
Comparison Between Graph Databases and RDF Engines for Modelling
Epidemiological Investigation of Nosocomial Infections
Lorena Pujante-Otalora
1
, Manuel Campos
1,2
, Jose M. Juarez
1
and Maria-Esther Vidal
3
1
MedAILab, Department of IT and Systems, University of Murcia, Campus Espinardo, Murcia, 30100, Spain
2
Murcian Bio-Health Institute, IMIB-Arrixaca, El Palmar, Murcia, 30120, Spain
3
Leibniz University of Hannover and L3S Research Center and TIB Leibniz Information Centre for Science and
Technology, Hannover, Germany
Keywords: Graph Database, Knowledge Graphs, Epidemiology, Benchmark, Neo4j, GraphDB.
Abstract: We have evaluated the performance of property and knowledge graph databases in the context of
spatiotemporal epidemiological investigation of an infection outbreak in a hospital. Specifically, we have
chosen Neo4j as graph database, and GraphDB for knowledge graphs defined following RDF and its extension
RDF*. We have defined a domain model describing a hospital layout and patient movements. For
performance comparison, we have created ten graphs with different sizes based on MIMIC-III, implemented
three epidemiological queries in Cypher, SPARQL and SPARQL* and defined three benchmarks that measure
the execution time and main memory consumption of the three queries in each graph and database engine.
Our research suggests that query complexity is a more determinant factor than graph size in the performance
of the query executions. Neo4j presents better times and memory consumption than GraphDB for simple
queries, but GraphDB is more efficient when traversing big subgraphs. Between RDF and RDF*, RDF* offers
a more compact and human-friendly modelling and a better performance of the query execution.
1 INTRODUCTION
In recent years, we have witnessed an exponential
growth in data to be stored and processed at any area,
what is called “big data”. This vast amount of data
with no rigid schemes and generally stored across
multiple machines promoted some new type of
databases, encompassed in the term NoSQL
databases (Leavitt, 2010). Graph databases are
NoSQL databases that represent data as property
graphs, providing an efficient storage and
management of highly interconnected data. The
interest in modelling data as property graphs has
increased in recent years due to their advantages in
network analysis and the semantic web. Related to
this context, we also find knowledge graphs, a
semantic web technology that links massive and
cross-domain data used on tasks like information
retrieval, hidden linkage identification, and
knowledge-driven decision support (Li et al., 2022).
Unlike graph databases, which present different
strategies to model the internal representation of the
graphs, knowledge graph databases usually represent
data as a set of triples in the form of <subject,
predicate, object> following RDF (Resource
Description Framework).
In the field of medicine and healthcare, both
property and knowledge graphs are increasingly used
in tasks such as validation of diagnoses (Gu et al.,
2022; Yin et al., 2021), prediction of patient clinical
pathways (Trevena et al., 2022), suggestions of
treatment plans(LIU et al., 2022) and drugs (Rivas &
Vidal, 2021) and integration of heterogeneous
healthcare data and electronic health records for a
common storage (Freedman et al., 2020) and its
interoperability (Kiourtis et al., 2019). We think a
graph can be a valuable tool for modelling a hospital
and the patients’ movements during their stays. Thus,
we could examine the connections between patients
and medical staff through the hospital layout over
time. In (Pujante-Otalora et al., 2023), we studied
how graphs are used in spatiotemporal
epidemiological research.
In this paper, we empirically evaluate the
performance of property and knowledge graphs to
solve spatio-temporal queries that correspond to tasks
of the epidemiological research of hospital infectious
outbreaks. In particular, the knowledge graphs are
Pujante-Otalora, L., Campos, M., Juarez, J. and Vidal, M.
Comparison Between Graph Databases and RDF Engines for Modelling Epidemiological Investigation of Nosocomial Infections.
DOI: 10.5220/0012319900003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 2, pages 23-36
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
23

based on triples and delimited by RDF 1.1 (RDF 1.1
Concepts and Abstract Syntax, n.d.) and its extension
RDF* (RDF-Star and SPARQL-Star, n.d.). To this
aim, we have designed a domain model for hospitals
focused on spatially and temporally describing the
patients' movements during their stay. We have also
generated graphs of different sizes, taking as a source
the freely-available database MIMIC-III (Johnson et
al., 2016), which comprises information about
patients admitted to critical care units at a large
tertiary care hospital. We have chosen Neo4j as the
graph database engine for the property graphs and
GraphDB for the knowledge graphs. For the
evaluation, three queries that represent three tasks
from the epidemiological research process have been
defined and implemented in Cypher -Neo4j query
language-, SPARQL 1.1 (SPARQL Protocol and RDF
Query Language)(SPARQL 1.1 Query Language,
n.d.) and its extension SPARQL*. These queries can
be defined as reachability and pattern-matching
graph-based tasks and have different levels of
complexity (number of required and optional paths,
length of the paths, number and type of the variables
to retrieve). Finally, we discuss the results of our
evaluation and outline the different conditions that
benefit the studied engines.
To summarize, our contributions are as follows:
i) Design of a domain model that describes a
hospital layout and the movements of the
inpatients as a log of all their events.
ii) Description of three epidemiological
research queries in terms of graph tasks and
their paths.
iii) An empirical study of the performance of
property graphs and knowledge graphs
based on the queries.
This paper is organised as follows: Section 2
summarizes related works. Section 3 describes the
graph database engines (Section 3.1), the domain
model (Section 3.2), the queries (Section 3.3), the
datasets (Section 3.4), the storage use (Section 3.5)
and the benchmarks (Section 3.6). Experimental
results and their discussion are in Section 4.
Conclusions are in Section 5.
2 RELATED WORK
In the past few years, several works that aim to
compare graph databases and knowledge graph
databases have been published. These comparisons
focus on the performance of the database, mainly
measured as the execution time of a set of queries that
address different graph tasks (adjacency, reachability,
pattern matching, shortest path, etc.) over real-world
or synthetic datasets of various sizes. The graph tasks,
the query strategy and the datasets affect the results
of the studies. (Hohenstein & Jergler, 2019) studied
fairness in the most applied methods in graph
database comparisons.
These comparative works often focus solely on
property or knowledge graph databases. In the case of
knowledge graph databases, GraphDB and Stardog
are among the RDF engines that appear the most and
with the best results. (Atemezing & Amardeilh, 2018)
studied the bulk loading, scalability and query
execution time of four RDF engines (GraphDB,
Oracle 12c, Stardog, Virtuoso) over two real-world
datasets from the EU Publications Office. (Bellini &
Nesi, 2018) compared the execution time of a set of
ten RDF engines against a Smart City RDF
Benchmark dataset.
In the case of graph databases, Neo4j is
commonly compared, presenting promising results.
(Wang et al., 2020) compared three graph databases
(Neo4j, TigerGraph, TuGraph) against some
synthetic social networks. (Ferro & Sinico, 2018)
compared three graph databases (ArangoDB, Neo4j,
OrientDB) and a relational database (PostgreSQL)
against a dataset based on an Italian Business
Register. (Monteiro et al., 2023) compared the
execution time and RAM and CPU usage of four
graph databases (JanusGraph, Nebula Graph, Neo4j,
TigerGraph) against data from the LDBC Social
Network Benchmark
0F0F.
Works that compare graph databases with RDF
engines are less common. (Kovács et al., 2019)
compared the execution times of two graph databases
(Neo4j, JanusGraph) with an RDF engine
(Blazegraph) using a dataset loaded with information
from Wikidata. (Li et al., 2022) compared the
execution time of four triple stores (GraphDB,
Virtuoso, RDF4j, Fuseki) with a graph database
(Neo4j) and an Ontology-Based Data Access
database (Ontop) against a dataset of topological data.
Our work differs from this last one in that their
queries represent geometrical operations for spatial
analysis of polygons based on points, while our
spatial representation of a hospital is based on relative
relations (next to, opposite, inside) of its architectural
elements (room, corridor, floor). Besides, we consider
the temporal dimension.
HEALTHINF 2024 - 17th International Conference on Health Informatics
24

3 DATA AND METHODS
3.1 Graph Database and RDF Engine
Neo4j is a database engine that relies on labelled
property directed multigraphs, which are
implemented in native structures. That is, the
underlying structure of the database has been
designed to store data in the form of nodes and edges.
Neo4j presents a schema-less and implements its own
query language, Cypher. It provides an API REST
and official drivers for several programming
languages.
GraphDB is a graph database used to store and
manage knowledge graphs that can perform semantic
inferencing. It is fully compliant with RDF 1.1 and
SPARQL 1.1, and it is one of the few commercial
graph databases that entirely supports their extensions
RDF* and SPARQL*. GraphDB implements the
RDF4J framework interfaces in pursuit of
compatibility with RDF serialization formats and
easy application integration. GraphDB offers an
official API REST for administration tasks but not
querying the database.
It is noteworthy that both databases are
implemented in Java. Some newer graph databases
implemented in C++ (e.g., TigerGraph) may offer
faster results.
3.2 Domain Model
Our model aims to represent patients’ movements
during their hospital stay. This model focuses on both
the temporal and the spatial dimensions. The spatial
dimension consists of a hierarchy of locations that
goes from bed to building, passing through rooms,
corridors, areas and floors. There are also spatial
relations between some locations of the same level in
the hierarchy, such as being next to or opposite some
other location. For example, a room can be next to
another, but not opposite a corridor. The spatial
dimension also covers the hospitalization units and
services that care for the patients during their stay.
The locations form the physical spatial dimension,
and the units and services are the logical spatial
dimension. For the temporal dimension, there is a log
with the hospital episodes of each patient, where each
episode represents an hospital stay and consists of a
set of all its events. An event represents any
movement of the patient in the hospital (a stay in a
room, a surgical operation, taking radiography, etc.)
or other actions, such as getting the results of a
microbiological test. Each episode and event has a
start and end date and time. Events may occur in a
time moment (both dates and times will be the same)
or interval. Some of them are connected to a location,
so they are the union point between temporal and
spatial dimensions. Events can also be connected to
the hospitalization units that cared for the patient.
In the model, some relations have properties. That
is the case of all the relations from the spatial
dimension, which have a weight to represent how
close the two locations are. The relation that connects
a microbiological test with the microorganism found
also has a property to mark if the microorganism is
multidrug-resistant. The right side of Figure 1 shows
the entire domain model. Given the lack of detailed
information on the dataset about the hospital layout
and what happened during the hospitalizations, we
have used a reduced version of the domain, shown on
the left side of Figure 1. The explanation of the
complete model is out of the scope of this work.
The implementation of our domain model differs
from property graph to knowledge graph. Neo4j
presents a schema-less model where nodes and edges
can belong to one or several classes that act like labels
since they do not restrict the number and datatype of
the properties of its nodes and edges. Moreover, nodes
and edges hold their own information in the form of
properties. When translating the domain model to an
RDF ontology, we find that it is an abstract
representation that does not differentiate data from
metadata since it is based on the concept of statement:
a triple made of a subject, a predicate and an object.
The subject denotes a resource defined by a URI
(Uniform Resource Identifier), and the predicate
denotes a property or relation of the subject. The object
is the value of the data property or the URI of another
resource (the subject of one statement can be the object
of another). The class (or classes) of a resource is also
the object of a statement. In a graph representation,
subjects and objects would be the nodes, and predicates
would be the edges between them.
A significant difference between a Neo4j graph
and an RDF knowledge graph is how implement
edges with properties. In RDF it is not possible to
make a statement where an edge is a subject. There
are four approaches to solve this problem: standard
reification, n-ary relations, singleton properties and
named graphs. We have chosen standard reification,
which consists in creating a new class to represent the
edges as resources (nodes), making possible
statements that have them as subjects and their data
properties as objects. On the other hand, RDF* allows
making statements about other statements. So it is
possible to define a statement that represents that two
resources are linked, which is the subject of another
statement that adds a property to the relation.
Comparison Between Graph Databases and RDF Engines for Modelling Epidemiological Investigation of Nosocomial Infections
25
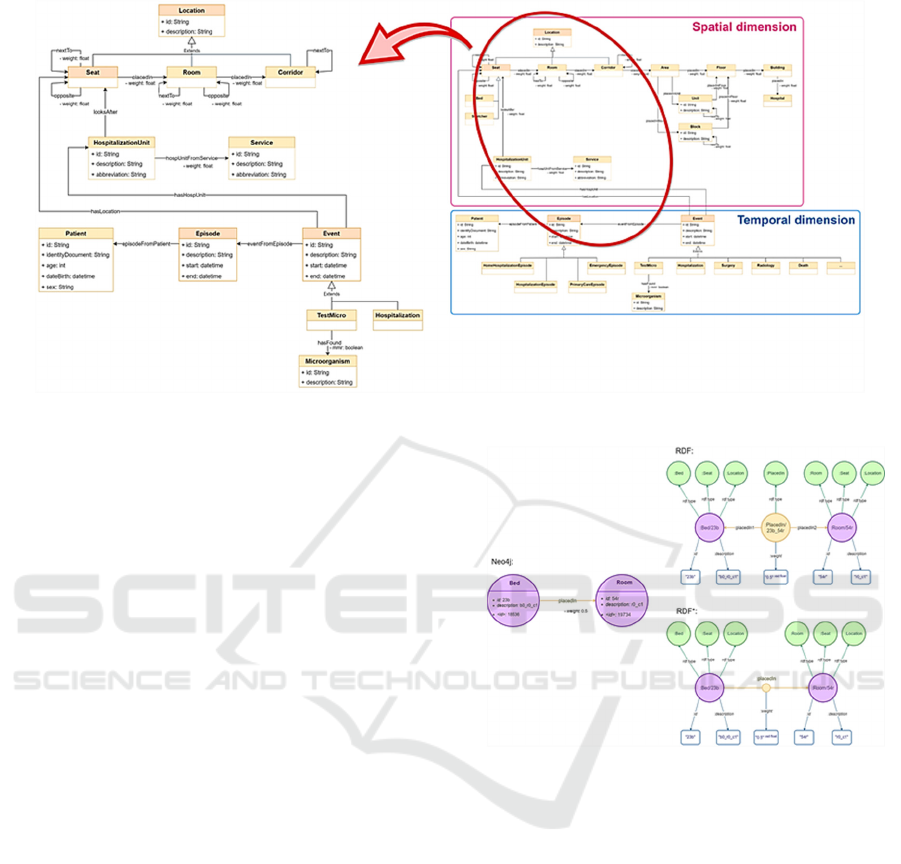
Figure 1: Domain model.
Figure 2 shows a graphical representation in each
data model of the edge placedIn between a Bed and a
Room, which has the property weight. From here on,
we will refer to Neo4j nodes and RDF and RDF*
resources as “nodes of individuals” and to the nodes
with the name of the classes and the value of the data
properties as “nodes of literals”.
3.3 Query Definition
We defined three queries representing relevant
clinical tasks for detecting and studying an infection
outbreak. All these queries could be described as
reachability and pattern-matching graph-based tasks,
where the paths are temporarily and semantically
conditioned. Tables 1, 2 and 3 show the description
and characteristics of every query. Figure 3 shows a
schematic representation of each query. Each query is
defined by the following characteristics:
• A general description of the query as a graph-
based task.
• Number of required and optional paths, where
“required” means that the path must match with
the data and “optional” means that it is not
mandatory that all of these paths match, but at
least one must do it.
• Tentative number of steps (number of traversed
edges) that each path should have in a
pseudocode version of the query defined
directly over the domain model diagram. When
translated to Cypher, SPARQL and SPARQL*,
these paths and their length may change.
Figure 2: Graphical representation of the relation placedIn
between two nodes Bed and Room with their properties. In
Neo4j nodes, <id> property is an internal identifier. In
RDF* representation, the small yellow node symbolises the
statement that links the purple nodes and on which a new
statement is added to link to it the weight property.
3.4 Dataset Characteristics
We have generated ten graphs of different sizes from
MIMIC-III. We have used admissions and transfers
tables to get the stays and ward movements of the
patients, services table to get the services responsible
for the patients and microbiologyevents table to get
their positive microbiological tests.
Since the information provided by MIMIC-III
only partially covers the classes and relations of our
domain model, we have extended it by creating a
fictitious hospital with a simple layout.
In the physical spatial dimension, MIMIC-III
works with wards. For each ward, we have created a
HEALTHINF 2024 - 17th International Conference on Health Informatics
26
Table 1: Description of Query 1.
Query 1 (Q1)
Clinical
task
Outbreak detection.
Description
Given a Service and a Microorganism,
find all the Patients hospitalised under
that Service who had an infection of the
given Microorganism. The search is
within a time interval.
Characteristics
Graph-based
task
Find the nodes that
are the union points
between two nodes.
Required paths: 2
Description
Number of
steps
- From the given
Microorganism to
connected Patients
3
- From the given Service
to connected Patients
4
Optional paths: 0
set of rooms with a maximum of ten beds. There must
be sufficient beds in each ward to encompass the
maximum number of simultaneous hospitalised
patients if we compacted all its hospitalisations in just
one year. Then, these beds are homogenously
distributed between the rooms.
We organised the rooms into four groups
(surgical, medical, mixed and neonatal) according to
the type of service that attends the most
hospitalisations in their wards. All the rooms are
located on a single floor and distributed in two
parallel main corridors. These corridors are divided
into several shorter corridors, each with a maximum
of twenty rooms. Our fictitious hospital has 156
Surgical rooms with 1,413 beds, 23 Medical rooms
with 179 beds, 56 Mixed rooms with 510 beds and 4
Neonatal rooms with 13 beds. Figure 4 is a schematic
representation of the hospital layout where we can see
the four groups of rooms and their distribution. For
the logical spatial dimension, we have added to each
service several hospitalization units, such that no
hospitalization unit attends more than 1,500
hospitalisations.
Note that the specific design for this example did
not impact the resolution of the queries, and other
distributions would be possible since the data model
allows for more complex designs while the classes
used are the same.
To evaluate the scalability of the queries at the
maximum extent with the data available, it is
necessary to generate graphs of different sizes. We
generated every graph to comprise the whole MIMIC-
Table 2: Description of Query 2.
Query 2 (Q2)
Clinical
task
Outbreak detection from the index patient.
The index patient is the first patient that
catches the attention of the researchers.
This patient may not be the first person
infected.
Description
Given a Patient and a Microorganism, find
all the Patients spatially connected with
them and who had an infection of the given
Microorganism. The search is within a time
interval.
Characteristics
Graph-based
task
Find all the connected
nodes to two nodes
through some specific
paths.
Required paths: 2
Description
Number
of steps
- From the given Patient to all
the Seats they have been
3
- From the Patients found in
the optional paths to the given
Microorganism
3
Optional paths: 6
Description
Number
of steps
- From the Seats to their
connected Locations via
placedIn until Area
1-3
- From the Rooms to other
adjacent Rooms
1
- From the Areas to other Areas
belonging to the same Logic
zone (group of areas, not
necessarily contiguous, with
some common characteristics)
2
- From all the Locations found
in the previous paths to their
connected Seats
1-3
- From the Seats of the
previous path to their
connected Patients
3
- From the Events of the given
Patient to the Hospitalization
Units that cared for them.
Then, from these
HospitalizationUnits to other
connected Patients
4
III dataset in one year and then picked sets of events
(with their linked episodes, patients and
microorganisms) such that it goes from around
Comparison Between Graph Databases and RDF Engines for Modelling Epidemiological Investigation of Nosocomial Infections
27
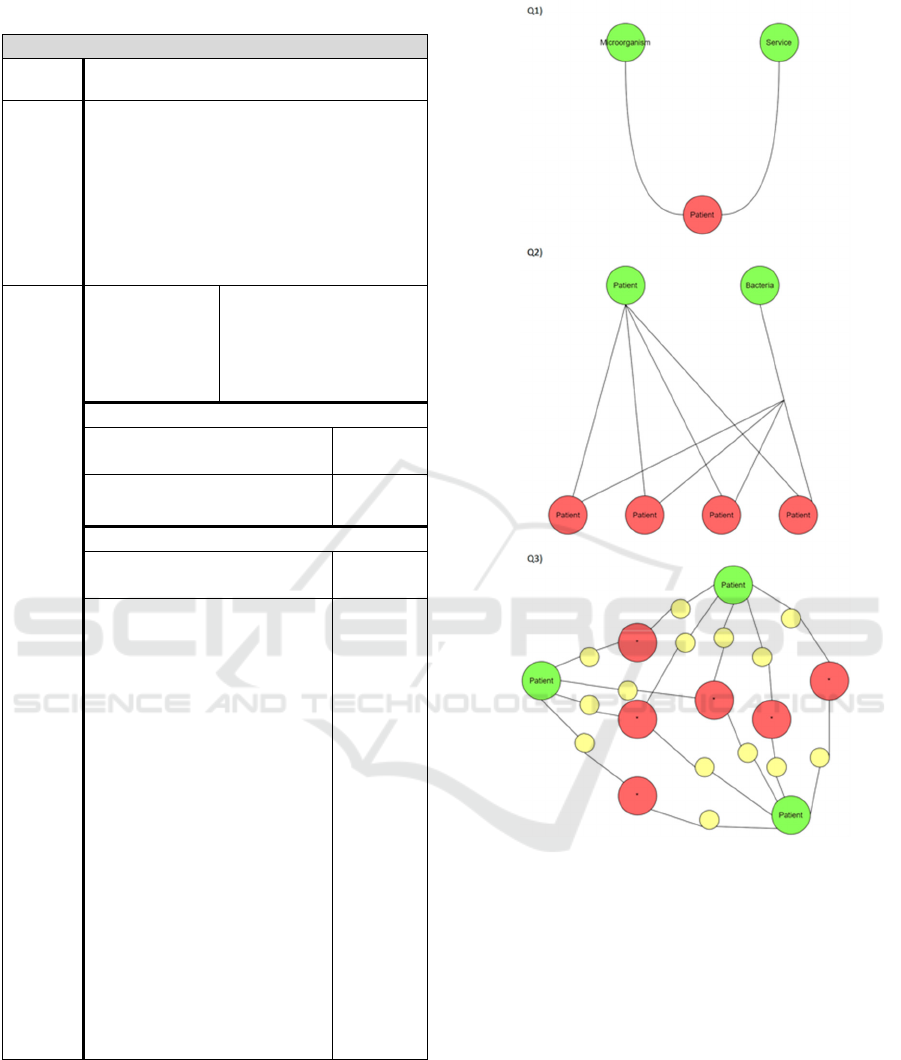
Table 3: Description of Query 3.
Query 3 (Q3)
Clinical
task
Investigation of contagion sources
Description
Given a set of Patients and a period of
time, find the shared Locations,
Hospitalization Units, Services and
Microorganism between them. That is,
we want to find what connects
patient_1 with patient_2, patient_1 with
patient_3, patient_2 with patient_3,
etcetera.
Characteristics
Graph-based
task
Find the nodes that are
the union points
between a given set of
nodes through some
specific paths.
Required paths: 1
Description
Number
of steps
From the given Patients to
all their Events
2
Optional paths: 6
Description
Number
of steps
- From each Event to every
other Event passing through
their:
• shared Hospitalization
unit.
2
• shared Service.
4
• shared Locations. That
is, first from the Event to
its connected Seat and
from this to its higher-
level Locations, and
then the inverse path.
2-8
• shared Locations,
through Logic zone.
10
• adjacent Rooms.
5
- From each TestMicro to
every TestMicro passing
through their shared
Microorganisms.
2
25,000 events up to 475,000 events in MIMIC-III
with successive increments of 50,000 events.
In total, we have ten graphs called G1 to G10,
being G1 the smaller and G10 the bigger. The hospital
layout and number of hospitalization units and
services are the same in all the graphs. Hence, there
is a different density of events per location.
Figure 3: Schematic representation of each query. Green
nodes represent the initial nodes given as a parameter, and
red nodes represent the result nodes after traversing the
defined required and optional paths, represented with the
black lines. In the case of Q3, red nodes have an asterisk
because they can be of any class since they are the union
between some initial nodes. Yellow nodes represent all the
nodes of the traversed paths, which are returned with the
red ones.
Table 4 shows for each graph the number of nodes
of the classes that most significantly impact the
execution of the queries. These are the most
numerous classes, and their nodes are often the start
or end of the queries or a connection between paths.
Table 5 shows the main “general graph features” of
each graph, differentiating between each graph
representation.
HEALTHINF 2024 - 17th International Conference on Health Informatics
28
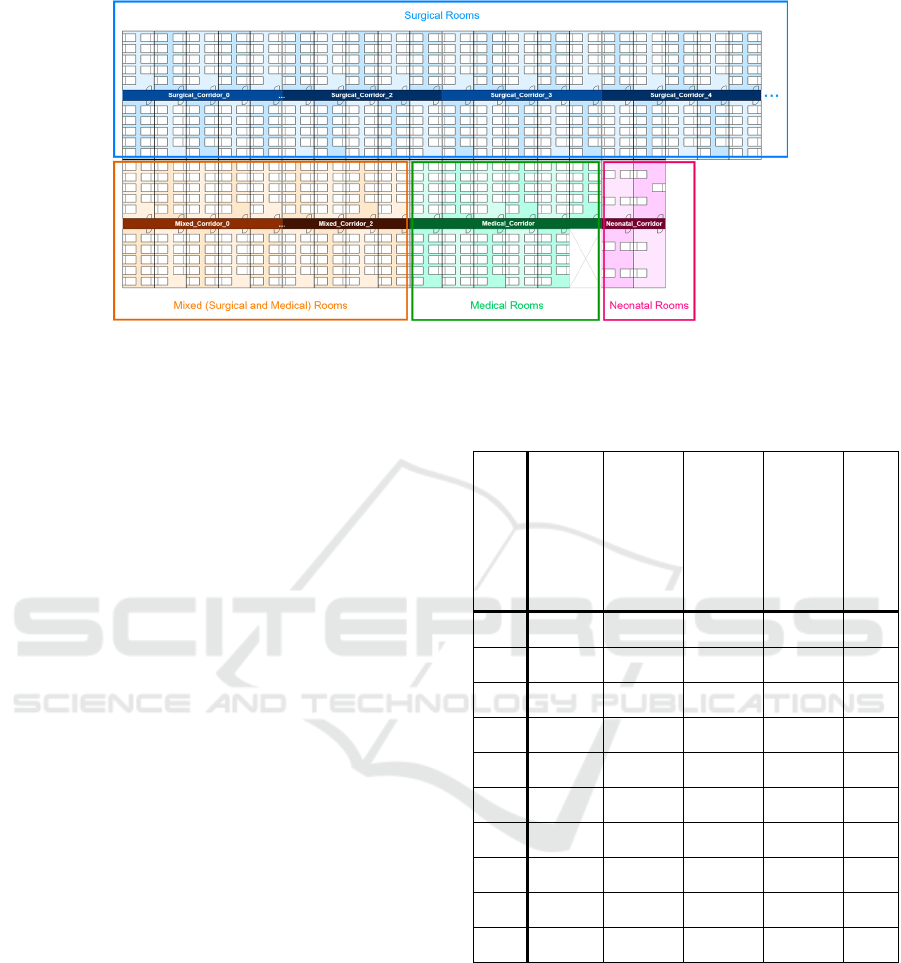
Figure 4: Schematic representation of our MIMIC-III hospital room layout. To simplify this figure, each corridor (except
Neonatal_Corridor) has half the number of rooms actually assigned, and only a portion of the Surgical corridors are shown.
Regarding the specific domain, a big hospital in
Spain would be the Complex University Hospital of La
Coruña in Galicia with about 1,300 beds. It registers
about 37,000 admissions yearly according to the
National Institute of Statics of Spain. This number is
near the 38,000 patients in the most extensive graph.
3.5 Storage Use
Concerning the disk space required to store the
graphs, we have observed a noteworthy difference
between Neo4j and GraphDB, which could be critical
for the scalability of large graphs. To optimise the data
storage, Neo4j uses linked lists of fixed-size records to
store each data type (nodes, edges, properties).
GraphDB combines an entity pool (files where all the
entities -URIs, literals, RDF* triples- are stored as 32-
bit or 40-bit internal IDs) with two main indexes,
subject-predicate index and predicate-object index.
Figure 5 shows how much disk space each graph
needs to be stored in each database engine. As
expected, the implementations in RDF* of the graphs
are a bit lighter than in RDF. The difference between
these two technologies goes between 6.2% and 14.4%
and increases with the size of the graph. It is worth
mentioning that each database usually represents an
increase of about 55 MB in RDF and RDF*. In Neo4j,
this increase goes from 5 MB in the last graphs to 12
MB in the first ones.
An interesting side finding was that although the
number of nodes and edges has been multiplied by 17,
the size to store the graphs has only been increased
around 7 times in RDF and 6.5 times in RDF*. In
contrast, G10 is 20 times bigger than G1 in Neo4j.
These findings could suggest that future studies
would need to analyse at which point graphs stored in
Neo4j are bigger than in GraphDB.
Table 4: Classes with a high impact on the execution of the
queries.
Graph
# Patients
# Events
# Events with
Location
# TestMicro
# Microorganisms
G1 1,903 25,050 8,054 16,722 135
G2 5,702 75,039 24,612 49,560 197
G3 9,028 125,200 39,458 84,351 219
G4 14,027 173,307 61,862 109,379 227
G5 14,114 225,214 63,139 159,633 257
G6 15,471 275,625 71,090 201,674 288
G7 17,244 324,776 77,864 243,646 298
G8 20,032 375,211 89,471 281,996 303
G9 27,276 425,141 122,393 298,212 304
G10 38,066 474,922 169,435 299,870 304
3.6 Benchmark Characteristics
In this work, we analysed the performance of the
database engines when executing the queries defined.
We set two representative measures:
• Execution Time: it represents the time
between sending the query request to the
API and receiving its answer.
• Maximum Main Memory: it is the amount of
main memory required to execute the task.
We got the maximum main memory by
Comparison Between Graph Databases and RDF Engines for Modelling Epidemiological Investigation of Nosocomial Infections
29
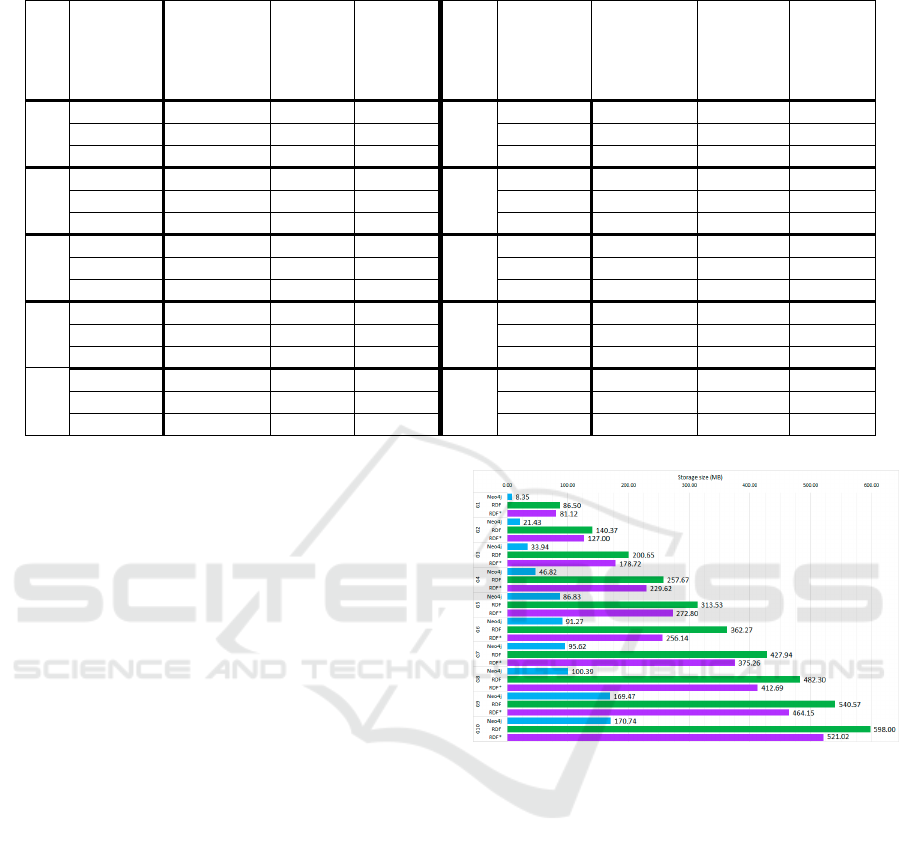
Table 5: General graph features. RDF and RDF* are deployed in GraphDB. The numbers are represented in thousands.
Graph
Database
Engine
# Nodes of
individuals
# Nodes
of
literals
# Edges
Graph
Database
Engine
# Nodes of
individuals
# Nodes
of
literals
# Edges
G1
Neo4j 31 0 66
G6
Neo4j 312 0 643
RDF 54 49 230 RDF 519 431 2,286
RDF* 31 49 208 RDF* 312 431 2,079
G2
Neo4j 90 0 186
G7
Neo4j 365 0 750
RDF 145 134 648 RDF 614 495 2,689
RDF* 90 134 593 RDF* 365 495 2,440
G3
Neo4j 148 0 305
G8
Neo4j 422 0 866
RDF 238 215 1,064 RDF 710 566 3,107
RDF* 148 215 974 RDF* 422 566 2,819
G4
Neo4j 208 0 430
G9
Neo4j 490 0 1,008
RDF 323 303 1,473 RDF 793 663 3,544
RDF* 208 303 1,358 RDF* 490 663 3,240
G5
Neo4j 260 0 534
G10
Neo4j 565 0 1,168
RDF 425 362 1,885 RDF 870 774 3,997
RDF* 260 362 1,720 RDF* 565 774 3,692
using a background process that measures
the main memory consumption of the
database engine while the query is running.
Then, we pick the maximum value.
From here on, we will refer to the execution time
as “time” and to the maximum main memory
consumption as “memory”.
We have configured three benchmarks to study the
performance of Neo4j and GraphDB. Each
benchmark evaluates one of the proposed queries in
the ten graphs (G1, G2…). For each graph, we have
defined 12 sets of values representing each month of
the year. These 12 sets are requested three times.
Then, we calculated the mean of the time and memory
values.
For each query, we have selected the patient,
microorganism or service that is in the third quartile
of the nodes of its class with more connections. In
queries requiring a set of patients, we always used 15.
These patients have been chosen randomly between
those with more connected patients than the third
quartile. We consider that 15 is a high enough number
to represent a significant outbreak.
To ensure that cache memory is not preserved
between different processes, each query request is
executed in cold cache.
Regarding the execution features, we have
worked with Neo4j CE 4.4.18 and GraphDB Free
10.2.0. We have implemented all the queries in
Cypher, SPARQL and SPARQL*. The requests have
been made locally. For the communication with the
Figure 5: Storage size of each graph on each database
engine.
databases we have used Neo4j Python Driver(Neo4j
Python Driver, 2023)
1F1F for Neo4j and
SPARQLWrapper(SPARQLWrapper, 2022)2F2F Python
library for GraphDB. The experiments were run on a
PC with an Intel i9-12900K, 16GB RAM and 1TB
M2 drive, running a 64-bit Linux Ubuntu 22.04.
4 EXPERIMENTAL RESULTS
AND DISCUSSION
4.1 Query 1
Q1 consists of the intersection of two short required
paths with a fixed length. It can be observed in
Figures 6a and 6b that Neo4j performs better in time
HEALTHINF 2024 - 17th International Conference on Health Informatics
30
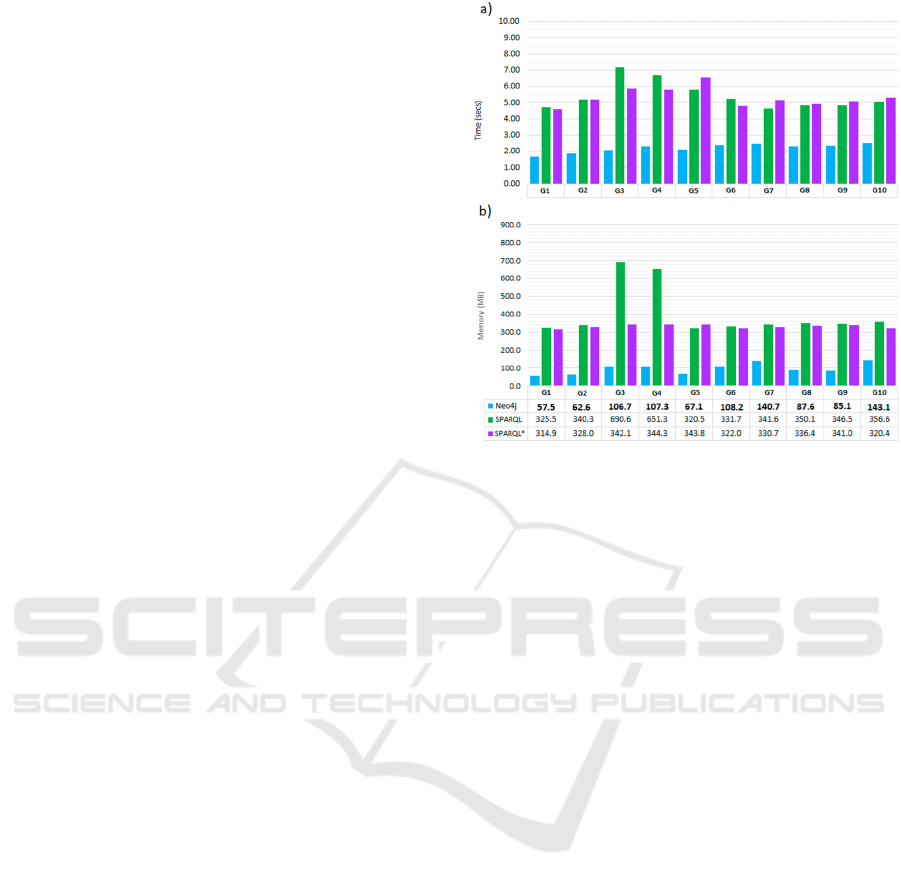
and memory in all the graphs. Another finding is that
SPARQL* requires a less time and memory than
SPARQL for almost all the graphs.
We can also observe that both the time and the
memory remain stable for all the graphs even though
the proportions of TestMicro per Microorganism and
Events per Service increased significantly. The only
exception is found in G3 and G4, where the memory
in SPARQL doubles that of the rest of the graphs. This
exception will be present in the rest of the queries and
might be related to a change in the query schedule,
which can be corroborated by the curves depicted by
the time series in SPARQL and SPARQL*. They
grow slightly and stay higher until G5, then descend,
and finally remain stable until the last graph.
The time behaviour of the three engines can be
explained if we assume that the number of results
(Patient nodes) of a query is related to the number of
edges and nodes traversed. For each Patient node, it
is necessary to traverse a path to its connected
Microorganism nodes and other path to its connected
Services. For example, if none of the Microorganism
nodes is the requested one, then the paths to the
Service nodes will not be traversed.
In Table 6, we have calculated for every graph the
average number of results (number of distinct Patient
nodes returned) for each month's set. Then, we have
divided it between the average time of Q1 in that
graph to measure how much time is needed to get a
result. This table shows that the number of results
does not increase with the graph’s size (total number
of nodes and edges) and that the time per result
depicts similar curves to the execution time.
4.2 Query 2
Q2 is a query that traverses a complete tree from a
Patient node to all the Patient nodes connected with
it through some optional paths over the entire domain
model. Note that the proportions of Events per Bed
and Events per HospitalizationUnit are high and
increase in each graph, implying an “explosion” of
paths to traverse. Thus, Q2 is a suitable query to
understand better how the engines behave when
traversing large subgraphs.
The period used as a parameter in the query is 30
days, which is a period even larger than the usually
considered in the initial investigation of an outbreak.
Figures 7a and 7b show the time and memory of
executing Q2. Regarding time, SPARQL and
SPARQL* are faster than Neo4j (except for the three
Figure 6: Comparison of Neo4j, SPARQL and SPARQL*
for Q1 executions in the graphs G1 to G10. a) Execution
time comparison in seconds. b) Maximum main memory
consumption comparison in MB. In memory chart, for
every graph the smallest value is marked in bold.
most minor graphs), and the difference grows with the
size of the graphs. If we observe Neo4j time, we can
differentiate two rises. The first one goes from G1 to
G5. The second one starts on G7 and does not seem
to have a maximum nearby.
We have calculated the average time per result as
in Q1, where results are the Patient nodes connected
to the starting one. The results are in Table 7, where
we can see that the time curves also happen in the
time per result series. But note that in the first hill, the
time per result grows with the number of results,
while in the last rise, the number of results remains
around 40, and the time per result only increases. We
suspect that from G5 to G7, there must be a change in
the strategy selected by the query scheduler.
In SPARQL and SPARQL*, time and memory
present curves with similar shapes and values to those
we got in Q1. Table 7 shows two similar behaviours
in the average time per result whose start and end
graphs coincide. This fact supports the supposition
that the query scheduler of GraphDB starts to change
the strategy with graphs with a size similar to G4
(around 500,000 nodes and 1,300,000 edges), and this
change is effective when the graph has a size similar
to G6 (around 1,000,000 nodes and 2,000,000 edges).
Comparison Between Graph Databases and RDF Engines for Modelling Epidemiological Investigation of Nosocomial Infections
31
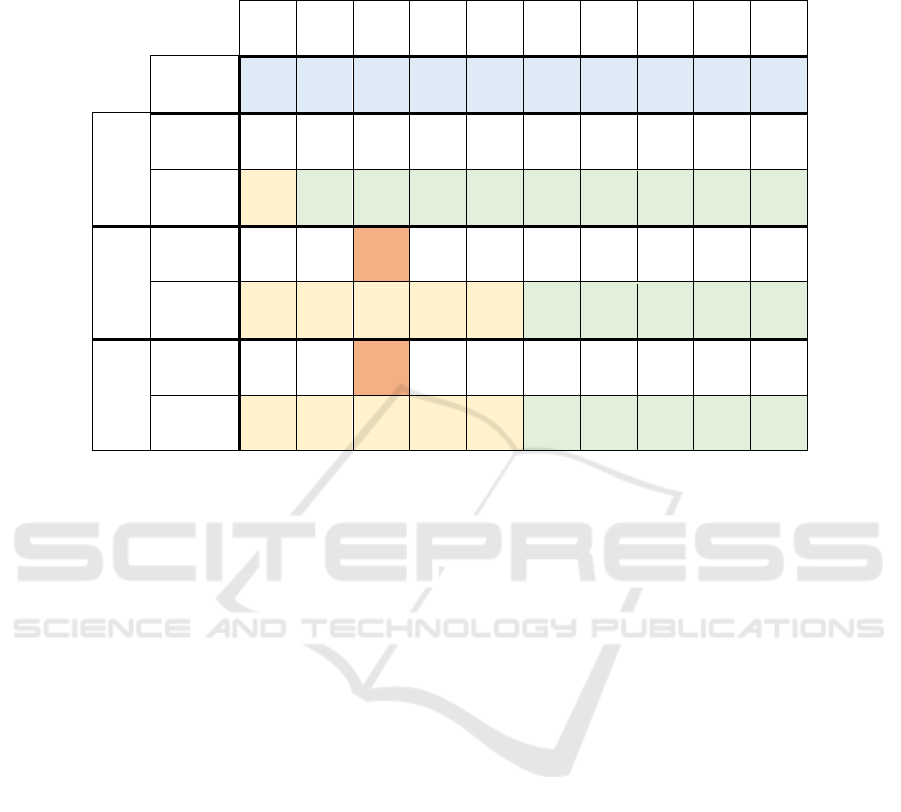
Table 6: Comparison of Neo4j, SPARQL and SPARQL * for Q1 executions in the graphs G1 to G10 of the average time to
get a result. We also show the average time of the query in each graph (Figure 6-a). We have marked the groups of
neighbouring graphs with similar behaviour in yellow and green. We have also marked the time relative maximum in orange.
G1 G2 G3 G4 G5 G6 G7 G8 G9 G10
Avg
#Results
14 25 26 31 34 38 36 36 37 38
Neo4j
Avg time
(secs)
1.69 1.85 2.07 2.29 2.09 2.40 2.47 2.30 2.33 2.50
AvgTime/
AvgRes
0.12 0.07 0.08 0.07 0.06 0.06 0.07 0.06 0.06 0.07
SPARQL
Avg time
(secs)
4.73 5.19
7.16 6.68 5.77 5.21 4.62 4.82 4.83 5.04
AvgTime/
AvgRes
0.33 0.21 0.28 0.22 0.17 0.14 0.13 0.13 0.13 0.13
SPARQL *
Avg time
(secs)
4.59 5.19
5.88 5.80 6.55 4.78 5.15 4.91 5.08 5.31
AvgTime/
AvgRes
0.32 0.21 0.23 0.19 0.19 0.13 0.14 0.14 0.14 0.14
4.2.1 Query 2: Version that Returns the
Solution Paths
Given the aim of Q2, it could be interesting that in
addition to the Patient nodes, it also returned the
traversed paths from the starting Patient to the Patient
nodes in the tree’s leaves. We have defined a modified
version to return the effective paths so we can study
if “saving” the paths along the query execution
implies a significant increase in time and memory.
Figures 7c and 7d show the time and memory of
executing the modified version of Q2. Neo4j time and
memory series show the same curves but with
increases of around 130% in time and 120% in
memory. SPARQL and SPARQL* present very
different behaviours between time and memory. Like
Neo4j, memory keeps the same curve with a slight
increase of around 105%. On the contrary, in time, we
can find that from G1 to G5, the query is about 125%
slower, and for the rest, the increase in time is around
155%.
Neo4j has a more significant memory
consumption than SPARQL and SPARQL*, possibly
due to how the queries are built. In Neo4j, it has been
necessary to add explicit instructions to save the
traversed paths as lists in new variables that must be
kept in memory. These instructions must also be
responsible for the overtime in this version of Q2. In
the case of SPARQL and SPARQL*, we did not need
to use explicit instructions for saving paths. However,
we had to add a new query connected to the original
one with a UNION sentence to keep the paths
between the Location nodes. This new query should
not imply a relevant cost in memory since the nodes
and edges it traverses should be already cached.
However, traversing them again and joining the
results of both queries should mean overtime.
4.3 Query 3
Q3 traverses the trees from 15 given Patients (15 is a
fixed value parameter, as commented on above) to
their Events. Then, some optional paths are defined to
search for the union nodes between the Events from
different Patients. Thus, it is a query where the
exploration of the graph is not as crucial as the
repetitive passage over a set of nodes and edges.
Therefore, the relation between time and memory
should differ from the other queries.
As we can see in Figures 8a and 8b, that is the case
of Neo4j. We find that the maximum memory is
around 120MB (with some exceptions in G3 and G4
that might be related to the strategy of the query
scheduler), a low number compared with those in Q2.
However, time in all the graphs is similar to that of
the biggest graphs in Q2.
HEALTHINF 2024 - 17th International Conference on Health Informatics
32
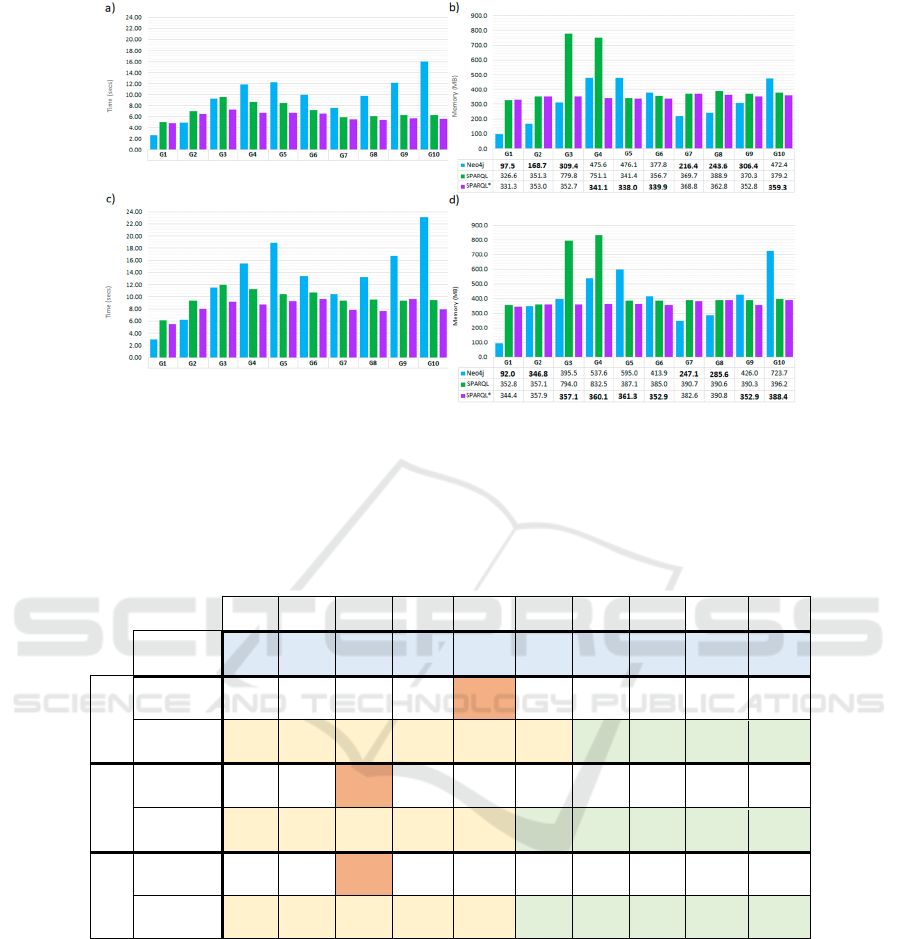
Figure 7: Comparison of Neo4j, SPARQL and SPARQL* for Q2 executions in the graphs G1 to G10. a) Execution time
comparison in seconds for original Q2. b) Maximum main memory consumption usage comparison in MB for original Q2.
c) Execution time comparison in seconds for alternative Q2. d) Maximum main memory consumption comparison in MB for
alternative Q2. In memory charts, for every graph the smallest value is marked in bold.
Table 7: Comparison of Neo4j, SPARQL and SPARQL* for original Q2 executions in the graphs G1 to G10 of the average
time to get a result. We also show the average time of the query in each graph (Figure 7-a). We have marked the groups of
neighbouring graphs with similar behaviour in yellow and green. We have also marked the time relative maximum in orange.
G1 G2 G3 G4 G5 G6 G7 G8 G9 G10
Avg
# Results
13 26 29 33 34 43 40 40 36 41
Neo4j
Avg time
(secs)
2.62 4.86
9.25 11.86
12.21 9.94 7.60 9.76 12.14 16.08
AvgTime/
AvgRes
0.20 0.19 0.31 0.36 0.36 0.23 0.19 0.25 0.34 0.39
SPARQL
Avg time
(secs)
4.99 6.98
9.51 8.60 8.42 7.20 5.82 6.08 6.23 6.21
AvgTime/
AvgRes
0.37 0.27 0.32 0.26 0.25 0.17 0.14 0.15 0.17 0.15
SPARQL*
Avg time
(secs)
4.78 6.40
7.26 6.67 6.70 6.55 5.43 5.37 5.66 5.61
AvgTime/
AvgRes
0.36 0.25 0.25 0.21 0.20 0.15 0.13 0.14 0.16 0.14
This difference between time and memory could
be explained because the nodes and edges between
the Events are charged in memory the first time they
are accessed and remain cached for the rest of the
accesses.
In the case of queries in SPARQL and SPARQL*,
they present memory consumptions with values
similar to those in Q1 and Q2, while the time is much
longer than in any other query.
Before analysing time, it is necessary to comment
that for the experiments, 15 Patients were chosen
between those in the quarter of Patients with more
connected Patients. However, it does not mean that
there must exist connections between all 15 Patients.
What is more, in this work, a bigger graph does not
have to mean more connection between a set of
patients. In Table 8, we can observe that the average
number of results (the number of different paths that
connect a Patient with another) does not increase with
the size of the graphs. In fact, the maximum number
of results is in G7, and the minimum is in G4. The
time series of Neo4j follows the direction of the
number of results: when the number of results
Comparison Between Graph Databases and RDF Engines for Modelling Epidemiological Investigation of Nosocomial Infections
33
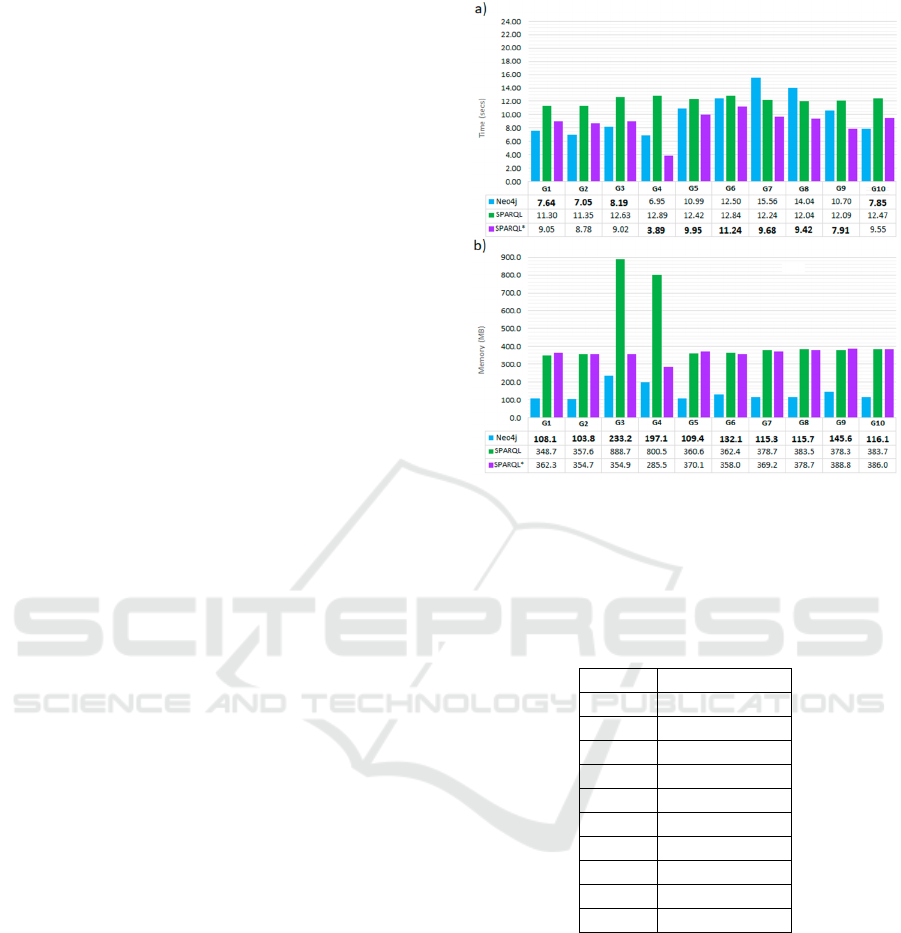
increases, time increases too, and the same when the
number of results decreases.
Curiously, SPARQL* presents a curve more similar
to Neo4j than SPARQL. In fact, the time differences
between SPARQL and SPARQL* stand out in this
query compared to the rest: here, SPARQL presents
times between 115% and 153% bigger than SPARQL*.
This significant difference may be because the majority
of the computation in this query is in traversing edges
with properties, which in SparQL* are represented
with a single edge (actually a statement of a statement),
while SparQL requires one “node of individuals” and
two edges (see Figure 2). Thus, we can judge that the
“edges with properties” from SPARQL* speed up
traversing the graph. Under this assumption, we can
observe that in Q1, where the edges with properties are
barely traversed, the time difference is not so
pronounced, and even in some graphs, SPARQL* is
slower than SPARQL.
4.4 General Aspects
Our experiments suggest that there are common
characteristics between queries related to time and
memory.
First, in Neo4j, there is a strong relation between
time and memory that is perceptible in Q1 and Q2,
but not in Q3. Q1 and Q2 consist of exploring a tree
in which traversing new edges and nodes is the main
computing effort. Contrarily, Q3 works with a limited
set of nodes traversing different combinations of
edges between them. These opposite behaviours give
us a clue about how Neo4j manages memory: as new
edges and nodes are traversed, they are saved in
memory and not deleted until the end of the query (as
long as there is enough memory) to avoid the costly
work of fetching data from disk.
Concerning Neo4j, it is shown that memory usage
does not depend so much on the total size of the graph
but rather on the subgraph to be traversed.
In the case of GraphDB, SPARQL and SPARQL*
present a memory usage similar, but clearly different
from Neo4j. We can observe that in all the
benchmarks, the memory is between 325MB and
400MB (with the exceptions in G3 and G4) and
increases or decreases slightly depending on the
query complexity and the data to save during the
execution (for example, the number of classes of
nodes and edges to return). The size of the graph also
affects the memory usage in GraphDB. We can find
clues about this in Q2 and Q3, where memory
gradually increases from G1 to G10 although the
traversed subgraphs are not necessarily bigger
because they are in a bigger graph.
Figure 8: Comparison of Neo4j, SPARQL and SPARQL*
for Q3 executions in the graphs G1 to G10. a) Execution
time comparison in seconds. b) Maximum main memory
consumption comparison in MB. In memory chart, for
every graph the smallest value is marked in bold.
Table 8: Average number of results of Q3 executions in the
graphs G1 to G10.
Graph Avg #Results
G1 1683
G2 1340
G3
1598
G4 1060
G5 2453
G6 3030
G7
4138
G8 3715
G9 2595
G10 1517
The results strongly suggest Neo4j and GraphDB
work with different memory usage schemes. But to
better understand GraphDB memory usage, it would
be necessary to measure its memory consumption
during query execution and analyse how it changes
from the start to the end. This way, we could confirm
our supposition that memory consumption is stable
throughout the execution and that the maximum
memory is not a peak like in Neo4j.
Regarding time, the results yielded some
interesting findings regarding Neo4j and GraphDB.
While in Neo4j, time changes depending on the type
of query and the size of the subgraph to traverse, in
HEALTHINF 2024 - 17th International Conference on Health Informatics
34

SPARQL and SPARQL*, time oscillations are lighter.
In Q1 and Q2, they start from a high minimum base
time (around 5 seconds, as opposed to Neo4j’s 2
seconds), but the maximum does not exceed more
than three times the minimum, in contrast to the
maximum times of Neo4j, which can multiply the
minimum by eight.
5 CONCLUSIONS AND FUTURE
WORK
In this research work, we evaluated the performance
of property and knowledge graphs based on triple
stores in the context of spatiotemporal
epidemiological investigation of an infection
outbreak in a hospital. Specifically, we chose Neo4j
as the graph database engine, which has its own data
model and query language, Cypher; and for
knowledge graphs, we used RDF and RDF* as
standard technologies to define the graphs, SPARQL
and SPARQL* languages to query data and GraphDB
database to store the graphs. We defined a domain
model that describes a hospital layout, the
organisation of its healthcare workers and the path of
its patients as a log of all their events. We stated three
queries that are steps of the epidemiological
investigation process and designed a benchmark to
evaluate the queries' time execution and memory in
ten graphs of different sizes whose data come from
the open-data clinical dataset MIMIC-III.
Our experiments provide convincing evidence
that neither in Neo4j nor GraphDB time and memory
are influenced by the total size of the graph, but for
other factors like the complexity of the query (number
of required and optional paths and their length), the
size of the traversed subgraph and if the goal of the
query is retrieving the leaves of a tree or do additional
paths between a set of nodes. Neo4j's performance
highly depends on these factors. For simple queries
and small traversed subgraphs, time and memory can
be half of those of GraphDB. On the other hand, time
and memory on GraphDB present minimum values
much higher than Neo4j but with less aggressive
growth. Thus, SPARQL and SPARQL* will scale
better than Neo4j for queries that need to traverse big
subgraphs.
Our research suggests that the extensions
provided by RDF* with respect to RDF, such as the
addition of the statements about other statements that
allows defining edges with properties, seem
promising for a more complete, compact and
comprehensible modelling. Furthermore, our
experiments suggest that RDF* offers a better
performance of the query execution and less storage
size for the graphs.
It is worth mentioning that RDF and RDF* are
W3C standards and not proprietary formats, as it is
Neo4j. This offers the advantage of greater flexibility
and solidity when choosing between possible
additional tools to work with.
To further compare property graph and knowledge
graph technologies in our context, we plan to define
queries for the rest of the epidemiological
investigation process tasks and, especially, queries to
execute general graph tasks (shortest path,
community detection) over our domain. It would also
be necessary to generate synthetic data that covers the
whole domain model.
ACKNOWLEDGEMENTS
This work was partially funded by the CONFAINCE
project (Ref: PID2021-122194OB-I00), supported
by the Spanish Ministry of Science and
Innovation, the Spanish Agency for Research
(MCIN/AEI/10.13039/501100011033) and, as
appropriate, EFRD A way of making Europe; and by
the GRALENIA project (Ref: 2021/C005/00150055)
supported by the Spanish Ministry of Economic
Affairs and Digital Transformation, the Spanish
Secretariat of State for Digitization and Artificial
Intelligence, Red.es and by the NextGenerationEU
funding. This research was also partially funded by
Fundación Séneca, Región de Murcia (Spain) (Ref:
21460/FPI/20).
REFERENCES
Atemezing, G. A., & Amardeilh, F. (2018). Benchmarking
commercial RDF stores with publications office
dataset. Lecture Notes in Computer Science (Including
Subseries Lecture Notes in Artificial Intelligence and
Lecture Notes in Bioinformatics), 11155 LNCS, 379–
394. https://doi.org/10.1007/978-3-319-98192-5_54
Bellini, P., & Nesi, P. (2018). Performance assessment of
RDF graph databases for smart city services. Journal of
Visual Languages and Computing, 45, 24–38.
https://doi.org/10.1016/J.JVLC.2018.03.002
Ferro, N., & Sinico, L. (2018). Graph Databases
Benchmarking on the Italian Business Register. CEUR
Workshop Proceedings, 2161. https://ceur-ws.org/Vol-
2161/paper43.pdf
Freedman, H., Miller, M. A., Williams, H., & Stoeckert, C.
J. (2020). Scaling and querying a semantically rich,
electronic healthcare graph. CEUR Workshop
Comparison Between Graph Databases and RDF Engines for Modelling Epidemiological Investigation of Nosocomial Infections
35

Proceedings, 2807. https://ceur-ws.org/Vol-2807/paper
C.pdf
SPARQLWrapper. (2022). https://github.com/RDFLib/
sparqlwrapper
Gu, Z., Yang, X., Jia, W., Xu, C., Yu, P., He, X., Chen, H.,
& Lin, Y. (2022). StrokePEO: Construction of a
Clinical Ontology for Physical Examination of Stroke.
Proceedings - 2022 9th International Conference on
Digital Home, ICDH 2022, 218–223.
https://doi.org/10.1109/ICDH57206.2022.00041
Hohenstein, U., & Jergler, M. (2019). Database
performance comparisons: An inspection of fairness.
DATA 2019 - Proceedings of the 8th International
Conference on Data Science, Technology and
Applications, 243–250. https://doi.org/10.5220/00079
26602430250
Johnson, A. E. W., Pollard, T. J., Shen, L., Lehman, L. H.,
Feng, M., Ghassemi, M., Moody, B., Szolovits, P.,
Anthony Celi, L., & Mark, R. G. (2016). MIMIC-III, a
freely accessible critical care database. Scientific Data,
3(1), 160035. https://doi.org/10.1038/sdata.2016.35
Kiourtis, A., Mavrogiorgou, A., Menychtas, A.,
Maglogiannis, I., & Kyriazis, D. (2019). Structurally
Mapping Healthcare Data to HL7 FHIR through
Ontology Alignment. Journal of Medical Systems,
43(3). https://doi.org/10.1007/S10916-019-1183-Y
Kovács, T., Simon, G., & Mezei, G. (2019). Benchmarking
graph database backends - What works well with
wikidata? Acta Cybernetica, 24(1), 43–60.
https://doi.org/10.14232/ACTACYB.24.1.2019.5
Leavitt, N. (2010). Will NoSQL Databases Live Up to Their
Promise? Computer, 43(2), 12–14. https://doi.org/10.11
09/MC.2010.58
Li, W., Wang, S., Wu, S., Gu, Z., & Tian, Y. (2022).
Performance benchmark on semantic web repositories
for spatially explicit knowledge graph applications.
Computers, Environment and Urban Systems, 98.
https://doi.org/10.1016/J.COMPENVURBSYS.2022.1
01884
LIU, D., WEI, C., XIA, S., & YAN, J. (2022). Construction
and application of knowledge graph of Treatise on
Febrile Diseases. Digital Chinese Medicine, 5(4), 394–
405. https://doi.org/10.1016/J.DCMED.2022.12.006
Monteiro, J., Sá, F., & Bernardino, J. (2023). Experimental
Evaluation of Graph Databases: JanusGraph, Nebula
Graph, Neo4j, and TigerGraph. Applied Sciences
(Switzerland), 13(9). https://doi.org/10.3390/APP1309
5770
Pujante-Otalora, L., Canovas-Segura, B., Campos, M., &
Juarez, J. M. (2023). The use of networks in spatial and
temporal computational models for outbreak spread in
epidemiology: A systematic review. Journal of
Biomedical Informatics, 143, 104422. https://doi.org/
10.1016/J.JBI.2023.104422
RDF 1.1 Concepts and Abstract Syntax. (n.d.). Retrieved
September 26, 2023, from https://www.w3.org/TR/rdf
11-concepts/
RDF-star and SPARQL-star. (n.d.). Retrieved September
25, 2023, from https://www.w3.org/2021/12/rdf-
star.html
Rivas, A., & Vidal, M. E. (2021). Capturing Knowledge
about Drug-Drug Interactions to Enhance Treatment
Effectiveness. K-CAP 2021 - Proceedings of the 11th
Knowledge Capture Conference, 33–40.
https://doi.org/10.1145/3460210.3493560
SPARQL 1.1 Query Language. (n.d.). Retrieved September
26, 2023, from https://www.w3.org/TR/sparql11-
query/
Trevena, W., Lal, A., Zec, S., Cubro, E., Zhong, X., Dong,
Y., & Gajic, O. (2022). Modeling of Critically Ill Patient
Pathways to Support Intensive Care Delivery. IEEE
Robotics and Automation Letters, 7(3), 7287–7294.
https://doi.org/10.1109/LRA.2022.3183253
Neo4j Python driver. (2023). https://neo4j.com/docs/
getting-started/languages-guides/neo4j-python/
Wang, R., Yang, Z., Zhang, W., & Lin, X. (2020). An
empirical study on recent graph database systems.
Lecture Notes in Computer Science (Including
Subseries Lecture Notes in Artificial Intelligence and
Lecture Notes in Bioinformatics), 12274 LNAI, 328–
340. https://doi.org/10.1007/978-3-030-55130-8_29
Yin, Y., Li, G. Z., Wang, Y., Zhang, Q., Wang, M., & Zhang,
L. (2021). Study on construction and application of
knowledge graph of TCM diagnosis and treatment of
viral hepatitis B. Proceedings - 2021 IEEE International
Conference on Bioinformatics and Biomedicine, BIBM
2021, 3906–3911. https://doi.org/10.1109/BIBM526
15.2021.9669760
HEALTHINF 2024 - 17th International Conference on Health Informatics
36
