
Fetal Health Classification Using One-Dimensional Convolutional Neural
Network
Anton Johan R
¨
oscher
a
and Dustin van der Haar
b
Academy of Computer Science and Software Engineering, University of Johannesburg, Kingsway Avenue and
University Rd, Auckland Park, South Africa
Keywords:
1D-CNN, CTG, Deep Learning.
Abstract:
Within the medical field, machine learning has the potential to allow doctors and medical professionals to make
faster, more accurate diagnoses, empowering specialists to take immediate action. Early diagnosis and preven-
tion of fetal health conditions can be achieved based on the biomarker data derived from the cardiotocography
signals. The study proposes using a one-dimensional convolutional neural network for fetal health classifica-
tion and compares it to conventional machine learning algorithms. A one-dimensional convolutional neural
network is shown to outperform traditional machine learning algorithms in both data sets (CTU-CHB and
UCI), with an accuracy of 89% - 94%.
1 INTRODUCTION
Artificial intelligence for early diagnosis of medi-
cal cases is invaluable to saving lives and preventing
other chronic patient conditions due to late diagnosis.
Several machine-learning models have been applied
successfully in cancer detection and diagnosis (Simes,
1985; Maclin et al., 1991; Cicchetti, 1992), tumor
classification and malignant cases through X-Ray ex-
amination (Bocchi et al., 2004). Artificial intelligence
allows doctors and medical professionals to diagnose
faster and more accurately empowering specialists to
take immediate actions based on the biomarker data
provided to the model.
Cardiotocography (CTG) is used during prenatal
and birth with the intention that the status of a foetus
can be classified as normal, suspect, or pathological.
The classifications are based on derived features from
the Fetal Heart Rate (FHR) and Urinary Contractions
(UC) for a given signal and are outlined in the Car-
diotocograph Interpretation and Response (car, 2020)
as well as the International Federation of Gynecology
and Obstetrics (FIGO) consensus guidelines on intra-
partum fetal monitoring (de Campos et al., 2015).
The main objective of this study is to build a com-
puterised model that will, to a certain precision, clas-
sify different cases of intrapartum-related conditions.
a
https://orcid.org/0000-0001-5374-5418
b
https://orcid.org/0000-0002-5632-1220
The study aims to address the shortage of the cur-
rent models that cannot classify suspected cases well
(C
¨
omert et al., 2016; Sundar et al., 2012) by compar-
ing various models and their macro accuracy for clas-
sifications. The contribution of this research to the
classification of CTG signals is to compare the per-
formance results of five supervised machine learning
models to improve Accuracy, F1-Score, Precision and
Recall across both the UCI and CTU data sets.
The single-classifier machine-learning models K-
Nearest Neighbours, Support Vector Machine, and
Decision Tree are considered based on their success
in previous studies. A decision tree is extended to a
Random Forest ensemble method to reduce the spread
of predictions and create a more robust model. Lastly,
a multi-layer perceptron (MLP) is used due to its pre-
dictive capabilities originating from its hierarchical
neuron structure. MLPs can solve problems stochas-
tically, allowing for solutions to complex problems to
be approximated accurately.
The outline of the paper is structured as follows.
Section 2 indicates related work in fetal cardiac clas-
sification and the best-performing machine learning
models. Section 3 specifies the data sets. Section 4
discusses the results on different data sets, with sec-
tion 5 concluding the research study.
Röscher, A. and van der Haar, D.
Fetal Health Classification Using One-Dimensional Convolutional Neural Network.
DOI: 10.5220/0012322300003654
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 13th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2024), pages 671-678
ISBN: 978-989-758-684-2; ISSN: 2184-4313
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
671

2 RELATED WORKS
Existing works has seen the following machine learn-
ing models being used: K-means, Decision Trees,
Random Forests, Multi-Layer Perceptron, Radial Ba-
sis Function Support Vector Machines, and Extreme
Learning Machines (Comert and Kocamaz, 2017;
Arif et al., 2020; Ayres-de Campos et al., 2005;
Chamidah and Wasito, 2015) due to their excellent
accuracy in evaluation and classifications of CTG sig-
nals. A study done by Ogasawara et al. (Ogasawara
et al., 2021) claims that a deep neural network-based
classification consisting of three convolutional layers
outperform conventional algorithms in accuracy and
precision for the same time window.
Kanika et al. (Agrawal and Mohan, 2019) as-
sert that a decision tree and a support vector machine
achieved above 90% accuracy, rivalling a deep neural
network model. Likewise, Sihem et al. (NITA et al.,
2018) believe that random forest is better suited for
CTG predictions due to its lower chance of overfitting
a model to cardiac-centred data. In 2017, Anish et al.
(Batra et al., 2017) focused on cardiotocography anal-
ysis using decision trees, support vector machines,
random forests and neural network machine learn-
ing algorithms by comparing their overall accuracy
through a confusion matrix. Anish et al. concluded
that most machine learning algorithms were similar
in accuracy (> 90%), with a decision tree achieving
the highest accuracy between these methods with a
95% accuracy.
The use of deep learning algorithms is a well-
established approach (Francis et al., 2022; Liang and
Li, 2021; Sahin and Subasi, 2015) to the classifica-
tion of fetal cardiotocography signals and overall fe-
tal well-being. Sai and Qia used a two-layer Convolu-
tional Neural Network for the classification of a CTG
signal (Chud
´
a
ˇ
cek et al., 2014) and evaluated it against
seven trained models.
The utilisation of ensemble models leads to not
only better performance in multi-class classification
but provide more accurate results over conventional
models (Rosly et al., 2018) within the medical appli-
cations.
3 METHODS
3.1 Data Sets
The CTU-CHB Intrapartum Cardiotocography
Database (Chud
´
a
ˇ
cek et al., 2014) consists of 552
intrapartum CTG recordings, acquired between 2009
and 2012 at the obstetrics ward of the University
Hospital in Brno, Czech Republic (Chud
´
a
ˇ
cek et al.,
2014). The ground truth labels of this data set were
not provided, and a manual classification based on
the umbilical artery pH balance, with a threshold
lower than 7.25, a low Apgar score at 5 minutes and
an increase in heart accelerations were used. An
increase in heart accelerations is a strong indicator
of a well oxygenated foetus, and, in conjunction
with pH and Apgar scores from the FIGO guidelines
(Sehgal et al., 2017) and a study done by Allanson
et al. (.ER et al., 2016), are the most distinguishing
features between normal and pathological outcomes.
The UCI dataset contains 2,126 fetal cardio-
grams, which have been subjected to feature extrac-
tion (Chud
´
a
ˇ
cek et al., 2014) and classified by three
professional obstetricians (de Campos et al., 2015;
Kadhim and Abed, 2020). The depictions of the
classes are (Sehgal et al., 2017; C et al., 2012): Nor-
mal where all morphological features fall within the
reassuring category. Suspect where some morpholog-
ical features fall within one of the non-reassuring cat-
egories, while the remainder resides within the reas-
suring category. Pathological in the case where two
or more morphological features fall within multiple
non-reassuring categories. In both datasets a notable
class imbalance exists, the CTU dataset feature dou-
ble the amount of pathological outcomes versus nor-
mal and suspect. In the UCI dataset the normal class
is heavily oversampled, constituting of 78% of the en-
tire dataset.
3.2 Data Pre-Processing
The CTU dataset required extensive pre-processing
to extract the best representation of the recorded sig-
nal. The figures below display the heart’s beats per
minute, which has been sampled at a frequency of
4Hz to provide four data points every second.
Figure 1: Raw unfiltered CTG Signal from the CTU data
set.
As seen in Figure 1, the raw CTG signal is sub-
ject to gaps where the heart rate incorrectly drops to
0; for that reason, a 20 minute window of the signal
was extracted to stay consistent across signals. Based
on the FIGO guidelines, a reading of 30 minutes is
required for the assessment of a CTG signal and ex-
ICPRAM 2024 - 13th International Conference on Pattern Recognition Applications and Methods
672

tended if the FHR pattern seems suspicious (S1G,
2014) . A sliding window was used to determine a
valid, stable starting point by ensuring that a change
of 10 bpm is not achieved for a minimum of 5 sam-
ples. Gaps shorter than five samples were then filled
with the mean value of the valid segment, and the
signal is interpolated using the Hermite spline inter-
polation mathematical function. The sample outliers
were then addressed, such that all values reside within
80 ≤ N ≤ 200, in line with FIGO Guidelines.
Figure 2: Filtered and interpolated CTG Signal after pro-
cessing.
3.3 Feature Extraction
Morphological Features refer to an organ’s shape,
structure, and functional characteristics. The FIGO
Guidelines focus on visually defined macroscopic
features of the fetal heart, along with numerous other
morphological measures (Sehgal et al., 2017) The
morphological features that were used in this study
are as follows: the mean FHR Baseline heart rate
in which the presence of accelerated and deceler-
ated heart rate is not present; the mean FHR heart
rate for the valid segment; the Number of Deceler-
ations in which the FHR slows down, as a result of
the hormones released from the parasympathetic flow
(Resnik et al., 2018), by more than 15 bpm over a 10-
second window; Number of Accelerations in which
the FHR speeds up due to the hormones released
from the sympathetic flow (Resnik et al., 2018), by
more than 15 bpm over a 15-second window. In ad-
dition to these features, the mean absolute deviation
is computed for the average distance between each
data point and the mean FHR value. After the mor-
phological features have been extracted, the data is
no longer constrained to the time domain but instead
considered as a representation of the most prominent
identifiers within the 20-minute extracted signal. The
non-morphological features used were present in the
accompanying signal meta-data and contained sup-
plementary information such as the pH value, Apgar
scores, foetus weeks, Meconium and Hypertension.
3.4 Model Selection and
Implementation
The choice of machine learning models used in this
research study, is drawn from the comprehensive re-
view of the literature in this field. The following mod-
els and their respective implementations are listed be-
low:
1. K Nearest Neighbours (KNN). The KNN algo-
rithm employed Euclidean distance calculation as
its metric to assess the proximity of data points.
2. Decision Tree. The implemented decision tree al-
gorithm is based on the Classification And Re-
gression Trees (CART) algorithm. The imple-
mentation utilises the Gini index as the splitting
criteria and incorporates bagging to generate mul-
tiple subsets of samples and training multiple de-
cision tree instances.
3. Random Forest. The random forest algorithm
used the same structure as the decision trees with
pruning to ensure trees are less suceptible to over-
fitting.
4. Support Vector Machine. One-vs-One classifier
consisting of N classes and N =
N(N−1)
2
binary
models are trained, with the sample classified as
the most voted class. A One-vs-One classifier was
used to mitigate potential class imbalances along
with a linear kernel to avoid overfitting and the
impact of potential outliers.
5. One-Dimensional Convolutional Neural Network
(1D-CNN). The traditional one-dimensional con-
volutional neural network consists of three main
layers - a convolutional layer, a pooling layer and
a fully connected layer.
The convolutional layer is responsible for the
computation of various features from the input
data by applying a mathematical convolution op-
eration to produce a feature map before being
pooled to downsample the feature map and reduce
computational costs.
Employing a scalable hyperparameter optimisa-
tion framework, various 1 Dimensional CNN con-
figurations were tested for their overall accuracy,
with the following topology being the most accu-
rate. One key factor during the training of the one-
dimensional convolutional neural network was the
learning rate, considered the contributing factor
to the convergence of the model. Using the loss
graph against each training step indicated the piv-
otal section where the loss decreased the fastest
as well as the learning rate used at that step. This
Fetal Health Classification Using One-Dimensional Convolutional Neural Network
673
learning rate was determined to be 0.001 and was
selected for subsequent model training iterations.
Further to the learning rate, the convolutional and
dense layers were diversified. Different com-
binations of accuracies were evaluated through
sparse categorical cross-entropy as the loss func-
tion. Whilst tuning hyperparameters, an early stop
with a patience of 20 epochs and model check-
pointing after each epoch was implemented for
model evaluation. Each tested configuration var-
ied in layers and neurons and trained for 100
epochs to establish the most accurate model topol-
ogy. The model parameters are as follows:
1. Three one-dimensional convolutional layers of
filter and kernel size 8, 6, and 3, respectively,
are responsible for feature extraction.
2. One dimensional max pooling layer of pooling
size 3 is responsible for reducing the dimen-
sionality.
3. One layer to flatten the feature map to a one-
dimensional input for the fully connected lay-
ers.
4. Three fully connected dense layers with 132
and 68 neurons and a relu activation function
for the first two layers and the final layer con-
sisting of 3 neurons and the softmax function.
The simplicity of this CNN network topology re-
duces the chance of the model overfitting during
training whilst ensuring the least impact on the
performance degradation.
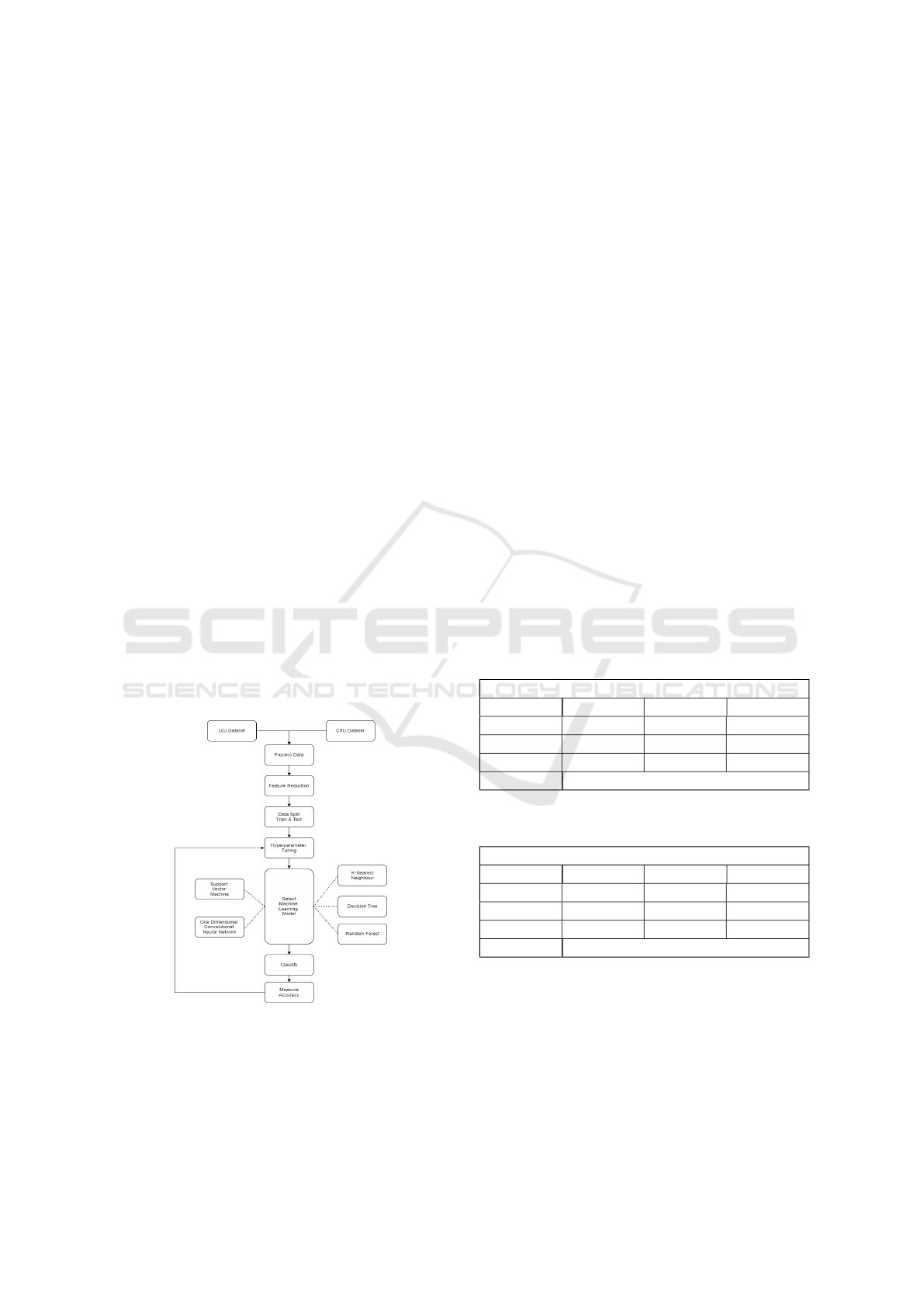
Figure 3: Applied machine learning methodology.
Figure 3 shows the outline of the steps taken, sub-
stituting different machine algorithms and tuning the
hyperparameters of each of these to obtain the best
accuracy.
Each model was evaluated using a confusion ma-
trix obtained in a one-vs-all manner where the Pre-
cision, Recall, and F-1 score values were computed
for each class separately. Each evaluation metric was
calculated for the respective classification classes, and
the overall performance of a multi-class classification
model could be determined by summarising the micro
value of each class, based on different hyperparame-
ters.
4 RESULTS AND DISCUSSION
An initial objective of the project was to evaluate and
compare the accuracy of different machine-learning
algorithms when classifying fetal cardiac conditions.
The accuracy of both 80/20 and 70/30 training/test set
configurations were evaluated. The larger training set
was chosen to ensure an ample test set for model eval-
uation. Evaluation metrics used within similar studies
(Ogasawara et al., 2021; Bernardes, 2022; C et al.,
2012; Arif et al., 2020) are Accuracy, F1-Score, Pre-
cision, and Recall with the following class mappings:
N=Normal, S=Suspect, P=Pathological
4.1 K Nearest Neighbours (KNN)
Table 1: K Nearest Neighbours performance metrics on the
CTU-CHB Interpartum data set.
K=5
Precision Recall F1-Score
N 84 % 74 % 78 %
S 82 % 78 % 80 %
P 82 % 88 % 85 %
Accuracy 82.17 %
Table 2: K Nearest Neighbours performance metrics on the
UCI data set.
K=5
Precision Recall F1-Score
N 85 % 72 % 78 %
S 85 % 75 % 79 %
P 80 % 90 % 85 %
Accuracy 88.06%
Table 1 presents the summary statistics for the K
Nearest Neighbours algorithm applied to the CTU-
CHB Interpartum data set. The K Nearest Neighbours
machine learning model could consistently and accu-
rately form a robust decision boundary and correctly
classify samples with a marginal increase in accuracy
(≈ 0.3 − 0.8) between 7 and 11 neighbour considera-
tions. Hakan and Abdulhamit also reported a similar
ICPRAM 2024 - 13th International Conference on Pattern Recognition Applications and Methods
674
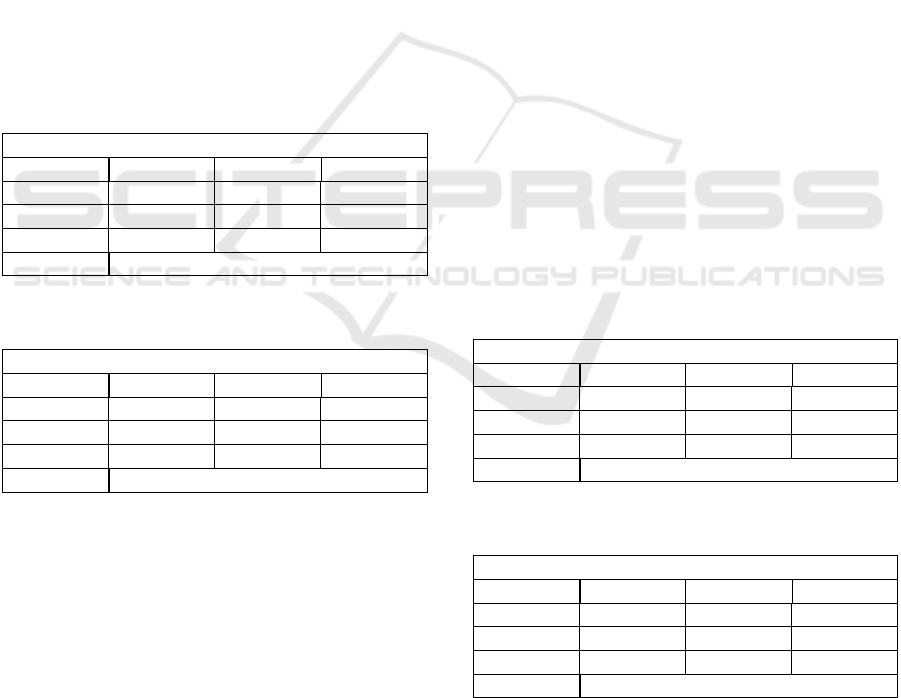
finding (Sahin and Subasi, 2015), albeit with higher
accuracy.
Table 2 shows strong evidence of an accurate K
Nearest Neighbours classifier, with an overall accu-
racy of 88.06%, lower than previously reported by
Hakan and Abdulhamit (Sahin and Subasi, 2015) on
the same data set. It could therefore be hypothesised
that the K Nearest Neighbour algorithm accurately
classifies fetal cardiac state as clustered samples are
homogeneous in terms of their features. The K Near-
est Neighbours algorithm inherently doesn’t make as-
sumptions on data distributions and the effectiveness
of the algorithm might indicate that the algorithm is
able to clearly identify complex and non-linear rela-
tionships latent in the data. A comparison between
the two results of the respective data sets, reveals that
the K Nearest Neighbours is an effective fetal cardiac
classification algorithm, given clear decision bound-
ary separations between clusters can be determined.
4.2 Decision Tree
Table 3: Decision Tree performance metrics on the CTU-
CHB Interpartum data set.
Max Depth=5
Precision Recall F1-Score
N 88 % 69 % 77 %
S 78 % 80 % 79 %
P 81 % 88 % 84 %
Accuracy 81.39 %
Table 4: Decision Tree performance metrics on the UCI
data set.
Max Depth=5
Precision Recall F1-Score
N 92 % 94 % 93 %
S 56 % 57 % 57 %
P 89 % 72 % 80 %
Accuracy 86.89 %
The results obtained from the analysis of decision
trees on fetal cardiac classification are summarised in
Table 3 and Table 4 for both the CTU-CHB Interpar-
tum and UCI Repository data sets, respectively.
It is encouraging to compare the findings of this re-
search study with those of other studies (Batra et al.,
2017; Sahin and Subasi, 2015; Rosly et al., 2018) who
found the accuracy of decision trees in fetal cardiac
classification to be highly accurate. The single most
striking observation to emerge from the data compar-
ison was the ability of the model to resist bias in un-
dersampled classes and maintain accuracy across dif-
ferent data sets. These results further support the hy-
pothesis that multiple perpendicular splits to the fea-
ture axes, combined, are capable of inferring com-
plex non-linear relationships within the entire feature
space to determine an effective decision boundary.
Surprisingly in both instances, changes in the maxi-
mum depth of the tree did not yield a noticeable im-
provement in the overall accuracy of the classifier.
In the current study, comparing the results of the
decision tree classifier for the two different data sets
showed the model is flexible in the number of fea-
tures used for classification, with a mean difference
of 3% in accuracy between varying tree parameters.
These results seem to be consistent with other re-
search, which found that the decision tree algorithm is
scalable to larger data sets whilst preserving its clas-
sification accuracy (Arif et al., 2020).
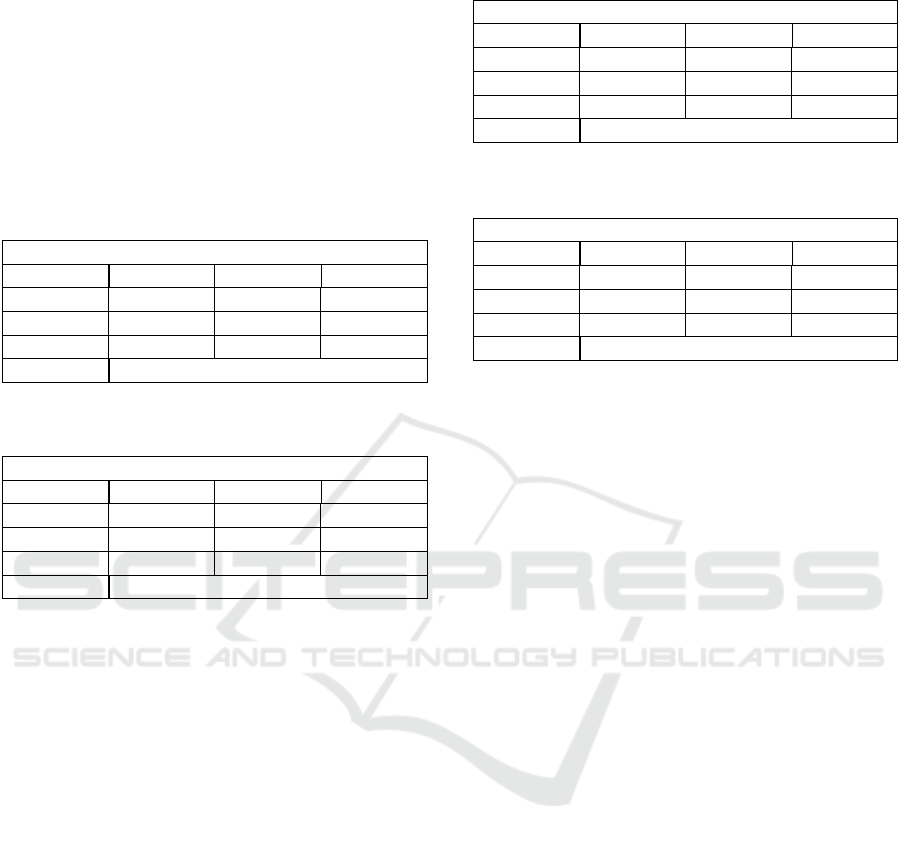
4.3 Random Forest
The findings illustrated below are consistent with that
of similar studies (Sahin and Subasi, 2015; Batra
et al., 2017) who noted that the accuracy of the Ran-
dom Forest algorithm and decision tree differ by no
more than 10% when utilising the same tree structure.
However, as indicated by Sihem et al. (NITA et al.,
2018), the accuracy of a Random Forest is propor-
tional to the number of trees present and the max-
imum depth of each tree. This is confirmed when
a forest of 325 trees with a maximum depth of 5 is
constructed, and a resulting accuracy of 80.87% and
88.90% are obtained respectively.
Table 5: Random Forest performance metrics on the CTU
data set.
Trees=325, Max Depth=5
Precision Recall F1-Score
N 82 % 73 % 77 %
S 78 % 78 % 78 %
P 82 % 86 % 84 %
Accuracy 80.87 %
Table 6: Random Forest performance metrics on the UCI
data set.
Trees=325, Max Depth=5
Precision Recall F1-Score
N 93 % 95 % 94 %
S 63 % 62 % 63 %
P 91 % 74 % 82 %
Accuracy 88.90 %
The marginal increases between the decision tree
and random forest classifiers are most likely attributed
to the limited data complexities and may not exploit
the advantages of a random forest to the fullest ex-
tent. The compact nature of the dataset cannot be
Fetal Health Classification Using One-Dimensional Convolutional Neural Network
675
dismissed, as it may exhibit constraints in terms of di-
versity and limited instances for classification classes.
These constraints can impact the generalising capabil-
ities of the random forest classifier. Unlike previous
studies, this study has been unable to demonstrate that
the ensemble method is better suited for fetal cardiac
classification, so that these findings may be limited.
4.4 Support Vector Machine
Table 7: Support Vector Machine performance metrics on
the CTU data set.
Func=OVO, Kernel=Linear
Precision Recall F1-Score
N 80 % 75 % 77 %
S 81 % 74 % 77 %
P 82 % 88 % 85 %
Accuracy 81.13 %
Table 8: Support Vector Machine performance metrics on
the UCI data set.
Func=OVO, Kernel=Linear
Precision Recall F1-Score
N 91 % 97 % 94 %
S 67 % 56 % 61 %
P 95 % 69 % 80 %
Accuracy 88.90 %
As seen from Table 7 and Table 8, the Linear
kernel and One-vs-One (OVO) performed well in
both data sets when classifying fetal cardiac state.
This outcome is contrary to a previous study which
only achieved an overall accuracy of ≈ 63% with-
out standardisation techniques of a 2-class diagnosis
(Nahiduzzaman et al., 2019). The results obtained in
this study for a linear Support Vector Classifier (SVC)
are in alignment with a similar study where a SVC
was successful in the classification of the fetal cardiac
state (Chamidah and Wasito, 2015) when given a fea-
ture space that is linearly separable. When we com-
pare the results of the SVM to that of the K Nearest
Neighbour, it can be seen that there exists linear hy-
perplanes that can correctly identify decision bound-
aries between the various classes using a One-vs-One
approach as opposed to a One-vs-All approach.
4.5 One-Dimensional Convolutional
Neural Network
The results, as shown in Table 9 and Table 10, re-
veals that a one-dimensional Convolutional Neural
Network is highly effective at determining classifica-
tions across both CTG data sets while accounting for
Table 9: One-dimensional convolutional neural network
performance metrics on the CTU data set.
Epochs=3000, Training Size=70%
Precision Recall F1-Score
N 95 % 82 % 88 %
S 99 % 96 % 97 %
P 91 % 98 % 95 %
Accuracy 93.79 %
Table 10: One-dimensional convolutional neural network
performance metrics on the UCI data set.
Epochs=3000, Training Size=70%
Precision Recall F1-Score
N 93 % 95 % 94 %
S 72 % 60 % 65 %
P 79 % 85 % 82 %
Accuracy 89.21 %
potential bias resulting from a lack of a large data set.
Per the present results, previous studies (Batra
et al., 2017; Sahin and Subasi, 2015; Ogasawara et al.,
2021; Liang and Li, 2021) have demonstrated that an
artificial neural network is better able to discern rela-
tionships that might be overseen by conventional ma-
chine learning algorithms based on the network topol-
ogy used. This theory is clearly highlighted when the
accuracy is compared to a relatively simple classifica-
tion model such as the K Nearest-Neighbour model,
where an increase in accuracy can be observed but
is limited to the data set and features used. This in-
crease in accuracy highlights the potential of the 1D
CNN being able to discern and capture local patterns
that are of importance within the signal data to extract
important features.
One such network topology is a Long-Short Term
Model and should be considered in future research
due to its excellent performance on time-sensitive
data, such as in the CTU data set. It can therefore
be assumed that Artificial Neural Networks outper-
form conventional algorithms when classifying fetal
cardiac state, with the possibility of time dependen-
cies being highlighted and compared to traditional al-
gorithms for the same period.
This study supports evidence from previous ob-
servations that deep learning architectures are excel-
lent at function approximation for learning represen-
tations of data through weights and biases. The re-
sults displayed in Tables 9 and 10 match those ob-
served in earlier studies where deep learning methods
outperformed conventional machine learning meth-
ods (Chamidah and Wasito, 2015; Ogasawara et al.,
2021; Batra et al., 2017) for classification. More-
over, it was noted that an increase in the training
data size (80/20) resulted in an increase in accuracy
ICPRAM 2024 - 13th International Conference on Pattern Recognition Applications and Methods
676

(> 4%) for the same model topology, albeit small
data sets for an Artificial Neural Network domain.
In both data set instances, the CNN could infer the
presence of an under-sampled class better than con-
ventional models and effectively distinguish between
them This is a rather significant outcome and has not
yet been described by previous studies from Sia, and
Qia (Liang and Li, 2021) and Wafaa et al. (Alsag-
gaf et al., 2020), where oversampling was addressed
using the synthetic minority oversampling technique
(SMOTE).
This study aims to expand the domain of deep
learning methods applied to fetal cardiotocography
classification, by providing an additional Convolu-
tional Neural Network topology, that is one dimen-
sional, and effective at classifying signal data. This
network topology differs from the previous Convo-
lutional Neural Networks used (Liang and Li, 2021)
as well as previous Multi-layer Perceptron topolo-
gies (Batra et al., 2017) and Long-Short Term Mem-
ory (LSTM) topologies (Chamidah and Wasito, 2015;
Ogasawara et al., 2021).
5 CONCLUSION
Fetal cardiotocography signals are exclusively used to
determine the cardiac state of a foetus during preg-
nancy. This study set out to compare machine learn-
ing algorithms applied to the classification of fe-
tal cardiac state to determine the most suitable ap-
proaches. The K Nearest Neighbours, Decision Tree,
and Random Forest algorithms results corroborated
the findings in previous studies (Batra et al., 2017;
Sahin and Subasi, 2015; Alsaggaf et al., 2020) with
similar accuracies on a shorter signal window. The
study noted the importance of features provided to
machine-learning classifiers and are inline with more
recent findings (Zhong et al., 2022) where the base-
line FHR, accelerated and decelerated FHR patterns
observed play a vital role in the analysis of foetus
heart rate. Results observed from the CNN applica-
tions on both datasets raise thought provoking ques-
tions regarding the nature and extent of neural net-
works and their ability to uncover latent relation-
ships, even in smaller datasets where diversity could
be small. This idea is further strengthened by the fact
that 20-minute signal excerpt is used over the previ-
ously used 30-minute signal excerpt by Ogasawara et
al. (Ogasawara et al., 2021). Moreover, a smaller set
of self-extracted signal features were used for the 1-
Dimensional CNN for which the model could still dis-
cern relationships to classify foetus state accurately.
A further study focusing on relationships between
fetal cardiac features and one-dimensional convolu-
tional neural networks are encouraged with different
network topologies.
In conclusion, the study successfully achieved ac-
curate classification of fetal cardiac states based on
features provided to various machine learning algo-
rithms. This study indicated that conventional ma-
chine learning algorithms are well suited for fetal
cardiac classification with a one-dimensional convo-
lutional neural network being best at discerning re-
lationships between different classes and, therefore
classify samples more accurately than conventional
machine learning methods. Together these results
provide important insights into successful machine-
learning fetal cardiac classification with the impor-
tance of deep learning methods for future research.
REFERENCES
(2014). S1-guideline on the use of CTG during preg-
nancy and labor. Geburtshilfe und Frauenheilkunde,
74(08):721–732.
(2020). Ctg interpretation and response - the royal women’s
hospital.
Agrawal, K. and Mohan, H. (2019). Cardiotocography anal-
ysis for fetal state classification using machine learn-
ing algorithms. pages 1–6.
Alsaggaf, W., C
¨
omert, Z., Nour, M., Polat, K., Brdesee,
H., and To
˘
gac¸ar, M. (2020). Predicting fetal hypoxia
using common spatial pattern and machine learning
from cardiotocography signals. Applied Acoustics,
167:107429.
Arif, M., Ahmed, R., Sadia, U., Tultul, S., and Chakma, R.
(2020). Decision tree method using for fetal state clas-
sification from cardiotography data. Journal of Ad-
vanced Engineering and Computation, 4.
Ayres-de Campos, D., Costa-Santos, C., Bernardes, J.,
Group, S. M. V. S., et al. (2005). Prediction of neona-
tal state by computer analysis of fetal heart rate trac-
ings: the antepartum arm of the sisporto® multicentre
validation study. European Journal of Obstetrics &
Gynecology and Reproductive Biology, 118(1):52–60.
Batra, A., Chandra, A., and Matoria, V. (2017). Car-
diotocography analysis using conjunction of machine
learning algorithms. In 2017 International Confer-
ence on Machine Vision and Information Technology
(CMVIT). IEEE.
Bernardes, J. (2022). Computerized analysis of car-
diotocograms in clinical practice physiopathological
and clinical studies. Journal of Perinatal Medicine,
0(0).
Bocchi, L., Coppini, G., Nori, J., and Valli, G. (2004). De-
tection of single and clustered microcalcifications in
mammograms using fractals models and neural net-
works. Medical engineering & physics, 26(4):303–
312.
Fetal Health Classification Using One-Dimensional Convolutional Neural Network
677

C, S., M.Chitradevi, M., and Geetharamani, G. (2012).
Classification of cardiotocogram data using neural
network based machine learning technique. Interna-
tional Journal of Computer Applications, 47(14):19–
25.
Chamidah, N. and Wasito, I. (2015). Fetal state classi-
fication from cardiotocography based on feature ex-
traction using hybrid k-means and support vector ma-
chine. In 2015 International Conference on Advanced
Computer Science and Information Systems (ICAC-
SIS). IEEE.
Chud
´
a
ˇ
cek, V., Spilka, J., Bur
ˇ
sa, M., Jank
˚
u, P., Hruban, L.,
Huptych, M., and Lhotsk
´
a, L. (2014). Open access in-
trapartum CTG database. BMC Pregnancy and Child-
birth, 14(1).
Cicchetti, D. V. (1992). Neural networks and diagnosis in
the clinical laboratory: state of the art. Clinical chem-
istry, 38(1):9–10.
Comert, Z. and Kocamaz, A. (2017). Comparison of ma-
chine learning techniques for fetal heart rate classifi-
cation.
C
¨
omert, Z., Kocamaz, A. F., and G
¨
ung
¨
or, S. (2016). Car-
diotocography signals with artificial neural network
and extreme learning machine. In 2016 24th Signal
Processing and Communication Application Confer-
ence (SIU), pages 1493–1496. Ieee.
de Campos, D. A., Spong, C. Y., and and, E. C. (2015).
FIGO consensus guidelines on intrapartum fetal mon-
itoring: Cardiotocography. International Journal of
Gynecology & Obstetrics, 131(1):13–24.
.ER, A., .T, W., .CRH, W., .O, T., and JE, D. . (2016).
Umbilical lactate as a measure of acidosis and pre-
dictor of neonatal risk: a systematic review. An In-
ternational Journal of Obstetrics and Gynaecology,
124(4):584–594.
Francis, F., Wu, H., Luz, S., Townsend, R., and Stock, S.
(2022). Detecting intrapartum fetal hypoxia from car-
diotocography using machine learning. In 2022 Com-
puting in Cardiology (CinC), volume 498, pages 1–4.
Kadhim, N. J. A. and Abed, J. K. (2020). Enhancing the
prediction accuracy for cardiotocography (CTG) us-
ing firefly algorithm and naive bayesian classifier. IOP
Conference Series: Materials Science and Engineer-
ing, 745(1):012101.
Liang, S. and Li, Q. (2021). Automatic evaluation of fetal
heart rate based on deep learning. In 2021 2nd In-
formation Communication Technologies Conference
(ICTC). IEEE.
Maclin, P. S., Dempsey, J., Brooks, J., and Rand, J. (1991).
Using neural networks to diagnose cancer. Journal of
medical systems, 15(1):11–19.
Nahiduzzaman, M., Nayeem, M. J., Ahmed, M. T., and Za-
man, M. S. U. (2019). Prediction of heart disease us-
ing multi-layer perceptron neural network and support
vector machine. In 2019 4th International Conference
on Electrical Information and Communication Tech-
nology (EICT). IEEE.
NITA, S., BITAM, S., and MELLOUK, A. (2018). An en-
hanced random forest for cardiac diseases identifica-
tion based on ECG signal. In 2018 14th International
Wireless Communications & Mobile Computing Con-
ference (IWCMC). IEEE.
Ogasawara, J., Ikenoue, S., Yamamoto, H., Sato, M.,
Kasuga, Y., Mitsukura, Y., Ikegaya, Y., Yasui, M.,
Tanaka, M., and Ochiai, D. (2021). Deep neural
network-based classification of cardiotocograms out-
performed conventional algorithms. Scientific Re-
ports, 11(1).
Resnik, R., Lockwood, C. J., Moore, T., Greene, M. F.,
Copel, J., and Silver, R. M. (2018). Creasy and
resnik’s maternal-fetal medicine: Principles and
practice. Elsevier, 8 edition.
Rosly, R., Makhtar, M., Awang, M. K., Awang, M. I., Rah-
man, M. N. A., and Mahdin, H. (2018). Compre-
hensive study on ensemble classification for medical
applications. International Journal of Engineering
&Technology, 7(2.14):186.
Sahin, H. and Subasi, A. (2015). Classification of the car-
diotocogram data for anticipation of fetal risks using
machine learning techniques. Applied Soft Comput-
ing, 33:231–238.
Sehgal, A., Allison, B. J., Gwini, S. M., Miller, S. L.,
and Polglase, G. R. (2017). Cardiac morphology and
function in preterm growth restricted infants: Rele-
vance for clinical sequelae. The Journal of Pediatrics,
188:128–134.e2.
Simes, R. J. (1985). Treatment selection for cancer patients:
application of statistical decision theory to the treat-
ment of advanced ovarian cancer. Journal of chronic
diseases, 38(2):171–186.
Sundar, C., Chitradevi, M., and Geetharamani, G. (2012).
Classification of cardiotocogram data using neural
network based machine learning technique. Interna-
tional Journal of Computer Applications, 47(14).
Zhong, M., Yi, H., Lai, F., Liu, M., Zeng, R., Kang, X.,
Xiao, Y., Rong, J., Wang, H., Bai, J., and Lu, Y.
(2022). CTGNet: Automatic analysis of fetal heart
rate from cardiotocograph using artificial intelligence.
Maternal-Fetal Medicine, 4(2):103–112.
ICPRAM 2024 - 13th International Conference on Pattern Recognition Applications and Methods
678
