
Biodegradable Biodevices: A Design Approach Based on Cellular
Automaton
William Solórzano-Requejo
a
, Carlos Aguilar
b
, Gabriel Callejo and Andrés Díaz Lantada
c
Department of Mechanical Engineering, ETSI Industriales, Universidad Politécnica de Madrid,
c/ José Gutiérrez Abascal 2, 28006 Madrid, Spain
Keywords: Biodegradable Medical Devices, Biodegradable Materials, Degradation Modelling, Simulation of Medical
Devices, Cellular Automata.
Abstract: This innovative study introduces a comprehensive methodology to simulate the two-dimensional degradation
of biodegradable materials, a crucial aspect in biodevice design. Several PVA specimens of diverse shapes
were created, and their degradation was computationally modelled using cellular automaton. Deterministic
and probabilistic transition rules were explored to identify the most accurate in the simulation of PVA
degradation. The results highlight the effectiveness of the probabilistic exponential rule, derived from Markov
Chains, for reliable degradation simulation. Furthermore, this approach was successfully applied to the
analysis of specific medical devices, enabling a detailed in silico assessment of degradation patterns in
coronary stents, tissue engineering scaffolds and craniosynostosis implants. This methodology deepens our
fundamental understanding of degradation and provides valuable information for engineers and medical
professionals, facilitating the creation of devices that integrate optimally with surrounding biological tissues.
1 INTRODUCTION
Cellular automaton (CA) are discrete, local
dynamical systems that can be considered in several
ways: as a mathematical idealization of natural
systems, a discrete caricature of microscopic
dynamics, a parallel algorithm, or a discretization of
partial differential equations. From an engineering
perspective, CAs are networks composed of finite-
state machines, also known as cells, that operate
through localized interactions. These cells evolve
collectively, and the evolution of the system is
determined by the interaction of their individual
components (Dascălu, 2018).
In essence, CAs are conceptualized as sets of cells
arranged in grids. Over sequential steps or iterations,
these grids are dynamically transformed according to
specific predefined rules. During these iterations, the
state of each cell changes, influenced by both the
predetermined rules and the previous states of
neighbouring cells. If all cells are updated
simultaneously, the automaton is called synchronous.
a
https://orcid.org/0000-0002-2989-9166
b
https://orcid.org/0000-0003-0291-3041
c
https://orcid.org/0000-0002-0358-9186
On the other hand, if only one cell is updated per
iteration, it is called asynchronous (Agapie et al.,
2014; Dascălu, 2018).
Furthermore, CA can vary in terms of
randomness. A deterministic automaton has no
randomness; the evolution of the cells strictly follows
predetermined rules. However, if the cell to be
updated is chosen randomly and/or the local transition
rule involves probabilities, the automaton is classified
as probabilistic (Agapie et al., 2014; Dascălu, 2018).
In the 1950s, Konrad Zuse, Stanislav Marcin
Ulam and John von Neumann formulated the first
theories of CA in their strictest sense (Deutsch &
Dormann, 2017). These pioneers used CA to model
real-world phenomena, and von Neumann was
particularly inspired by the self-reproduction
capability observed in biological organisms (Von
Neumann & Burks, 1966). His fascination with the
inherent ability of living entities to create similar
beings led him to explore artificial life. This
exploration laid the foundation for the emergence of
artificial life as a distinct field of study, connecting it
34
Solórzano-Requejo, W., Aguilar, C., Callejo, G. and Díaz Lantada, A.
Biodegradable Biodevices: A Design Approach Based on Cellular Automaton.
DOI: 10.5220/0012313600003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 1, pages 34-41
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

with artificial intelligence and genetic algorithms.
These disciplines share a common thread of natural
inspiration, as each replicates certain characteristics
or constructive principles found in natural systems
(Dascălu, 2018).
In 1970, John Conway presented the Game of Life
automaton, a seemingly simple but profoundly
meaningful representation of the processes of birth
and death. This creation quickly became an icon in
the field of CA, capturing the public imagination with
its easily understood rules and captivating
simulations. The Game of Life demonstrated the
extraordinary ability of minimal, localized rules to
generate intricate, self-organizing patterns (Gardner,
1970). It served as a paradigmatic example of the
fundamental concept underlying CA: a basic, regular
structure capable of giving rise to a wide variety of
phenomena, even from initially chaotic states.
CA has been widely applied in diverse fields to
model dynamical systems that exhibit organized
behaviour, including areas such as statistical physics,
biology, medicine, ecology, and socioeconomic
interactions (Agapie et al., 2014). Interestingly, CAs
have also found utility in the medical device design
process. These automata have been used to represent
a patient's mental state by integrating information
from electroencephalogram analysis (Colafiglio
et al., 2023), for neuronal image segmentation
(Kalkhof et al., 2023), simulation of cells colonizing
tissue engineering scaffolds (Díaz Lantada et al.,
2023), optimization of biomimetic cell culture
systems (Ballesteros Hernando et al., 2019), and even
metamaterial design (Z. Liu et al., 2023).
Regarding the design of biodegradable and
bioabsorbable implants made from polymers or
metals, it is crucial to understand and quantify the
degradation of the material and how its behaviour
affects the biodesign. In this context, this article
delves into the detailed explanation of a methodology
that uses CA to two-dimensionally model the
degradation of polymeric specimens.
2 MATERIALS AND METHODS
2.1 Test Bench and Specimens
To investigate the influence of neighbourhood on CA
dynamics, various test specimens were utilized,
including circular, triangular, hexagonal,
quadrilateral, D and 4-shape geometries (Figure 1),
all with maximum dimensions of 30 x 30 x 2 mm for
the analysis of their two-dimensional degradation.
These probes were designed using Autodesk
®
Fusion
360 (Autodesk Inc., San Rafael, CA, USA).
Figure 1: PVA test specimens.
For the fabrication of the samples, Poly (vinyl
alcohol) (PVA) was chosen as the material due to its
rapid solubility in water, which facilitated the testing
process outlined in this research. This thermoplastic
material was purchased from Smart Materials 3D
(Pol. Ind. El Retamar, c/ Tomillo 7, 23680 Alcalá la
Real, Jaén, Spain) in form of monofilaments with a
1.75 mm diameter. The specimens were
manufactured using fused deposition modelling
(FDM), employing a Bambu Lab X1 Carbon Combo
3D printer equipped with a 0.4 mm diameter hardened
steel nozzle.
To generate the tool path for printing, Bambu
Studio (Austin, TX, USA), an open-source slicing
software based on Prusa Slicer, was employed. The
print parameters were consistently configured for all
printed specimens: a layer thickness of 0.2 mm, a
print speed of 40 mm/s, a bed temperature of 50 °C, a
nozzle temperature of 220 °C, two perimeters, and a
100% rectilinear infill pattern.
To investigate degradation, a 12.0-megapixel
high-resolution optical sensor with an f/1.5 lens and
an optical photo stabiliser was used to capture test
images at regular one-minute intervals and merge
them all into a video. To ensure consistent results and
avoid localised degradation, the samples were
carefully placed inside a glass container,
proportionally submerged in water. To prevent water
ingress from the top to the bottom, PETG discs were
used. This ensures that the degradation is two-
dimensional, specifically from the sides of the
specimen. The container was filled with 600 mL of
water, and maintained at a constant temperature of
27°C.
2.2 Image Processing
Frames were extracted from the video of each
experiment, revealing the appearance of bubbles due
to specimen degradation. Subsequently, it was
necessary to segment the PVA specimen, as this
segmentation allowed defining the CA transition
rules. To distinguish between the background, the
bubbles and the PVA in each frame, the K-Means
algorithm was used. This unsupervised algorithm
analysed the unlabelled image and grouped pixels
Biodegradable Biodevices: A Design Approach Based on Cellular Automaton
35

with similar characteristics. The result was a mask
divided into 3 zones (K = 3). Unfortunately, K-Means
was insufficient to extract the degraded PVA, so it
was complemented with Morphological Geodesic
Active Contour (MGAC). MGAC is especially
suitable for images with visible contours, even in
cases where these contours are noisy, cluttered, or
partially blurred, however, it requires preprocessing
to highlight these contours (Caselles et al., 1997). In
this case, K-Means segmentation was used to
highlight the contours, penalizing the areas belonging
to the background and the bubbles by multiplying
their pixel values by 0 and 0.5 respectively, and
highlighting the region with degraded material by
preserving their pixel values (Figure 2). In addition,
all images were aligned by changing their perspective
to avoid deformations that may result in erroneous
compression of the degradation. This whole process
was done in Python
®
3.7.14 (Phyton Software
Foundation) using OpenCV and scikit-image
libraries.
Figure 2: Image processing steps.
2.3 Cellular Automaton
CA is specified by a grid composed of cells, a set of
states characterizing the cells ( 𝜀), an interaction
neighbourhood (𝒩) and a rule (𝑅) which determines
the dynamics (Deutsch & Dormann, 2017). To model
the degradation of PVA samples, it is necessary to
define all these parameters since the complexity of
the model depends on them. Moreover, the higher
generality of the probabilistic CA (PCA) makes this
model more efficient compared to the deterministic
one (Agapie et al., 2014). In the following
subsections, each of the components of the CA will
be explained in detail.
2.3.1 Grid, Boundary Condition and States
A grid, denoted ℒ, is composed of a set of cells
represented by 𝑟. This grid ℒ establishes the spatial
framework in which the automaton operates and can
assume a finite or infinite nature. In our model, the
CA grid is the initial already processed grayscale
image captured after the experiments performed on
each sample (Figure 3A).
Figure 3: (A) Grids, (B) states and (C) neighbourhood of
the CA model.
In this scenario, the cells correspond to individual
pixels of the image. This discrete spatial
representation is crucial for CAs. In this case it is
possible to quantify the pixel dimensions, each pixel
measuring 50 x 50 µm. This accurate spatial analysis
is essential in the computational framework, because
if the pixel size dimensions change, the transition
rules could differ, and the model would have to be
recalibrated.
The CA model has two states: "live" (𝜀1),
which indicates the presence of PVA material in the
cell, and "dead" (𝜀0), which indicates the absence
of material or the presence of water causing PVA
degradation (Figure 3B). This simplifies the
complexity of the model, as it only considers these
two states.
Since the CA mesh is finite, it is necessary to
impose boundary conditions that specify the
interaction neighbourhood of the cells at the boundary
of the mesh. That boundary can be periodic, implying
that the mesh is closed, reflective, the state of the cells
at the mesh boundary is replicated, or fixed, one state
is defined for the entire boundary. In this case, the
fixed boundary was chosen and defined with the dead
state because it favours material degradation.
2.3.2 Neighbourhood and Rule
An interaction neighbourhood 𝒩
𝑟
defines the cells
that impact the state dynamics of a specific cell
𝑟
𝑖,𝑗
∈ℒ. In two-dimensional CA, two prevalent
types of neighbourhoods are widely used: the von
Neumann neighbourhood, encompassing the four
neighbouring cells along the vertical and horizontal
axes, and the Moore neighbourhood (𝒩
) comprising
both side neighbours and corner cells. When
considering degradation processes, all surrounding
cells influence the degradation of the central cell
(𝑟𝑖,𝑗), leading to the incorporation of the Moore
neighbourhood in CA degradation models (Figure
3C). This choice reflects the comprehensive influence
of adjacent and diagonal cells on the degradation
BIODEVICES 2024 - 17th International Conference on Biomedical Electronics and Devices
36
dynamics of the focal cell, providing a more accurate
representation of the system's behaviour.
𝒩
𝑖,
𝑗
𝑟
𝑖1,
𝑗
,𝑟
𝑖1,
𝑗
1
,𝑟
𝑖,
𝑗
1
,
𝑟
𝑖1,
𝑗
1
,𝑟
𝑖1,
𝑗
,𝑟
𝑖1,
𝑗
1
,
𝑟
𝑖,
𝑗
1
,𝑟
𝑖1,
𝑗
1
(1)
The rule can be probabilistic or deterministic. The
deterministic approach is simple: if a neighbouring
cell is dead, the central cell will be dead in that
iteration. In contrast, PCA introduces complexity.
According to this scenario, if 𝑛 cells surrounding the
central cell are dead, there is a chance of degradation
with a probability of 𝑃
. Clearly, as the number of
degraded cells around the central cell increases, the
probability of going from a living to a dead state
increase. How to obtain the probabilities in each case
is detailed in the Results section. Furthermore, in the
CA degradation model, all cells are updated
simultaneously, so it is a synchronous automaton.
3 RESULTS
3.1 Modelling of PVA Specimens
In the Materials and Methods section, all the
parameters involved in the creation of the CA for
modelling the degradation of the PVA were
described, except the rule that regulates the dynamics
of the system. In this case, it is interesting to compare
the deterministic approach with the probabilistic one;
the latter requires the transition probabilities as a
function of the neighbourhood, but because the
current state is the only one responsible for the next
state, CA becomes the perfect candidate for using
Markov Chains (MC) to regulate the probabilities
(Agapie et al., 2014).
First, it is important to understand the degradation
phenomena. For this purpose, an analysis of the
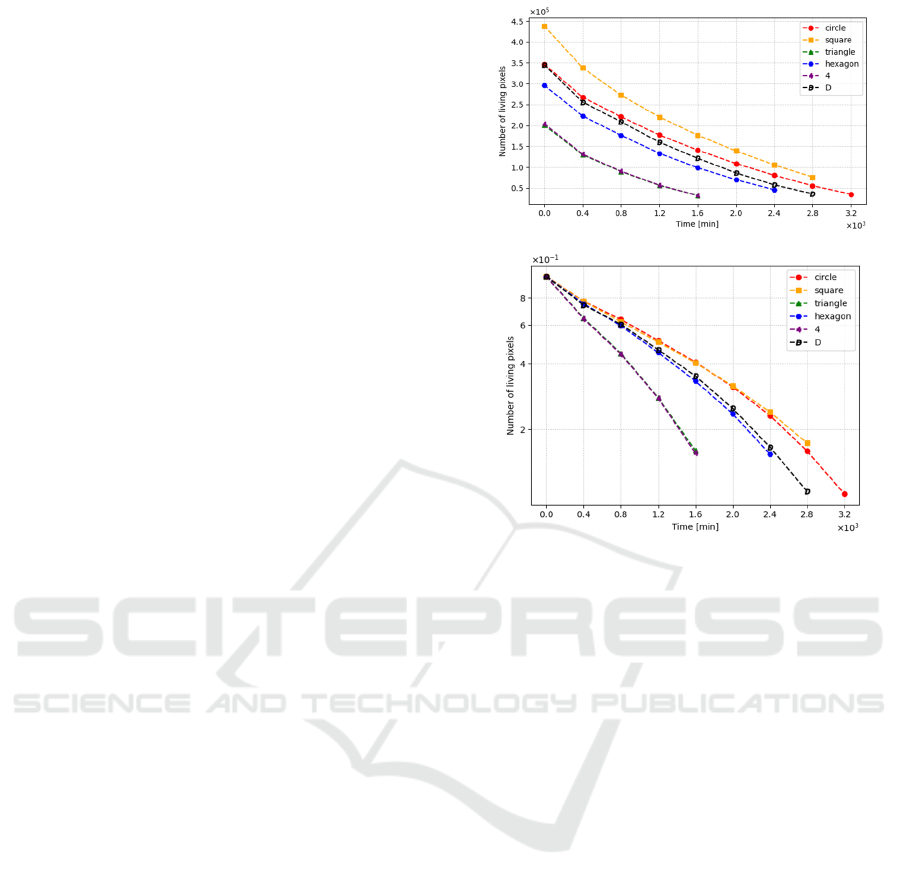
evolution of living pixels was carried out. Figure 4A
summarises the decreasing exponential trend of the
live pixels over time. To determine whether the
degradation rate is constant or not, the scale was
changed to exponential, and the number of live pixels
was normalised (Figure 4B). This graph demonstrates
two important aspects: the degradation rate is not
constant, and the geometry of PVA specimens
influences the degradation process because similar
shapes exhibit almost the same degradation rate. For
this reason, circular, triangular, and hexagonal
specimens were chosen to apply the MC model and
obtain the transition matrix, which defines the
probabilities of the CA.
(a)
(
b
)
Figure 4: Graphical representation of living pixel dynamics,
showing (A) dimensional and (B) dimensionless data.
MCs are stochastic processes characterised by a
finite set of states. For these processes, the transition
probabilities between states, denoted as 𝑝
, depend
exclusively on the current state of the system. These
probabilities are organised in a positive square
transition matrix 𝑃𝑝
,,
. In this context, the
"dead" state (𝜀0) is absorbing, since once the
system enters this state (𝑝
1), it can never leave.
By calculating the 𝑝
transition probability from
the ratio of the total number of cells to live cells, it is
evident that this MC is heterogeneous because the
transition matrix changes over time.
Therefore, the average value of the live-to-dead
transition probability was calculated for each of the
samples under study, considering only their
degradation information every 400 minutes until the
system is 80% degraded. Since the average transition
probability is 0.254, the following rule is established:
if the central cell is surrounded by at least one dead
cell, the degradation probability is 0.254 (𝑅
). In
addition, using the standard deviation information,
two probabilities were calculated: 0.17 and 0.41. Two
additional rules are defined considering that the
probability follows an exponential (𝑅
) or linear
(𝑅
) function, with 0.17 corresponding to the
Biodegradable Biodevices: A Design Approach Based on Cellular Automaton
37

probability of degradation if one of the neighbouring
cells is dead and 0.41 if six cells are dead.
In addition, two other rules are established using
linear (𝑅
__
) and exponential distributions
(𝑅
__
). The probability starts at 0.1 when one of
the cells is degraded and increases to 0.9 when six of
them are degraded. It is important to note that when
there are six degraded neighbours, it means that the
cell is attached to the specimen only by one live pixel,
and if there are seven dead cells, it implies that this
pixel is isolated. In both cases, it is determined that
the central cell will be degraded in that iteration.
Furthermore, all these rules are compared with the
deterministic one (𝑅
), which establishes that if at
least one of the neighbours is degraded, the central
cell will also be degraded. The whole CA was
programmed in Python
®
and the rules were applied if
the central cell is alive and if the transition
probability, which depends on the number of dead
neighbours, is greater than a random number.
Figure 5 compares the images obtained from the
experiments with the simulations of the CA with the
different rules. To determine the similarity between
the experimental images and those obtained through
the simulation, the mean square error (MSE) metric
was used, then, the iteration that presented the highest
similarity with the image of the degraded PVA
specimen at each moment was selected. This metric
also allowed us to quantify which of the rules
provides the most morphologically reliable
simulation. From the information provided by Figure
5 and the MSE analysis, it is concluded that the
exponential rule (𝑅
) obtained from the information
found in the Markov chain is the most reliable.
3.2 Simulation of Medical Devices
Degradation
Biodegradable implants offer several advantages for
medical professionals. First, they eliminate the need
for a second surgical intervention for removal, which
saves time and resources. In addition, biodegradation
can offer other significant advantages, for example,
rigid, non-biodegradable titanium implants can cause
problems such as refractures when removed, as the
bone has not been able to bear sufficient load during
the healing process. In contrast, biodegradable
implants can degrade slowly, gradually transferring
the load to the healing bone.
In tissue engineering, biodegradable scaffolds are
essential to provide adequate mechanical support for
damaged tissue and degrade gradually as new tissue
grows. However, the challenge lies in finding
materials with specific mechanical properties and
degradation rates for different tissues, as well as being
able to fabricate customized scaffolds with precise
pore interconnections (Y.-Y. Liu et al., 2023).
For coronary stents, a meticulous approach is
required to optimize mechanical properties and
degradation rate. These biodegradable stents offer a
promising advantage over traditional metallic stents
in that they dissolve completely in the body after a
period, thus reducing long-term adverse effects such
as restenosis. The ability to remove a foreign object
from the body after treatment of an obstruction is
especially attractive given current demographic
trends, which indicate that people are living longer
after a percutaneous coronary intervention procedure
(Tabares Ocampo et al., 2023). Traditionally,
biodegradable stents require a thicker strut compared
to conventional stents due to the weaker nature of the
degradable materials. However, a thicker strut leads
to worse patient outcomes.
As mentioned in both cases, it is essential to
consider the degradation of the medical device as an
essential design variable. This degradation is not only
dependent on the material and the environment, but
also on the geometry of the device and must be in
balance with tissue regeneration. The device must
disintegrate completely once it has fulfilled its
mechanical function. About this characteristic, the
degradation of the material leads to a loss of
properties that must be quantified. In some cases, this
loss may be beneficial, while in others it may be
detrimental. Therefore, controlling degradation
ensures a stable mechano-biological response.
To estimate in silico how the calibrated model
could aid in biodevice design, CT-like axial slices
were generated using Chitubox
®
v1.9. 0 (Chitubox,
Zhongcheng Future Industrial Park, Hangcheng
Avenue, Baoan District, Shenzhen, Guangdong,
China 518128) of a personalized coronary stent
exposed in the study by (Solórzano-Requejo et al.,
2023) and a tissue-engineered gyroid scaffold
designed using the open-source software MS Lattice
(Al‐Ketan & Abu Al‐Rub, 2021). Figure 6 shows the
slice degradation process for each of the medical
devices. By performing the conversion from
iterations to time, considering that 68 iterations equal
to approximately 400 minutes, the designer would be
able to analyze whether the implant degrades in the
right time frame and which areas are most prone to
degrade rapidly. Moreover, this is achieved at a very
low computational cost, as the CA model is very
optimal in that aspect.
BIODEVICES 2024 - 17th International Conference on Biomedical Electronics and Devices
38
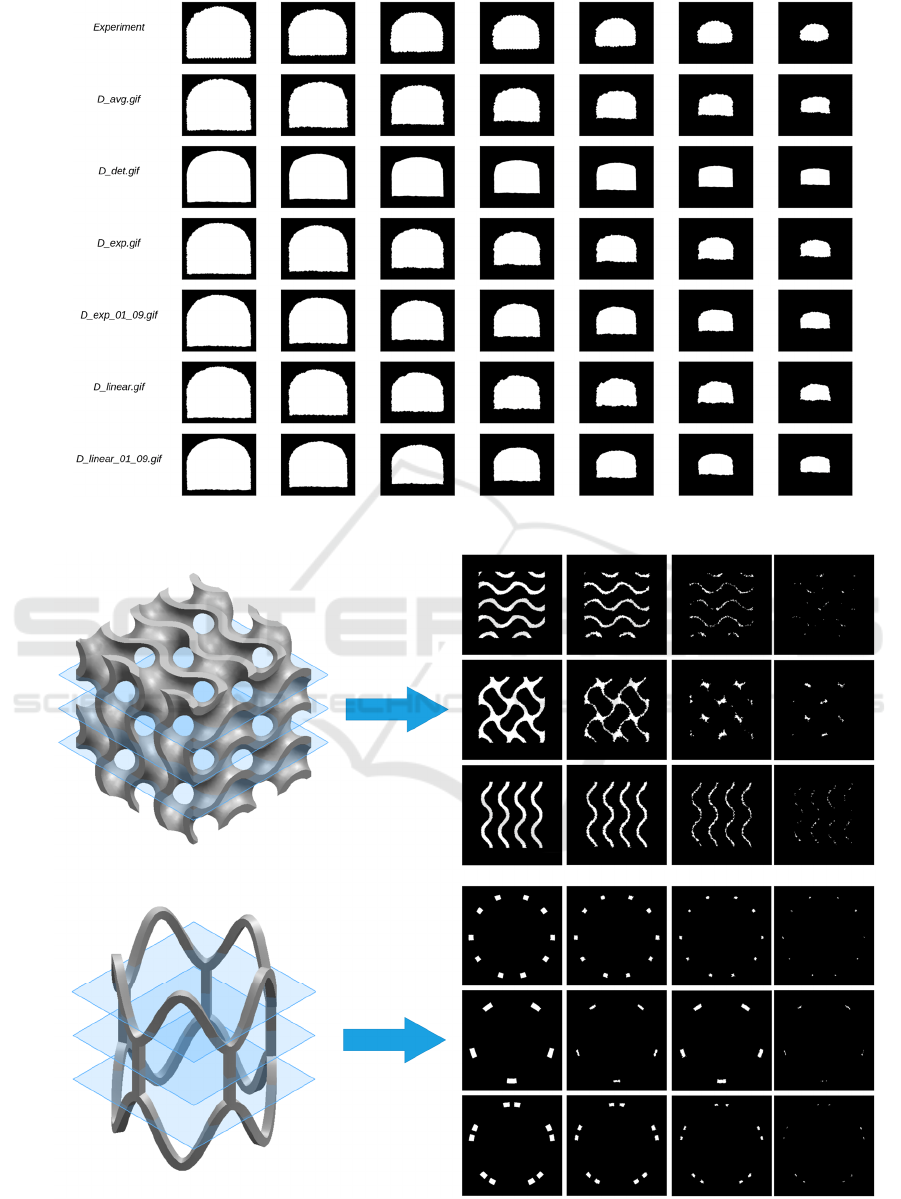
Figure 5: Comparison between experimental results and simulations with different rules for the D-shaped specimen.
Figure 6: PCA simulation of axial slices of a tissue engineering scaffold and coronary stent.
Biodegradable Biodevices: A Design Approach Based on Cellular Automaton
39
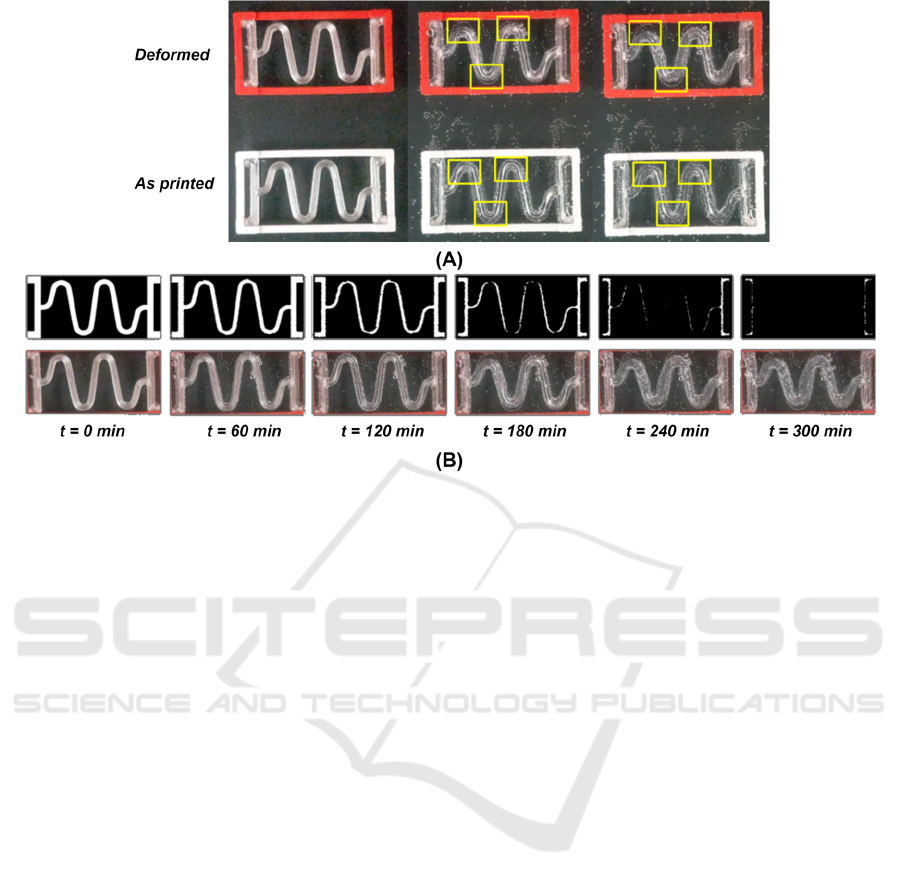
Figure 7: (A) Comparison of the degradation between two springs: one deformed by a framework during the degradation
process and the other printed directly with the deformed shape. (B) Simulation using the CA whose rules are influenced by
the stress state.
4 FUTURE PROPOSALS
In this study, a calibrated model capable of capturing
the complex two-dimensional interactions in medical
devices has been developed. However, to accurately
assess the degradation of these devices, it is essential
to adopt a three-dimensional perspective. By
connecting this model to finite element software, it is
possible to analyze how mechanical properties, such
as stiffness and creep, are affected by the progressive
degradation of the structure.
In the context of the mechanical state of the
material, it is crucial to consider how stresses and
strains impact the degradation rate. Therefore, it is
possible to use information obtained from a structural
analysis to define the rules of the CA model. These
rules should not only depend on the neighborhood,
but also on the stress state of the structure. More
mechanically loaded areas will experience faster
degradation compared to less stressed areas.
Additionally, in this study, experiments have been
carried out with PVA serpentine springs, which serve
as a prototype of implants for craniosynostosis,
aiming to promote brain growth. To test the influence
of the stress state, the degradation of two springs, one
deformed by a frame during degradation and the other
printed directly with the deformed shape obtained
from the finite element software as an STL file, was
compared. The results were revealing: the
degradation was noticeably more pronounced in the
curved regions that were highly deformed. Then, the
probabilities calibrated in our model were adjusted
as a function of the deformation of the springs
(Figure 7).
This finding suggests that it is possible to
hybridize both methodologies, using a three-
dimensional CA model within finite element software
meshed with hexahedral elements. In this approach,
each mesh element can represent a cell, and the
degradation probabilities would depend on the
specific stress state of each cell. This would allow
accurate simulation of how the material behaves
when the stress state varies, especially in
biodegradable biomedical devices under varying
loads.
5 CONCLUSIONS
This study presents a methodology for accurate
simulation of two-dimensional biodegradable
specimens, representing a significant advance in the
accurate simulation of degradation in medical
devices. Detailed results and meticulous comparisons
of CA rules have revealed that the exponential rule
BIODEVICES 2024 - 17th International Conference on Biomedical Electronics and Devices
40

(𝑅
) derived from Markov chains offers a highly
reliable simulation of PVA degradation.
This approach not only deepens our fundamental
understanding of degradation, which is influenced by
material, environment, and part geometry, but also
has practical applications in the design of
biodegradable medical devices. By simulating
devices such as coronary stent, tissue engineering
scaffold and prototypes of implants for
craniosynostosis, their degradation can be assessed in
silico, providing valuable information for engineers
and medical professionals.
In addition, this study has clearly pointed to future
research directions. The adoption of a three-
dimensional approach, integrating CA with finite
element models, promises to offer even more accurate
simulation, especially when considering the stress
state of each cell. This hybrid approach could unlock
new insights into how devices respond to different
loads, essential for the design of devices that integrate
optimally with the surrounding biological tissue.
ACKNOWLEDGEMENTS
This project has received funding from the European
Union’s Horizon Europe research and innovation
programme under grant agreement No 101047008
(BIOMET4D). Views and opinions expressed are
however those of the authors only and do not
necessarily reflect those of the European Union or the
European Innovation Council and SMEs Executive
Agency (EISMEA). Neither the European Union nor
the EISMEA can be held responsible for them.
REFERENCES
Agapie, A., Andreica, A., & Giuclea, M. (2014).
Probabilistic Cellular Automata. Journal of Computa-
tional Biology, 21(9), 699-708. https://doi.org/10.1089/
cmb.2014.0074
Al‐Ketan, O., & Abu Al‐Rub, R. K. (2021). MSLattice: A
free software for generating uniform and graded lattices
based on triply periodic minimal surfaces. Material
Design & Processing Communications, 3(6).
https://doi.org/10.1002/mdp2.205
Ballesteros Hernando, J., Ramos Gómez, M., & Díaz
Lantada, A. (2019). Modeling Living Cells Within
Microfluidic Systems Using Cellular Automata
Models. Scientific Reports, 9(1), 14886.
https://doi.org/10.1038/s41598-019-51494-1
Caselles, V., Kimmel, R., & Sapiro, G. (1997). Geodesic
Active Contours. International Journal of Computer
Vision, 22(1), 61-79. https://doi.org/10.1023/A:10079
79827043
Colafiglio, T., Lofù, D., Sorino, P., Festa, F., Di Noia, T.,
& Di Sciascio, E. (2023). Exploring the Mental State
Intersection by Brain-Computer Interfaces, Cellular
Automata and Biofeedback. IEEE EUROCON 2023 -
20th International Conference on Smart Technologies,
461-466. https://doi.org/10.1109/EUROCON56442.20
23.10198964
Dascălu, M. (2018). Cellular Automata and
Randomization: A Structural Overview. En R. Lopez-
Ruiz (Ed.), From Natural to Artificial Intelligence—
Algorithms and Applications. IntechOpen.
https://doi.org/10.5772/intechopen.79812
Deutsch, A., & Dormann, S. (2017). Cellular Automaton
Modeling of Biological Pattern Formation. Birkhäuser
Boston. https://doi.org/10.1007/978-1-4899-7980-3
Díaz Lantada, A., Urosa Sánchez, M., & Fernández
Fernández, D. (2023). In silico Tissue Engineering and
Cancer Treatment Using Cellular Automata and Hybrid
Cellular Automata-Finite Element Models:
Proceedings of the 16th International Joint Conference
on Biomedical Engineering Systems and Technologies,
56-63. https://doi.org/10.5220/0011742300003414
Gardner, M. (1970). Mathematical Games. Scientific
American, 223(4), 120-123.
Kalkhof, J., González, C., & Mukhopadhyay, A. (2023).
Med-NCA: Robust and Lightweight Segmentation with
Neural Cellular Automata. En A. Frangi, M. De
Bruijne, D. Wassermann, & N. Navab (Eds.),
Information Processing in Medical Imaging (Vol.
13939, pp. 705-716). Springer Nature Switzerland.
https://doi.org/10.1007/978-3-031-34048-2_54
Liu, Y.-Y., Blazquez, J. P. F., Yin, G.-Z., Wang, D.-Y.,
Llorca, J., & Echeverry-Rendón, M. (2023). A strategy
to tailor the mechanical and degradation properties of
PCL-PEG-PCL based copolymers for biomedical
application. European Polymer Journal, 198, 112388.
https://doi.org/10.1016/j.eurpolymj.2023.112388
Liu, Z., Fang, H., Xu, J., & Wang, K.-W. (2023). Cellular
automata inspired multistable origami metamaterials
for mechanical learning. https://doi.org/10.48550/
ARXIV.2305.19856
Solórzano-Requejo, W., Aguilar, C., Zapata Martínez, R.,
Contreras-Almengor, O., Moscol, I., Ojeda, C., Molina-
Aldareguia, J., & Diaz Lantada, A. (2023). Artificial
Intelligence and Numerical Methods Aided Design of
Patient-Specific Coronary Stents: Proceedings of the
16th International Joint Conference on Biomedical
Engineering Systems and Technologies, 37-45.
https://doi.org/10.5220/0011639000003414
Tabares Ocampo, J., Marín Valencia, V., Robledo, S. M.,
Upegui Zapata, Y. A., Restrepo Múnera, L. M.,
Echeverría, F., & Echeverry-Rendón, M. (2023).
Biological response of degradation products of PEO-
modified magnesium on vascular tissue cells,
hemocompatibility and its influence on the
inflammatory response. Biomaterials Advances, 154,
213645. https://doi.org/10.1016/j.bioadv.2023.213645
Von Neumann, J., & Burks, A. W. (1966). Theory of self-
reproducing automata. IEEE Transactions on Neural
Networks, 5(1), 3-14.
Biodegradable Biodevices: A Design Approach Based on Cellular Automaton
41
