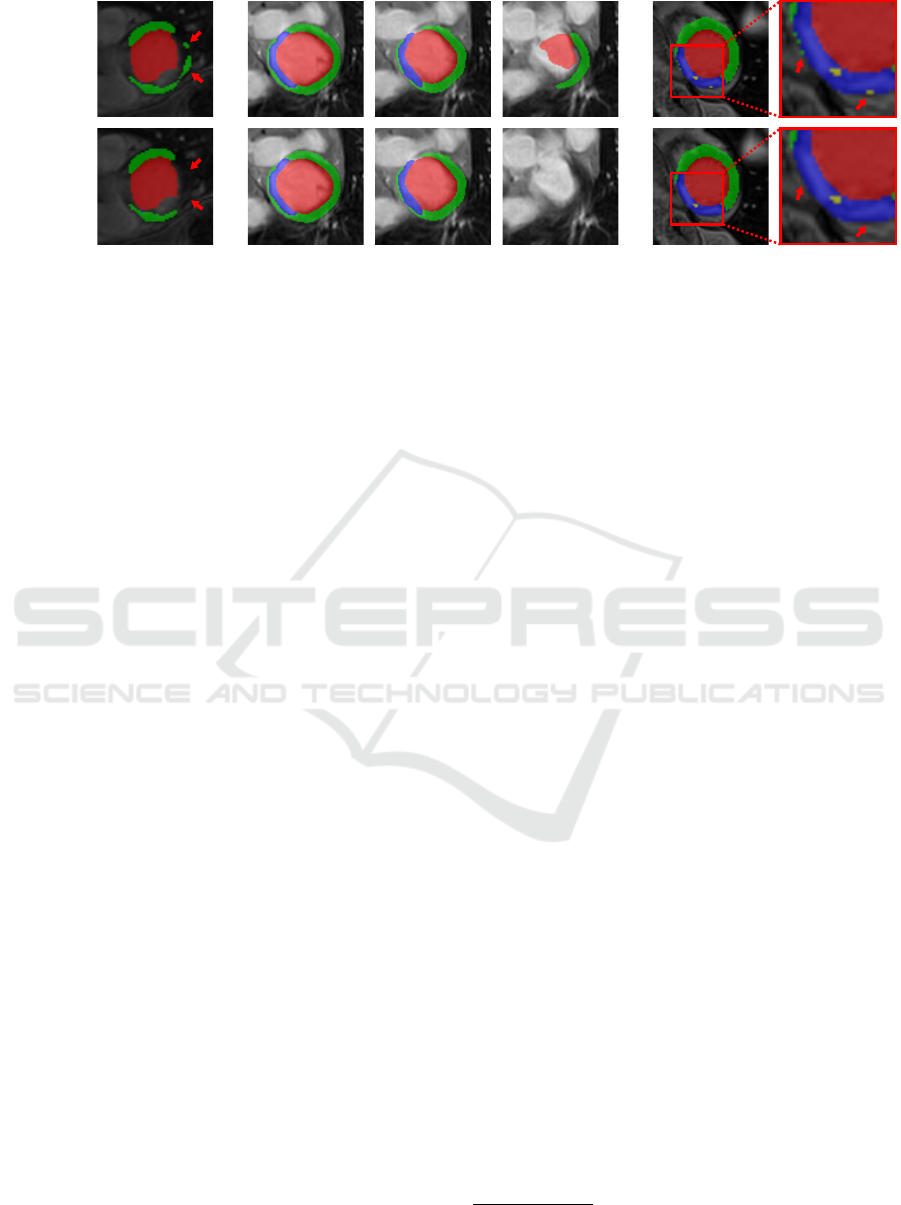
Prediction
Prediction
w Post-P.
Figure 4: CaRe-CNN predictions before (row 1) and after (row 2) post-processing. Images refer to the proposed disconnected
component removal (col. 1), the top-most slice removal (col. 2-4) and the outlier region replacement (col. 5-6). Red arrows
indicate regions of interest.
Due to the large slice thickness of the data, we also
perform a connected component analysis for every
in-plane 2D slice independently, following the same
steps as described for the 3D variant and using a 2D
4-connected kernel in-plane. The 2D strategy mostly
affects the topmost slice that still contains foreground
predictions and removes some smaller in-plane dis-
connected regions from that slice, see Fig. 4 (col. 1).
Next, we propose a top-most slice removal strat-
egy, where we compare the remaining foreground vol-
ume of the topmost slice that contains foreground pre-
dictions to the foreground volume of its neighboring
slice towards the hearts’ apex (i.e. the slice ’below’
the top-most slice). In case that the volume of the
topmost slice is less than half the neighboring slice’s
volume, the topmost slice is removed completely. An
example is shown in Fig. 4 (col. 4).
Lastly, an outlier region replacement strategy is
applied, where very small regions of a single label
are treated as outliers and are replaced if they are iso-
lated from larger regions of the same label. In the
first step of this strategy, isolated regions are identi-
fied by performing a connected component analysis
per label using a 3D 6-connected kernel. Any region
with a volume smaller than 0.1 ml is considered to
be an outlier and undergoes a correction step, where
the local neighborhood of each outlier voxel is ob-
served to select a new label for that voxel, see Fig. 4
(col. 5-6). Specifically, we obtain label votes from
all voxels within a 3D kernel of size 9 × 9 in-plane
and 5 out-of-plane, due to the large slice thickness.
This anisotropic kernel is sufficient as we perform a
weighting based on a 3D Gaussian with sigma value 2
that considers the actual physical distance of any can-
didate. Importantly, votes from voxels marked as out-
liers are not considered. Finally, the maximum of the
weighted votes indicates the most likely label for that
voxel, which is then used as the label for that voxel.
3 EXPERIMENTAL SETUP
3.1 Dataset
In this work, we used the publicly available dataset
from the MYOSAIQ challenge
2
, which was held in
conjunction with FIMH 2023. The aim of the MYO-
SAIQ challenge is to automatically segment four dif-
ferent cardiac structures from LGE MR images of pa-
tients after myocardial infarction. These structures
encompass the LV, MYO, MIT and MVO if present.
The dataset consists of 467 LGE MR images which
are split into 376 training and 93 test images. All im-
ages belong to one of three subgroups. The first sub-
group (D8) encompasses LGE images of 123 patients
with acute myocardial infarction up to eight days
after the infarction and originates from the MIMI-
cohort (Belle et al., 2016). The second subgroup (M1)
consists of LGE images of 204 patients, while the
third subgroup (M12) contains LGE images of 140
patients, which were respectively obtained one and 12
months after coronary intervention and are part of the
HIBISCUS-cohort. For every image in the training
dataset, a corresponding ground truth segmentation is
available. As the whole dataset consists of images
after myocardial infarction, all ground truth segmen-
tations in the dataset contain the LV, MYO and MIT
label. However, the MVO label is exclusive to the D8
subgroup and only present in roughly 66% of the D8
data. The in-plane physical resolution of the dataset
varies from 0.9 to 2.2 mm and averages at 1.57 mm.
Out-of-plane, the physical resolution varies from 5 to
8 mm.
2
https://www.creatis.insa-lyon.fr/Challenge/myosaiq/,
last accessed on October 8, 2023
CaRe-CNN: Cascading Refinement CNN for Myocardial Infarct Segmentation with Microvascular Obstructions
57

3.2 Data Augmentation
We augment training data using the training frame-
work from (Payer et al., 2017; Payer et al., 2019)
in 3D using random spatial and intensity transforma-
tions. Spatially, we perform translation (±20 vox-
els), rotation (±0.35 radians), scaling (first isotropi-
cally with a factor between [0.8,1.2], then per dimen-
sion with a factor between [0.9,1.1]) and elastic de-
formation (eight grid nodes per dimension, deforma-
tion values are sampled from ±15 voxels). For robust
intensity normalization of the MR images, the 10
th
and 90
th
percentile are linearly normalized to −1 and
1, respectively. After normalization, a random inten-
sity shift (±0.2) as well as an intensity scaling with
a factor between [0.6,1.4] is applied to the training
image before modulating intensity values per label by
an additional shift of (±0.2) and scaling with a factor
of [0.9, 1.1]. All augmentation parameters are sam-
pled uniformly from the respective value range. Im-
ages of the test set are not augmented, however, they
are robustly normalized identically to the training data
to ensure similar intensity ranges. To ensure consis-
tency of the physical dimensions across the dataset,
all training and test images are trilinearly resampled
to an isotropic spacing of 1 × 1 ×1 mm and an image
size of 128 × 128 × 128 voxel before being provided
to the CaRe-CNN model.
3.3 Implementation Details
At each stage of CaRe-CNN, a U-Net-like (Ron-
neberger et al., 2015) network architecture is em-
ployed in 3D which follows the same structure, see
Fig. 1. Similar to an encoder-decoder, the architecture
can be separated into a contracting and an expanding
path. Importantly, by using skip-connections, the out-
put of each level of the contracting path is concate-
nated to the input of the same level of the expand-
ing path in the channel dimension. At each of the
five levels of the contracting and the expanding path,
we use a single block consisting of two convolutions
with an intermediate dropout layer (Srivastava et al.,
2014), after which a pooling or an upsampling layer
is employed, respectively. Two respectively three ad-
ditional convolution layers are employed before and
after the U-Net-like network of each stage. All in-
termediate convolution layers use a 3 × 3 × 3 kernel
and 64 filters, while the last convolution layer of each
stage uses a 1 × 1 × 1 kernel and as many filters as
there are labels at the respective stage. He initializa-
tion (He et al., 2015) is used to initialize all weights
and the dropout rate is 0.1. We employ max pooling
layers and tri-linear upsampling layers with a kernel
size of 2 × 2 × 2. Leaky ReLU (Maas et al., 2013)
with a slope of 0.1 is used after intermediate convolu-
tion layers, while a softmax activation is used after the
last layer of each stage to compute the loss. As opti-
mizer, we employ Adam (Kingma and Ba, 2015) with
a learning rate of 0.001, use an Exponential Moving
Average strategy (Laine and Aila, 2016) with a decay
of 0.999 and train for 200, 000 iterations. For each
training iteration, we select one image with and one
without the MVO label which corresponds to a batch
size of 2 for the Stage 1 and Stage 2 models. To ensure
stable training, only images with the MVO label are
processed by the Stage 3 model, which results in an
effective batch size of 1 for that model. During the de-
velopment of our method, we trained our model only
on 2/3 of the training data and used the remaining
1/3 of the data as a validation set. For our submission
to the challenge, we trained CaRe-CNN on all train-
ing data and evaluation was performed on the hidden
test set. Final results were obtained by averaging the
prediction of a CaRe-CNN ensemble of 10 models on
the test set and 5 models on the validation set.
4 RESULTS
The quantitative evaluation is performed by compar-
ing our CaRe-CNN method to the other 17 partici-
pants of the MYOSAIQ challenge on the hidden test
set. For each participant, we obtained ten metric
scores for each label individually from the official
evaluation platform
3
, which is publicly available. The
used metrics respectively encompass the mean and
standard deviation of the Dice score (DSC) in percent
as well as the Hausdorff distance (HD) and average
symmetric surface distance (ASSD) in mm. Further-
more, the list of metrics includes the mean correlation
coefficient score (CC), mean absolute error (MAE),
limits of agreement (LOA) and the continuous ranked
probability score (CRPS).
In order to summarize the results, we computed
the mean score over the four labels for each metric
and present them in Table 1 for each participant. This
is also true for the standard deviation of DSC, HD and
ASSD, where we also computed the mean score over
the labels. The best score for each metric is given in
bold, while the second and third best metric scores
are shown in underlined blue and italicized orange,
respectively.
Table 2 presents quantitative results per label for
each metric to give some insight into the individual
scores. In the interest of space, we only provide the
3
https://codalab.lisn.upsaclay.fr/competitions/13631,
last accessed on October 8, 2023
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
58
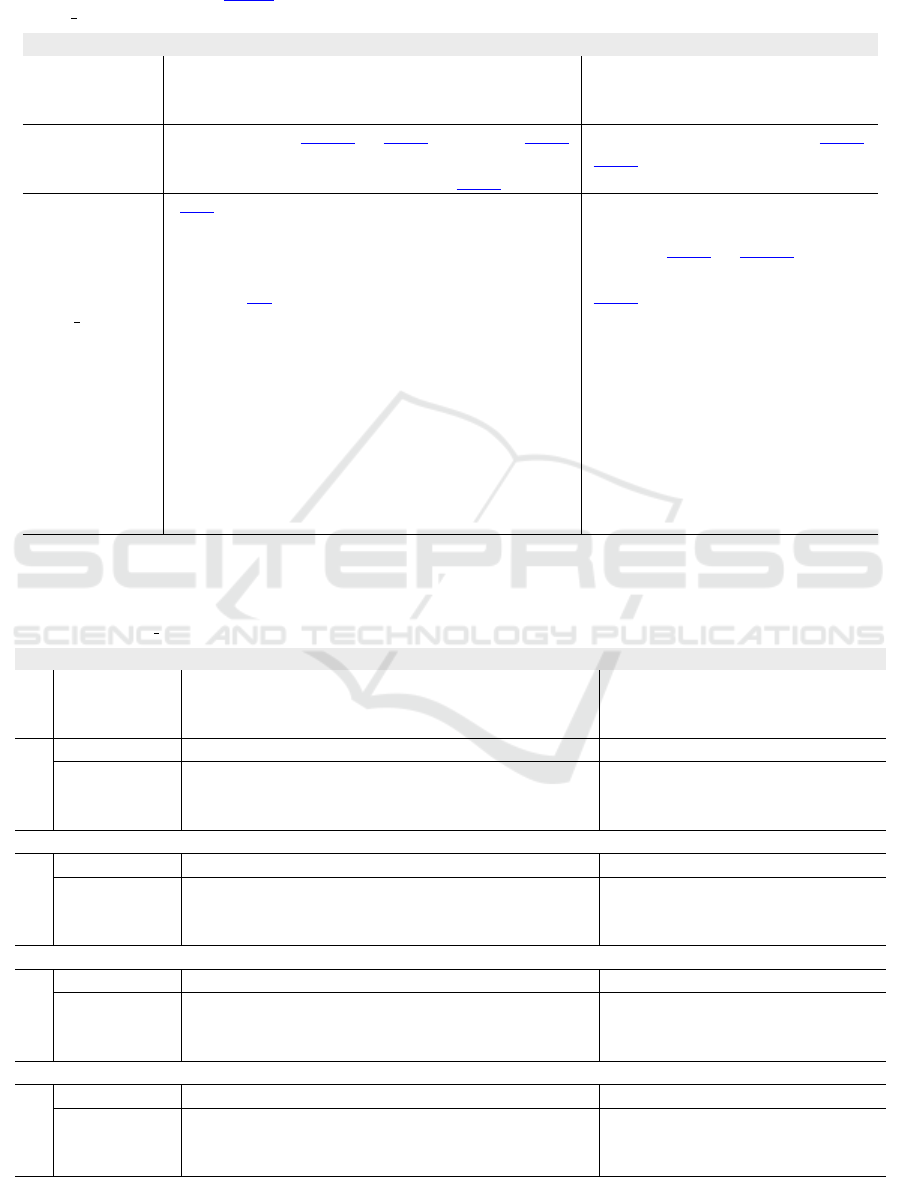
Table 1: Quantitative evaluation showing the mean score over all labels for ten metrics. The proposed CaRe-CNN is com-
pared to the other MYOSAIQ challenge participants. Invalid mean scores due to non-numeric results for at least one label
are indicated by -. The best, second and third best scores are highlighted.
†
Our teamname on the evaluation platform is
’ominous ocelot’.
‡
Abbreviation for ’luiskabongo-inheart’.
Mean over Labels
Team
DSC (%) HD (mm) ASSD (mm)
CC
(↑)
MAE
(↓)
LOA
(↓)
CRPS
(↓)
mean std mean std mean std
(↑) (↓) (↓) (↓) (↓) (↓)
gemr22 74.9 10.3 13.452 7.545 0.711 0.607 0.931 6.044 18.228 0.011
(proposed)
†
78.9 8.5 13.200 9.244 0.574 0.560 0.938 5.500 16.140 0.010
akaroui 75.4 10.0 13.779 8.392 0.697 0.689 0.929 5.827 17.644 0.035
Hairuiwang 75.6 9.8 14.538 9.965 0.711 0.673 0.936 5.810 18.219 0.038
azanella 75.1 10.6 13.483 7.195 0.724 0.685 0.905 5.842 18.337 0.010
KiwiYyy 74.6 12.3 13.771 7.626 0.734 0.702 0.930 5.754 17.247 0.010
hoanguyen93 74.3 11.0 13.905 9.009 0.744 0.758 0.904 5.924 19.297 0.010
nicoco 73.7 9.4 14.839 9.290 0.737 0.630 0.938 6.169 18.044 0.130
hang jung 73.7 9.9 15.063 8.752 0.835 0.766 0.907 6.415 19.802 0.014
Dolphins 73.4 11.9 15.711 10.061 0.754 0.675 0.911 6.578 20.556 0.042
rrosales 73.0 11.0 15.045 8.217 0.856 0.728 0.940 6.788 19.442 0.020
luiskabongo
‡
72.3 10.6 15.584 9.561 0.804 0.718 0.917 7.099 21.622 0.105
calderds 72.0 11.7 17.321 11.853 0.849 0.767 0.909 6.628 20.820 0.039
marwanabb 69.8 12.9 15.667 9.499 1.131 1.183 0.883 7.534 22.477 0.071
agaldran 69.3 19.6 15.947 10.926 - - 0.722 11.712 54.175 -
Erwan 65.5 12.3 20.502 8.416 1.200 1.022 0.853 7.204 22.358 0.012
farheenramzan 55.3 10.6 20.594 9.051 1.641 1.233 0.720 9.349 26.389 0.016
MYOSCANS - - - - - - - - - -
Table 2: Quantitative evaluation showing the individual label scores of the three best MYOSAIQ challenge participants for ten
metrics. ’Overall Best’ refers to the best score obtained by any participant and is used as an upper baseline for each label and
metric. The best score for each metric considering all 18 participants is highlighted in bold.
†
Our teamname on the evaluation
platform is ’ominous ocelot’.
Best 3 Methods per Label
Team
DSC (%) HD (mm) ASSD (mm)
CC
(↑)
MAE
(↓)
LOA
(↓)
CRPS
(↓)
mean std mean std mean std
(↑) (↓) (↓) (↓) (↓) (↓)
LV
Overall Best 93.7 2.8 6.406 2.013 0.392 0.233 0.980 6.881 17.121 0.012
gemr22 93.5 3.1 6.471 2.145 0.408 0.259 0.980 7.308 18.533 0.012
(proposed)
†
93.4 3.4 6.666 2.155 0.419 0.290 0.980 6.881 17.121 0.012
akaroui 93.7 3.0 6.406 2.013 0.392 0.264 0.978 7.313 18.768 0.012
MYO
Overall Best 82.2 4.1 11.753 5.712 0.390 0.211 0.967 7.891 22.251 0.013
gemr22 81.7 4.7 11.794 6.365 0.395 0.246 0.958 9.013 26.664 0.015
(proposed)
†
81.6 5.0 12.839 7.144 0.405 0.253 0.954 9.845 25.686 0.016
akaroui 82.2 4.7 12.214 6.711 0.390 0.259 0.964 8.463 24.263 0.014
MIT
Overall Best 68.4 16.1 16.746 12.482 0.924 1.310 0.855 4.044 17.866 0.007
gemr22 66.0 17.1 18.201 12.482 1.005 1.431 0.799 4.510 20.197 0.008
(proposed)
†
68.4 16.1 16.746 13.414 0.924 1.377 0.833 4.044 18.647 0.007
akaroui 65.8 17.4 19.790 14.751 1.092 1.666 0.789 4.582 20.873 0.008
MVO
Overall Best 72.0 9.5 14.539 5.682 0.547 0.321 0.995 1.231 3.106 0.003
gemr22 58.5 16.4 17.343 9.187 1.037 0.492 0.987 3.343 7.516 0.008
(proposed)
†
72.0 9.5 16.548 14.261 0.547 0.321 0.985 1.231 3.106 0.003
akaroui 59.9 15.0 16.705 10.092 0.913 0.566 0.984 2.950 6.671 0.106
CaRe-CNN: Cascading Refinement CNN for Myocardial Infarct Segmentation with Microvascular Obstructions
59
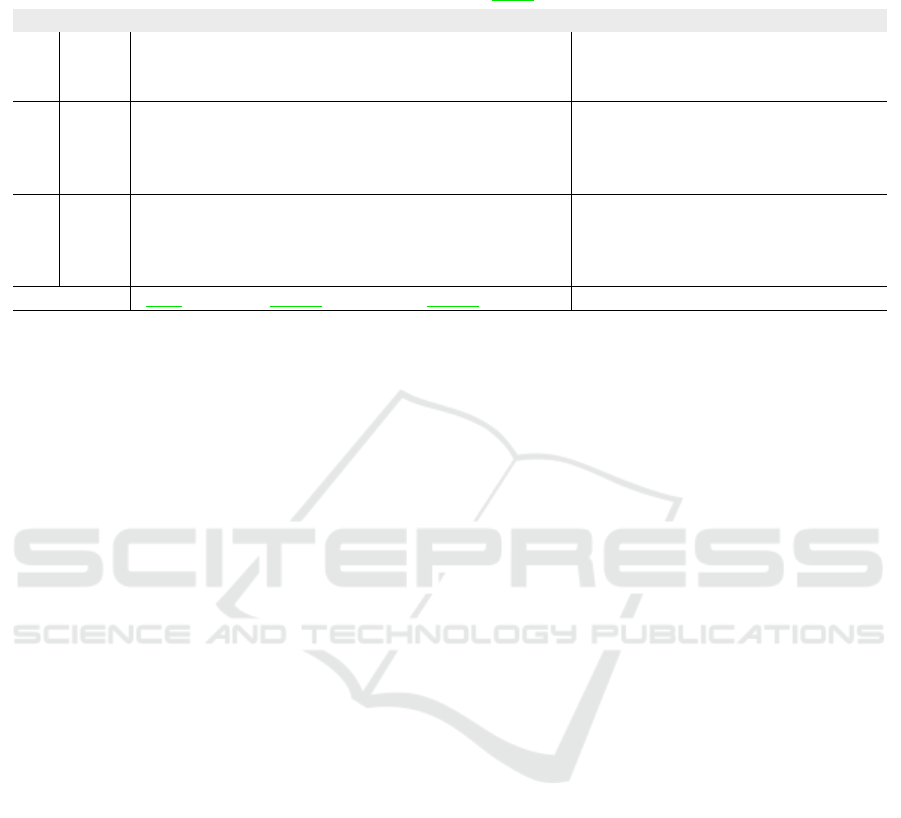
Table 3: Ablation of the proposed post-processing (PP) when applied to our CaRe-CNN ensemble predictions. Scores be-
fore (×) and after (✓) post-processing are shown for each label and ten metrics. The last row refers to the mean difference,
where improvements when using post-processing are highlighted in green, while declines are highlighted in red.
Ablation of Proposed CaRe-CNN Ensemble
PP Label
DSC (%) HD (mm) ASSD (mm)
CC
(↑)
MAE
(↓)
LOA
(↓)
CRPS
(↓)
mean std mean std mean std
(↑) (↓) (↓) (↓) (↓) (↓)
×
LV 93.4 3.3 6.892 2.157 0.422 0.294 0.980 6.837 17.215 0.011
MYO 81.6 4.8 12.088 6.611 0.400 0.238 0.957 9.944 25.271 0.016
MIT 68.5 15.9 16.892 13.46 0.901 1.346 0.837 4.000 18.491 0.007
MVO 71.7 10.0 17.569 13.853 0.576 0.329 0.985 1.210 3.104 0.003
✓
LV 93.4 3.4 6.666 2.155 0.419 0.290 0.980 6.881 17.121 0.012
MYO 81.6 5.0 12.839 7.144 0.405 0.253 0.954 9.845 25.686 0.016
MIT 68.4 16.1 16.746 13.414 0.924 1.377 0.833 4.044 18.647 0.007
MVO 72.0 9.5 16.548 14.261 0.547 0.321 0.985 1.231 3.106 0.003
Mean Diff. +0.1 0 -0.161 +0.223 -0.001 +0.009 -0.002 +0.003 +0.120 +0.000
scores for the three best performing methods in the
challenge as announced at the FIMH 2023 confer-
ence. Nevertheless, to indicate the overall best score
over all teams for each metric and label, we addition-
ally show the best score obtained by any participant
as an upper bound baseline. The best score for each
metric when considering all 18 challenge participants
is given in bold.
Table 3 shows an ablation of the proposed CaRe-
CNN ensemble with and without post-processing.
Again, the scores were obtained from the evaluation
platform of the challenge, where we submitted our
prediction results from the exact same models with
and without post-processing. We show the score for
each label and all metrics evaluated in the challenge.
The last row represents the mean difference between
the scores obtained with and without post-processing.
Underlined green numbers indicate an improvement
and red numbers refer to a decline in performance
when post-processing is applied compared to when it
is not.
The qualitative evaluation of our CaRe-CNN is
performed by visually inspecting the predictions. As
ground truth segmentations for the test set data are
hidden, we also present qualitative results of CaRe-
CNN trained on 2/3 and validated on 1/3 of the actual
training data for the MYOSAIQ challenge in Fig. 5 to
allow a comparison of our predictions to the ground
truth. Additionally, we provide qualitative results of
our final method submitted to the challenge on the
test set in Fig. 6, however, without publicly available
ground truth segmentations, the predictions are only
compared to the respective input images. Both figures
show three consecutive slices of two MR scans of pa-
tients after acute MI per subgroup (D8, M1, M12).
5 DISCUSSION
Quantitative Evaluation. The mean score over the
four labels presented in Table 1 shows, that on average
our method achieved the best score for eight out of
ten metrics. Other participants only outperformed our
method on the mean standard deviation of the HD as
well as the CC, where CaRe-CNN obtained a tied sec-
ond best score. Most notably, our method shows great
improvements compared to the other methods on the
DSC and ASSD scores. Specifically, with a mean
DSC of 78.9%, CaRe-CNN achieved an improvement
of 3.3% compared to the second best method with
75.6%. The same 3.3% window applied to the range
[75.6%,72.3%] encompasses the second up to the 12
th
best mean DSC score. Similarly, with a result of
0.574 mm on the ASSD score our method achieved
an improvement of 0.123 mm over the second best
ASSD score with 0.697 mm. The second up to the
10
th
best score lie within the same 0.123 mm window
of [0.697 mm,0.820 mm].
More details are provided in Table 2, where the
per label scores and the overall best score of any
method are shown. For the LV results, it can be ob-
served that our method obtained the best scores for
MAE and LOA, and obtained tied best scores with
other methods for the CC and CRPS. Moreover, the
shown DSC, HD and ASSD scores are all very close
to one another with our method achieving 93.4%
(best: 93.7%) DSC, 6.67 mm (best: 6.41 mm) HD
and 0.42 mm (best: 0.39 mm) ASSD. On MYO, our
method did not obtain the best score on any metric and
underperformed compared to the overall best score
most notably with 12.839 mm (best: 11.753 mm)
HD, 9.845 (best: 7.891) MAE and 25.686 (best:
22.251) LOA. Nevertheless, on other metrics like
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
60
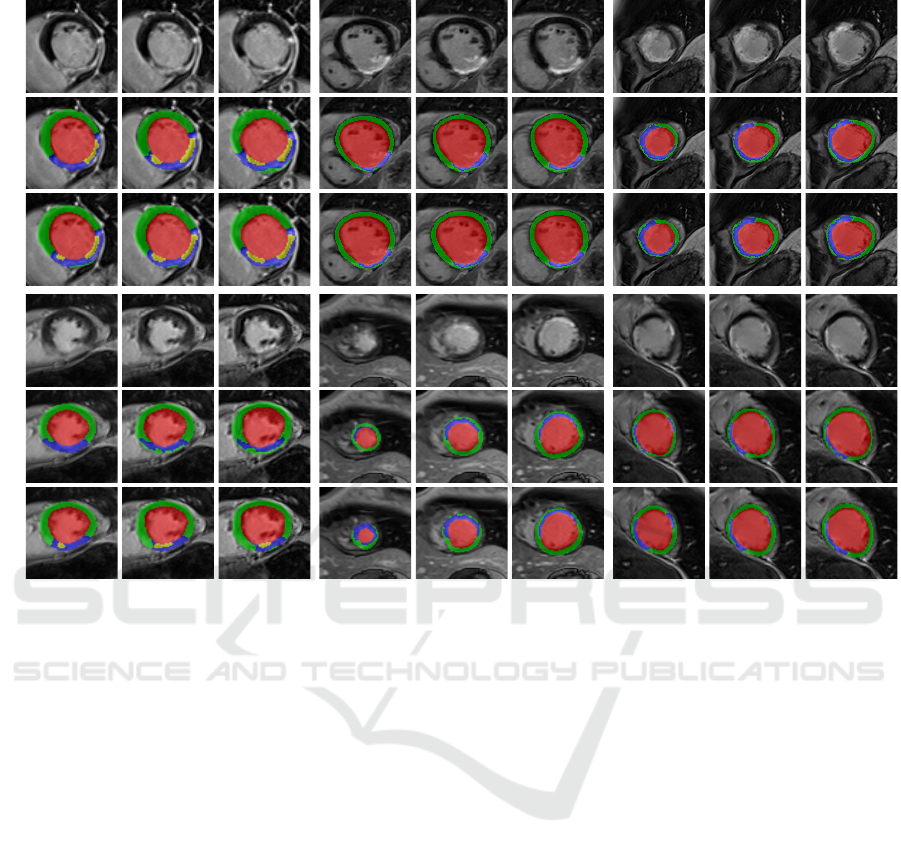
Image
GTPred.
Image
GTPred.
D8 M1 M12
Figure 5: Qualitative results of CaRe-CNN on the validation set. Columns refer to three consecutive slices of LGE MR scans
of patients after MI for the three subgroups: D8 (col. 1-3), M1 (col. 4-6) and M12 (7-9). Rows refer to scans of two separate
patients and show the image (rows 1, 4), ground truth (rows 2, 5) and prediction of CaRe-CNN (rows 3, 6).
DSC and ASSD our method remains competitive to
the other methods achieving 81.6% (best: 82.2%)
DSC and 0.405 mm (best: 0.390 mm) ASSD.
Compared to the other challenge participants,
our CaRe-CNN excelled when segmenting the diffi-
cult but clinically most relevant MIT and MVO la-
bels, where our method obtained the best score for
six and seven out of the ten metrics, respectively.
Among the three challenge winners, our method
achieved good improvements on the MIT label with
68.4% (+2.4%) DSC, 16.746 mm (−1.455 mm) HD,
0.924 mm (−0.081 mm) ASSD and 4.044 (−0.466)
MAE. Interestingly, our method underperformed on
the HD of the MVO label achieving a mean of
16.548 mm (best: 14.539 mm) and a standard devi-
ation of 14.261 mm (best: 5.682 mm). On other met-
rics, however, CaRe-CNN achieved great improve-
ments for the MVO label compared to the other two
challenge winners, namely 72.0% (+12.1%) DSC,
0.547 mm (−0.366 mm) ASSD, 1.231 (−1.719)
MAE and 3.106 (−3.565) LOA.
Post-Processing. When observing the training data
more closely, we noticed that the ground truth an-
notations of the heart labels towards the base of the
heart are not always complete. Most notably, how far
slices are labeled towards the base varies from image
to image, which is likely an artifact from the anno-
tation protocol. While such incomplete annotations
are not incorrect, they introduce a bias to the dataset
which is reflected by a machine learning model and
leads to some expected inconsistencies in the model
predictions. We mitigate these inconsistencies using
a series of post-processing steps to obtain more con-
sistent predictions and show that quantitative scores
for all metrics are almost unchanged in Table 3. The
most affected metric is the HD resulting in a mean
of 13.200 mm (mean difference: −0.161 mm) and
a standard deviation of 9.243 mm (mean difference:
+0.223 mm) after post-processing. This confirms our
expectation, that the top-most slice removal strategy
paired with the large slice thickness of 5.6 mm on av-
erage leads to the HD being the most affected met-
ric as it is defined as the maximum distance of any
voxel-pair of the same label between ground truth and
prediction. Nevertheless, the relative change over all
metrics averages to 0.6% when using post-processing,
which confirms that it can be safely applied in order to
CaRe-CNN: Cascading Refinement CNN for Myocardial Infarct Segmentation with Microvascular Obstructions
61
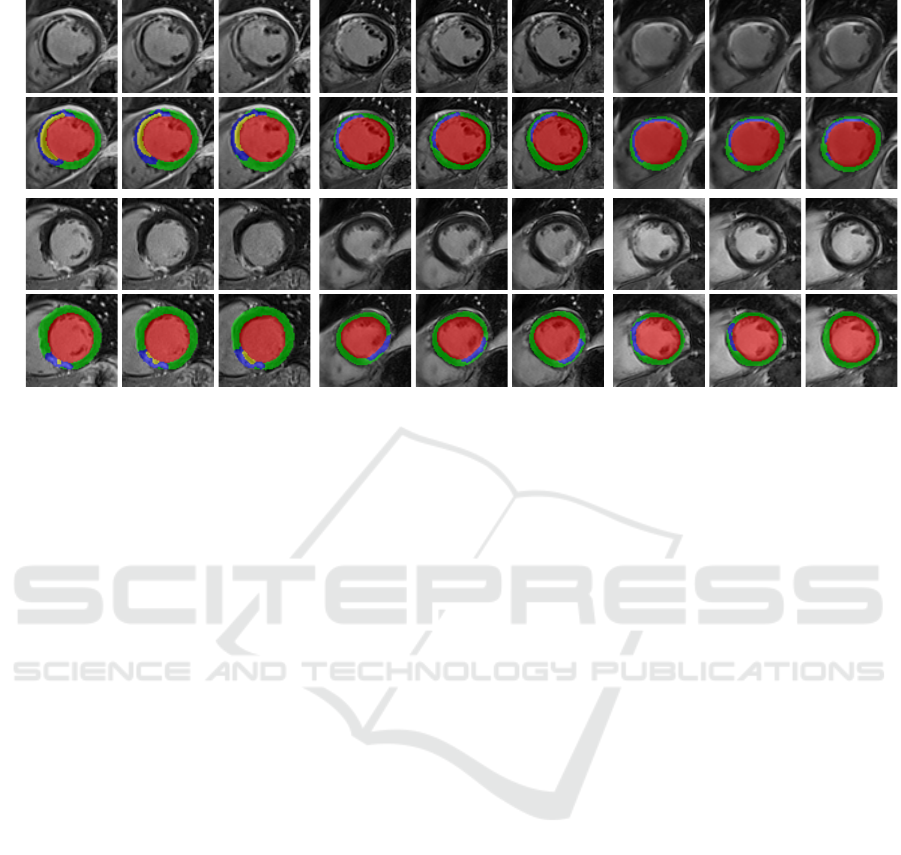
Image
Pred.
Image
Pred.
D8 M1 M12
Figure 6: Qualitative results of CaRe-CNN on the test set. Columns refer to three consecutive slices of LGE MR scans of
patients after MI for each subgroup: D8 (col. 1-3), M1 (col. 4-6) and M12 (7-9). Rows refer to scans of two separate patients
and show the image (rows 1, 3) and prediction of CaRe-CNN (rows 2, 4). Ground truth is not available for the test set.
improve the qualitative consistency of the predictions.
Qualitative Evaluation. The qualitative results on
the validation set in Fig. 5 confirm that most label
predictions are very close to the ground truth. On
closer inspection, however, some differences can be
spotted. For example, one of the two MVO regions is
predicted in one additional consecutive slice in con-
trast to the ground truth (D8, top), while the MIT la-
bel is overpredicted close to the apex (M1, bottom).
Also, an MVO label prediction for a patient without
MVO is visible (D8, bottom). Nevertheless, many re-
gions are predicted correctly, most notably even for
data where the wall is in parts only two to three vox-
els thick (M12, bottom). On the test set in Fig. 5,
qualitative results can only be compared to the LGE
MR image. Overall, the label predictions appear to be
realistic which is supported by our quantitative evalu-
ation, however, further confirmation needs to be per-
formed by an expert.
Challenges and Limitations. One major challenge
of correctly segmenting the structures of interest
arises from the limited resolution of the LGE MR
data in combination with the shape and small phys-
ical size of the structures, most notably the MIT and
MVO label. While the LV is comparatively easy to
segment due to its size and blob-like shape in 3D, the
f-MYO label that surrounds the LV averages to a mid-
diastolic thickness of 6.47 ± 1.07 mm in women and
7.90 ± 1.24 mm in men (Walpot et al., 2019) with-
out considering infarction. In a small cohort, (Khalid
et al., 2019) showed that during ejection, healthy wall
segments are roughly three times as thick (8.73 mm)
compared with infarcted wall segments (2.86 mm).
Furthermore, infarction might only affect some part
of the myocardial tissue in transmural direction such
that two or even all three of the f-MYO sublabels
(MYO, MIT and MVO) might be present across the
already thin wall. The in-plane resolution of the LGE
MR data with 1.57 mm on average paired with the
small physical size of some of the structures of inter-
est leads to a potential transmural thickness of only a
few voxels for these labels. Moreover, segmentation
models are inherently uncertain near the label bor-
ders and thus, prone to single voxel errors, which can
strongly affect the scores for small structures like the
MIT and MVO labels. The combination of these ef-
fects explains the disparity of the LV to the MIT and
MVO label scores for which CaRe-CNN achieved the
best score in six (MIT) and seven (MVO) out of ten
metrics among 18 challenge participants.
A remaining challenge arises from the MVO la-
bel predictions for some patients of the D8 subgroup,
where the label was not predicted when it should be
present or vice versa. Since the presence of MVO
is linked to an increased risk of adverse cardiovascu-
lar events (Hamirani et al., 2014; Rios-Navarro et al.,
2019), incorrect predictions of MVO might impact
clinical decision making if trusted blindly. While
manual verification by an expert is necessary, our
state-of-the-art predictions can alleviate the manual
workload to obtain correct segmentations of patient-
specific anatomy.
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
62

6 CONCLUSION
In this work we presented CaRe-CNN, a 3-stage
cascading refinement CNN, which segments cardiac
LGE MR images after MI. The cascading architec-
ture is designed to exploit the hierarchical label def-
inition of the data and is trained end-to-end fully
in 3D. Furthermore, we employed a series of post-
processing steps that improve the consistency of the
predictions by taking anatomical constraints into ac-
count. The proposed CaRe-CNN was submitted to
the MYOSAIQ challenge, where it ranked second out
of 18 participating teams and achieved state-of-the-art
segmentation results, most notably when segmenting
the difficult MIT and MVO labels. Due to great im-
provements over related work on the difficult but clin-
ically very relevant MVO label, our method obtained
the best score in eight out of ten metrics when com-
puting the mean over all labels. Precise segmenta-
tions of healthy and infarcted myocardial tissue after
MI allow patient-specific therapy planning and are an
important step towards personalized medicine. In our
future work, we plan to investigate uncertainty quan-
tification strategies to further improve CaRe-CNN for
future rounds of the MYOSAIQ challenge.
ACKNOWLEDGEMENTS
This research was funded by the InstaTwin grant
FO999891133 from the Austrian Research Promotion
Agency (FFG).
REFERENCES
Akkus, Z., Galimzianova, A., Hoogi, A., Rubin, D. L., and
Erickson, B. J. (2017). Deep Learning for Brain MRI
Segmentation: State of the Art and Future Directions.
Journal of Digital Imaging, 30:449–459.
Belle, L., Motreff, P., Mangin, L., Rang
´
e, G., Marcaggi,
X., Marie, A., Ferrier, N., Dubreuil, O., Zemour, G.,
Souteyrand, G., et al. (2016). Comparison of Im-
mediate with Delayed Stenting Using the Minimal-
ist Immediate Mechanical Intervention Approach in
Acute ST-Segment–Elevation Myocardial Infarction:
The MIMI Study. Circulation: Cardiovascular Inter-
ventions, 9(3):e003388.
Campos, F. O., Neic, A., Mendonca Costa, C., Whitaker,
J., O’Neill, M., Razavi, R., Rinaldi, C. A., Scherr,
D., Niederer, S. A., Plank, G., et al. (2022). An Au-
tomated Near-Real Time Computational Method for
Induction and Treatment of Scar-related Ventricular
Tachycardias. Medical Image Analysis, 80:102483.
Chen, C., Qin, C., Qiu, H., Tarroni, G., Duan, J., Bai, W.,
and Rueckert, D. (2020). Deep Learning for Cardiac
Image Segmentation: A Review. Frontiers in Cardio-
vascular Medicine, 7:25.
Chen, Z., Lalande, A., Salomon, M., Decourselle, T., Pom-
mier, T., Qayyum, A., Shi, J., Perrot, G., and Cou-
turier, R. (2022). Automatic Deep Learning-based
Myocardial Infarction Segmentation from Delayed
Enhancement MRI. Computerized Medical Imaging
and Graphics, 95:102014.
Esteva, A., Kuprel, B., Novoa, R. A., Ko, J., Swetter, S. M.,
Blau, H. M., and Thrun, S. (2017). Dermatologist-
level Classification of Skin Cancer with Deep Neural
Networks. Nature, 542(7639):115–118.
Fahmy, A. S., Rausch, J., Neisius, U., Chan, R. H., Maron,
M. S., Appelbaum, E., Menze, B., and Nezafat, R.
(2018). Automated Cardiac MR Scar Quantification
in Hypertrophic Cardiomyopathy Using Deep Con-
volutional Neural Networks. JACC: Cardiovascular
Imaging, 11(12):1917–1918.
Feng, S., Liu, Q., Patel, A., Bazai, S. U., Jin, C.-K., Kim,
J. S., Sarrafzadeh, M., Azzollini, D., Yeoh, J., Kim,
E., et al. (2022). Automated Pneumothorax Triaging
in Chest X-rays in the New Zealand Population Using
Deep-learning Algorithms. Journal of Medical Imag-
ing and Radiation Oncology, 66(8):1035–1043.
Gillette, K., Gsell, M. A., Prassl, A. J., Karabelas, E., Re-
iter, U., Reiter, G., Grandits, T., Payer, C.,
ˇ
Stern, D.,
Urschler, M., et al. (2021). A Framework for the Gen-
eration of Digital Twins of Cardiac Electrophysiology
from Clinical 12-leads ECGs. Medical Image Analy-
sis, 71:102080.
Graves, A., Mohamed, A.-r., and Hinton, G. (2013). Speech
Recognition with Deep Recurrent Neural Networks.
In Proceedings of the IEEE International Conference
on Acoustics, Speech and Signal Processing, pages
6645–6649.
Hamirani, Y. S., Wong, A., Kramer, C. M., and Salerno,
M. (2014). Effect of Microvascular Obstruction and
Intramyocardial Hemorrhage by CMR on LV Remod-
eling and Outcomes after Myocardial Infarction: A
Systematic Review and Meta-Analysis. JACC: Car-
diovascular Imaging, 7(9):940–952.
He, K., Zhang, X., Ren, S., and Sun, J. (2015). Delving
Deep into Rectifiers: Surpassing Human-Level Per-
formance on ImageNet Classification. In Proceedings
of the IEEE International Conference on Computer
Vision, pages 1026–1034.
Hellermann, J. P., Jacobsen, S. J., Gersh, B. J., Rodeheffer,
R. J., Reeder, G. S., and Roger, V. L. (2002). Heart
Failure after Myocardial Infarction: A Review. The
American Journal of Medicine, 113(4):324–330.
Khalid, A., Lim, E., Chan, B. T., Abdul Aziz, Y. F., Chee,
K. H., Yap, H. J., and Liew, Y. M. (2019). Assess-
ing Regional Left Ventricular Thickening Dysfunction
and Dyssynchrony via Personalized Modeling and 3D
Wall Thickness Measurements for Acute Myocardial
Infarction. Journal of Magnetic Resonance Imaging,
49(4):1006–1019.
Kim, R. J., Fieno, D. S., Parrish, T. B., Harris, K., Chen,
E.-L., Simonetti, O., Bundy, J., Finn, J. P., Klocke,
F. J., and Judd, R. M. (1999). Relationship of MRI
Delayed Contrast Enhancement to Irreversible Injury,
CaRe-CNN: Cascading Refinement CNN for Myocardial Infarct Segmentation with Microvascular Obstructions
63

Infarct Age, and Contractile Function. Circulation,
100(19):1992–2002.
Kim, R. J. and Manning, W. J. (2004). Viability Assess-
ment by Delayed Enhancement Cardiovascular Mag-
netic Resonance: Will Low-dose Dobutamine Dull the
Shine? Circulation, 109(21):2476–2479.
Kingma, D. P. and Ba, J. L. (2015). Adam: A Method for
Stochastic Optimization. In Proceedings of the Inter-
national Conference on Learning Representations.
Laine, S. and Aila, T. (2016). Temporal Ensembling for
Semi-Supervised Learning. In Proceedings of the In-
ternational Conference on Learning Representations.
Lalande, A., Chen, Z., Pommier, T., Decourselle, T.,
Qayyum, A., Salomon, M., Ginhac, D., Skan-
darani, Y., Boucher, A., Brahim, K., et al. (2022).
Deep Learning Methods for Automatic Evaluation
of Delayed Enhancement-MRI. The Results of the
EMIDEC Challenge. Medical Image Analysis,
79:102428.
Maas, A. L., Hannun, A. Y., and Ng, A. Y. (2013). Recti-
fier Nonlinearities Improve Neural Network Acoustic
Models. In Proceedings of the International Confer-
ence on Machine Learning, volume 30, page 3. At-
lanta, GA.
Moccia, S., Banali, R., Martini, C., Muscogiuri, G., Pon-
tone, G., Pepi, M., and Caiani, E. G. (2019). Develop-
ment and Testing of a Deep Learning-based Strategy
for Scar Segmentation on CMR-LGE Images. Mag-
netic Resonance Materials in Physics, Biology and
Medicine, 32:187–195.
Payer, C.,
ˇ
Stern, D., Bischof, H., and Urschler, M. (2017).
Multi-label Whole Heart Segmentation using CNNs
and Anatomical Label Configurations. In Interna-
tional Workshop on Statistical Atlases and Computa-
tional Models of the Heart, pages 190–198. Springer.
Payer, C.,
ˇ
Stern, D., Bischof, H., and Urschler, M. (2019).
Integrating Spatial Configuration into Heatmap Re-
gression based CNNs for Landmark Localization.
Medical Image Analysis, 54:207–219.
Payer, C.,
ˇ
Stern, D., Bischof, H., and Urschler, M. (2020).
Coarse to Fine Vertebrae Localization and Segmenta-
tion with SpatialConfiguration-Net and U-Net. In 15th
International Joint Conference on Computer Vision,
Imaging and Computer Graphics Theory and Applica-
tions (VISIGRAPP 2020) - Volume 5: VISAPP, pages
124–133.
Perin, E. C., Silva, G. V., Sarmento-Leite, R., Sousa, A. L.,
Howell, M., Muthupillai, R., Lambert, B., Vaughn,
W. K., and Flamm, S. D. (2002). Assessing My-
ocardial Viability and Infarct Transmurality with Left
Ventricular Electromechanical Mapping in Patients
with Stable Coronary Artery Disease: Validation by
Delayed-Enhancement Magnetic Resonance Imaging.
Circulation, 106(8):957–961.
Rios-Navarro, C., Marcos-Garces, V., Bayes-Genis, A.,
Husser, O., Nunez, J., and Bodi, V. (2019). Mi-
crovascular Obstruction in ST-segment Elevation My-
ocardial Infarction: Looking Back to Move For-
ward. Focus on CMR. Journal of Clinical Medicine,
8(11):1805.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-Net:
Convolutional Networks for Biomedical Image Seg-
mentation. In Proceedings of the International Con-
ference on Medical Image Computing and Computer-
Assisted Intervention, pages 234–241.
Rosenthal, M. E., Oseran, D. S., Gang, E., and Peter,
T. (1985). Sudden Cardiac Death following Acute
Myocardial Infarction. American Heart Journal,
109(4):865–876.
Schinkel, A. F., Poldermans, D., Elhendy, A., and Bax,
J. J. (2007). Assessment of Myocardial Viability
in Patients with Heart Failure. Journal of Nuclear
Medicine, 48(7):1135–1146.
Selvanayagam, J. B., Kardos, A., Francis, J. M., Wiesmann,
F., Petersen, S. E., Taggart, D. P., and Neubauer,
S. (2004). Value of Delayed-enhancement Cardio-
vascular Magnetic Resonance Imaging in Predicting
Myocardial Viability after Surgical Revascularization.
Circulation, 110(12):1535–1541.
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I.,
and Salakhutdinov, R. (2014). Dropout: A Sim-
ple Way to Prevent Neural Networks from Overfit-
ting. The Journal of Machine Learning Research,
15(1):1929–1958.
Van der Wall, E., Vliegen, H., De Roos, A., and Bruschke,
A. (1996). Magnetic Resonance Techniques for As-
sessment of Myocardial Viability. Journal of Cardio-
vascular Pharmacology, 28:37–44.
Walpot, J., Juneau, D., Massalha, S., Dwivedi, G., Rybicki,
F. J., Chow, B. J., and In
´
acio, J. R. (2019). Left Ven-
tricular Mid-diastolic Wall Thickness: Normal Values
for Coronary CT Angiography. Radiology: Cardio-
thoracic Imaging, 1(5):e190034.
Wroblewski, L. C., Aisen, A. M., Swanson, S. D., and
Buda, A. J. (1990). Evaluation of Myocardial Vi-
ability following Ischemic and Reperfusion Injury
using Phosphorus 31 Nuclear Magnetic Resonance
Spectroscopy in Vivo. American Heart Journal,
120(1):31–39.
Xu, C., Wang, Y., Zhang, D., Han, L., Zhang, Y., Chen,
J., and Li, S. (2022). BMAnet: Boundary Mining
with Adversarial Learning for Semi-supervised 2D
Myocardial Infarction Segmentation. IEEE Journal
of Biomedical and Health Informatics, 27(1):87–96.
Xu, C., Xu, L., Gao, Z., Zhao, S., Zhang, H., Zhang, Y., Du,
X., Zhao, S., Ghista, D., Liu, H., et al. (2018). Direct
Delineation of Myocardial Infarction without Contrast
Agents using a Joint Motion Feature Learning Archi-
tecture. Medical Image Analysis, 50:82–94.
Zabihollahy, F., White, J. A., and Ukwatta, E. (2018). My-
ocardial Scar Segmentation from Magnetic Resonance
Images using Convolutional Neural Network. In Med-
ical Imaging 2018: Computer-Aided Diagnosis, vol-
ume 10575, pages 663–670. SPIE.
Zhang, Y. (2021). Cascaded Convolutional Neural Network
for Automatic Myocardial Infarction Segmentation
from Delayed-enhancement Cardiac MRI. In Interna-
tional Workshop on Statistical Atlases and Computa-
tional Models of the Heart, pages 328–333. Springer.
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
64
