
A Comparison of Recurrent and Convolutional Deep Learning
Architectures for EEG Seizure Forecasting
Sina Shafiezadeh
1 a
, Marco Pozza
2 b
and Alberto Testolin
1,2 c
1
Department of General Psychology, University of Padova, Padova, Italy
2
Department of Mathematics, University of Padova, Padova, Italy
fi
Keywords:
Seizure Prediction, Epilepsy, Artificial Intelligence, Convolutional Neural Networks, Long Short-Term
Memory Networks, Electroencephalography, Signal Processing, Scalogram Images.
Abstract:
Many research efforts are being spent to discover predictive markers of seizures, which would allow to build
forecasting systems that could mitigate the risk of injuries and clinical complications in epileptic patients.
Although electroencephalography (EEG) is the most widely used tool to monitor abnormal brain electrical
activity, no commercial devices can reliably anticipate seizures from EEG signal analysis at present. Re-
cent advances in Artificial Intelligence, particularly deep learning algorithms, show promise in enhancing
EEG classifier forecasting accuracy by automatically extracting relevant spatio-temporal features from EEG
recordings. In this study, we systematically compare the predictive accuracy of two leading deep learning
architectures: recurrent models based on Long Short-Term Memory networks (LSTMs) and Convolutional
Neural Networks (CNNs). To this aim, we consider a data set of long-term, continuous multi-channel EEG
recordings collected from 29 epileptic patients using a standard set of 20 channels. Our results demonstrate
the superior performance of deep learning algorithms, which can achieve up to 99% accuracy, sensitivity, and
specificity compared to more traditional machine learning approaches, which settle around 75% in all evalu-
ation metrics. Our results also show that giving as input the recordings from all electrodes allows to exploit
useful channel correlations to learn more robust predictive features, compared to convolutional models that
treat each channel independently. We conclude that deep learning architectures hold promise for enhancing
the diagnosis and prediction of epileptic seizures, offering potential benefits to those affected by such invali-
dating neurological conditions.
1 INTRODUCTION
Epilepsy, a chronic neurological disorder that affects
approximately 50 million people worldwide of all
ages, is characterized by recurring seizures, often ac-
companied by loss of consciousness and control of
bladder or bowel function (World Health Organiza-
tion, 2023). EEG is considered a promising, non-
invasive clinical diagnostic method that could be used
to continuously monitor brain electrical activity, po-
tentially allowing for automatic detection or even
forecasting epileptic seizures in advance (Van Mierlo
et al., 2020). A reliable monitoring device would
allow patients to avoid dangerous situations or even
plan the administration of preventive treatments, such
as electrical stimulation or targeted drug delivery, thus
a
https://orcid.org/0000-0002-5462-4893
b
https://orcid.org/0009-0004-5798-0297
c
https://orcid.org/0000-0001-7062-4861
significantly improving their quality of life.
In recent years, the growing effectiveness of deep
learning methods in clinical diagnosis and disease
prediction (Liu et al., 2020; Calesella et al., 2021) has
reignited enthusiasm for harnessing machine learning
algorithms in the complex task of seizure forecasting
(Abbasi and Goldenholz, 2019). It should be noted
that seizure forecasting aims to anticipate an up-
coming seizure before it clinically manifests, making
this task much more challenging compared to seizure
detection, a simpler classification problem that re-
quires discriminating between normal brain activity
and seizure states. A common strategy to implement
predictive models involves extracting diverse descrip-
tive characteristics from EEG recordings, which are
then used to train machine learning algorithms to rec-
ognize time intervals close to an imminent seizure
(Assi et al., 2017). However, the most recent ap-
proaches exploit deep learning techniques, which can
Shafiezadeh, S., Pozza, M. and Testolin, A.
A Comparison of Recurrent and Convolutional Deep Learning Architectures for EEG Seizure Forecasting.
DOI: 10.5220/0012311800003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 1, pages 583-590
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
583
achieve higher accuracy by extracting more sophisti-
cated, non-linear features directly from the raw sig-
nals (Usman et al., 2020).
Several deep learning architectures have been pro-
posed to process EEG signals efficiently. Long
Short-Term Memory (LSTM) networks (Hochreiter
and Schmidhuber, 1997) are particularly effective in
learning temporal features from time series and have
thus been widely applied to the automatic analysis of
EEG recordings, with promising results also in some
seizure prediction tasks (Tsiouris et al., 2018; Varnos-
faderani et al., 2021). CNNs are instead effective in
discovering spatial features from images, but can also
be used to analyze windowed signals extracted from
time series (LeCun et al., 1995) and have thus been
successfully applied to EEG seizure prediction (San-
Segundo et al., 2019; Hussein et al., 2021). Moreover,
some authors have proposed to enhance the classic 2D
convolutional architecture by introducing 3D convo-
lutions, enabling the extraction of features from both
spatial and temporal dimensions and the discovery
of correlations between feature maps and contiguous
frames in the previous layer (Ji et al., 2012). In EEG
signal analysis, this allows to consider inter-channel
correlations, which can be particularly useful when
classifying preictal and interictal seizure states (Wang
et al., 2021; Ozcan and Erturk, 2019).
In this study, we compare the performance of
LSTMs, 2D CNNs, and 3D CNNs in the challeng-
ing task of seizure forecasting. For the LSTM, we di-
rectly fed the model the raw signals recorded from all
EEG channels, which should allow the network to ex-
ploit both temporal and inter-channel features to carry
out the prediction task. For the 2D CNN, we gave
images as input to the model representing the scalo-
gram of windowed EEG signals recorded from each
channel independently. For the 3D CNN, the input
is instead constituted by the image scalogram of all
channels, which similarly to the LSTM case should
allow the network to exploit inter-channel dependen-
cies besides spatio-temporal features extracted from
each individual scalogram.
We hypothesize that deep learning models would
achieve higher predictive performance compared to
standard machine learning algorithms, such as those
based on eXtreme Gradient Boosting (XGBoost)
(Shafiezadeh et al., 2023), and that neural architec-
tures that exploit inter-channel correlations such as
the LSTM and the 3D CNN would result in better
forecast accuracy.
2 METHODS
2.1 EEG Data Set
We used a data set of long-term, continuous multi-
channel EEG recordings collected by the Epilepsy
and Clinical Neurophysiology Unit at the Eugenio
Medea IRCCS Hospital in Conegliano, Italy. EEG
data were recorded from 29 epileptic patients (15
males and 14 females) at a sampling rate of 256 Hz.
We selected 20 common channels based on the inter-
national standard 10-20 EEG scalp electrode position-
ing system (see Fig. 1).
Each seizure event was characterized by four pos-
sible states: preictal, ictal, postictal, and interictal.
These states correspond to the periods before the on-
set of a seizure, the onset and conclusion of a seizure,
the period following a seizure, and normal brain ac-
tivity, respectively. In our data, the onset and termi-
nation of ictal states were manually identified using
video recorded data from video-EEG monitoring by
two expert clinicians. The seizure prediction task was
framed as a binary classification problem of diagnos-
ing preictal (30 minutes preceding a seizure) vs. in-
terictal (30 to 120 minutes preceding a seizure) states.
To guarantee the use of proper interictal time periods,
we excluded seizures occurring in recordings shorter
than 3 hours, resulting in a total of 93 seizures re-
tained for subsequent analysis. However, all seizures
were randomized together for cross-validation and di-
viding the data into train and test data points.
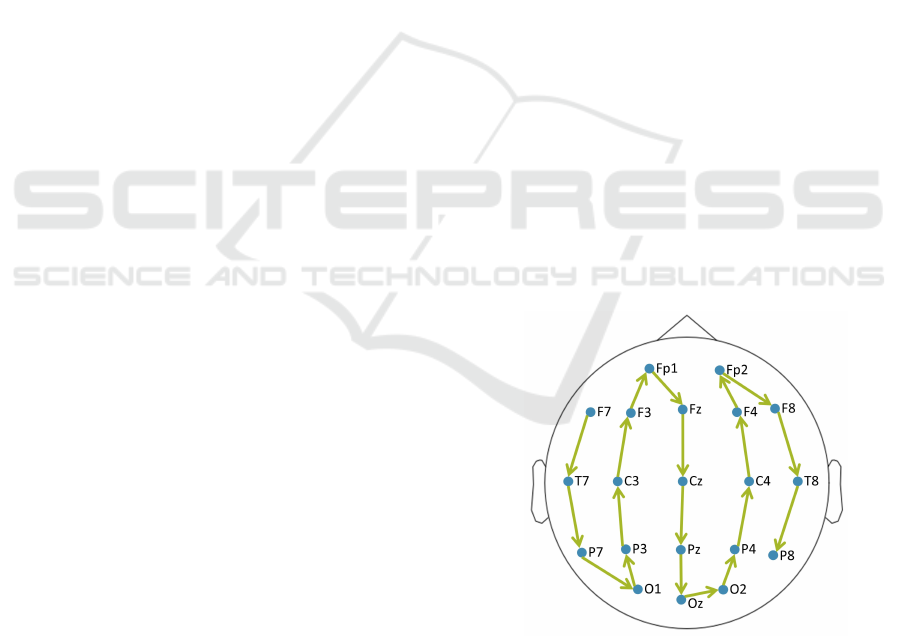
Figure 1: The 20 common channels were used in this study.
The channels ordering process initiates from the left tempo-
ral lobe (starting from F7) and extends to the right temporal
lobe (ending to P8). This deliberate sequence was designed
to formulate 3D inputs as a stack of 2D segments incorpo-
rating inter-channel correlations.
BIOSIGNALS 2024 - 17th International Conference on Bio-inspired Systems and Signal Processing
584

2.2 EEG Signal Pre-Processing
Several filtering procedures were applied to enhance
signal quality. Specifically, a 125 Hz low-pass fil-
ter and a 1 Hz high-pass filter were applied to retain
high-frequency signals relevant to abnormal brain ac-
tivities before seizures, while simultaneously remov-
ing DC offset and baseline fluctuations (Allen et al.,
1992; Arroyo and Uematsu, 1992). Moreover, to mit-
igate power line interference, two notch filters operat-
ing at 50 Hz and 100 Hz were incorporated in the pre-
processing pipeline (Niknazar et al., 2015; Thangavel
et al., 2021). EEG signals were finally downsampled
to 128 Hz, and the time series were normalized by
subtracting the average EEG reference computed on
each patient’s training data. All filtering operations
were performed using the MNE package within the
Python 3.8.5 version.
2.3 Scalogram Images
The input matrices for CNN models were created by
transforming the EEG time series into scalogram im-
ages, as proposed in recent studies (Yildiz et al., 2021;
Varlı and Yılmaz, 2023). To this aim, the Contin-
uous Wavelet Transform (CWT) was used to com-
pute multiple expansions and the wavelet’s time off-
set, yielding a variety of frequency values for ana-
lyzing continuous-time signals that can be used to
characterize energy density at a local time-frequency
within the transformation (Peng et al., 2002). A more
precise and flexible time–frequency resolution can
be obtained by considering the absolute value of the
CWT when producing scalogram images (Bostanov,
2004), which is particularly effective since it effec-
tively captures high- and low-frequency information
also in non-stationary signals like EEG (Falamarzi
et al., 2014).
More precisely, a function known as the main
wavelet performs the window’s role within the
wavelet transform; during the conversion process, this
main wavelet function is scaled and shifted, enabling
extensive time interval windowing for low frequen-
cies and compressed time interval windowing for high
frequencies (T
¨
urk and
¨
Ozerdem, 2019). The Mor-
let wavelet was employed for the continuous wavelet
transformations (Kareem and Kijewski, 2002). CWT
can be represented mathematically in continuous time
as in Equation (1), where W(s, τ), x(t), (t), s, and τ
represent the wavelet coefficients, the time signal, the
basic wavelet function conjugate, the scale, and the
position parameter, respectively:
W
x
(s,τ) =
1
√
s
Z
+∞
−∞
x(t)ψ
∗
(
t −τ
s
)dt (1)
In our study, scalogram images were created us-
ing sliding windows of 30 seconds with 50% over-
lap, resulting in greater accuracy compared to shorter
windows of 10 or 20 seconds. To improve computa-
tional efficiency during model training, the resulting
time-frequency scalogram images were resized to 96
× 96 pixels using cubic interpolation, following the
methodology adopted by (Ozdemir et al., 2021) and
(T
¨
urk and
¨
Ozerdem, 2019). Samples of resized scalo-
grams depicting distinct preictal and interictal states
are shown in Fig. 2.
2.4 Deep Learning Architectures
All models were implemented using the PyTorch
framework (version 1.13.0). Training and testing
phases were carried out using a virtual machine
equipped with an Nvidia V100 GPU allocated in the
Google Cloud Platform.
2.4.1 Hyperparameters Tuning
Model hyperparameters were iteratively refined using
a hierarchical strategy to attain the optimal configu-
ration using a reasonable amount of computing time.
For the LSTM model we tuned the hyperparameters
using the Optuna framework (Akiba et al., 2019); the
search space included the number of hidden layers (1,
2, 3), the size of hidden layers (32, 64, 128, 256),
learning rate (1e-5 to 0.1), batch size (16, 32, 64, 128,
256), and the dropout rates (0.1 to 0.9). The CNNs
hyperparameters were tuned in a specific order, start-
ing from the number of hidden layers (2, 3, 4), the
number of kernels (16, 32, 64), the kernel size (2, 3,
5), the learning rate (0.01, 0.001, 0.0001), the num-
ber of dropout layers (1, 2, 3), the dropout rate (0.1,
0.2, 0.5), batch size (16, 32, 64) and the use of pool-
ing layers to reduce the size of feature maps progres-
sively. The hyperparameters yielding the best perfor-
mance at each step were selected before progressing
to the next optimization step.
2.4.2 LSTM
The LSTM network received an input time series of 5-
second raw signals with a 3-second overlap, resulting
in an input size of 20 channels × 640 time points. The
final LSTM architecture was made up of two hidden
layers, with 32 units each. Two drop-out layers with
a drop-out rate of 0.1 and 0.3 were included between
the two LSTM layers and before the fully connected
layer, respectively. The final LSTM architecture is
represented in Fig. 3.
A Comparison of Recurrent and Convolutional Deep Learning Architectures for EEG Seizure Forecasting
585
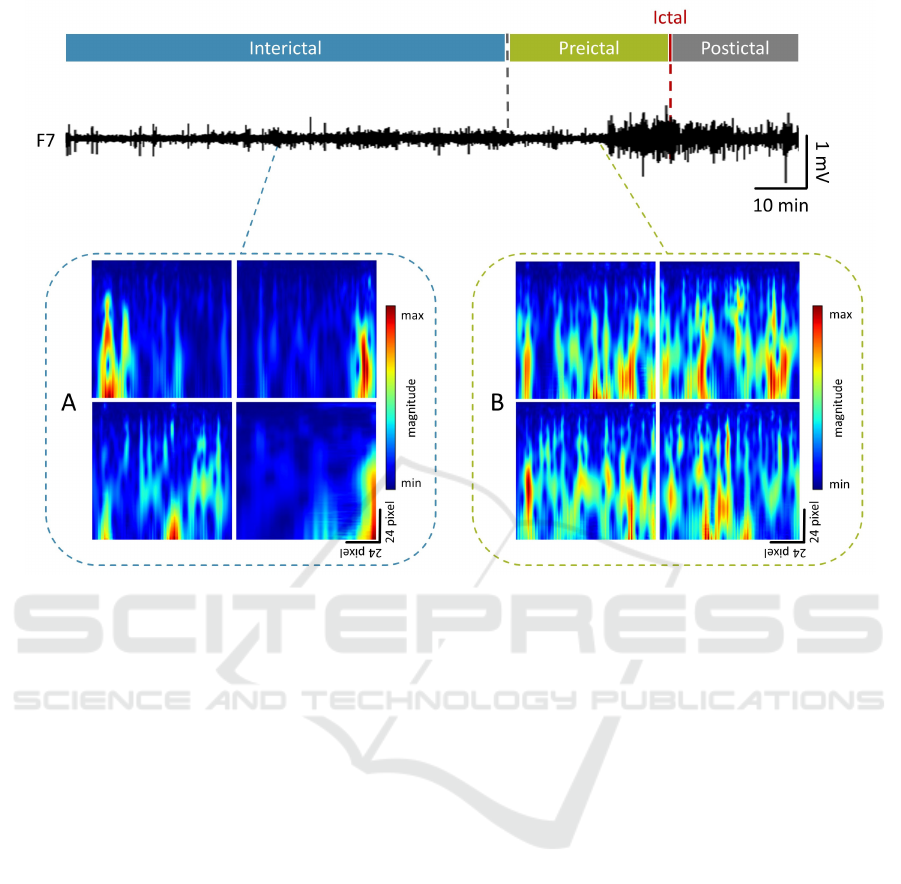
Figure 2: Example of a 145 min EEG signal trace recorded from channel F7 during a seizure event, segmented to identify
interictal vs. preictal states. Panel A shows four different sample images from the interictal state, while panel B shows four
different sample images from the preictal state.
2.4.3 2D CNN
For the 2D convolutional network, scalogram images
originating from each channel were treated indepen-
dently. Consequently, the input dimension for the
2D model was 96 × 96. The final architecture com-
prised three convolutional layers, with 16, 32, and 64
kernels, activated by Rectified Linear Unit (ReLU)
functions and complemented with batch normaliza-
tion layers. The optimal kernel size was 3 × 3 for
all layers, and a stride of 3 and padding of one pixel
was applied. To mitigate the risk of overfitting, two
dropout layers with a dropout rate of 0.1 were placed
after the third convolutional layer and the flatting
layer. The flattened layer was further processed by
an additional ReLU-activated dense layer comprising
128 units, and this layer was finally decoded by an
output layer that used a sigmoid activation function to
produce the model’s prediction.
2.4.4 3D CNN
To create the 3D input, a specific order of channels
from the left to the right temporal lobe was adopted:
F7, T7, P7, O1, P3, C3, F3, Fp1, Fz, Cz, Pz, Oz, O2,
P4, C4, F4, Fp2, F8, T8, and P8 (see Fig. 1). The
corresponding scalogram images were given as input
to the 3D model, resulting in a 20 × 96 × 96 input
shape. To allow for a fair comparison, the existing
2D CNN optimal architecture was subject to adapta-
tion solely in terms of kernel dimensions, transition-
ing from 2D to 3D. The final 3D architecture is repre-
sented in Fig. 4.
2.5 Model Evaluation
All models were trained for 100 epochs, and the train-
ing accuracy was recorded after each epoch in order
to track learning progress over time. Furthermore,
randomized cross-validation was carried out in each
fold by dividing the 93 seizures into 80% train and
20% test sets. However, during the five-fold cross-
validation implementation, the average accuracy of
testing data points after each fold was considered
to select the best model and compare three models.
Models’ performances were measured using 5-fold
cross-validation in terms of accuracy (ACC), sensi-
tivity (SEN), and specificity (SPE), which were com-
puted from true positives (TP), false positives (FP),
true negatives (TN), and false negatives (FN) as fol-
lows:
BIOSIGNALS 2024 - 17th International Conference on Bio-inspired Systems and Signal Processing
586
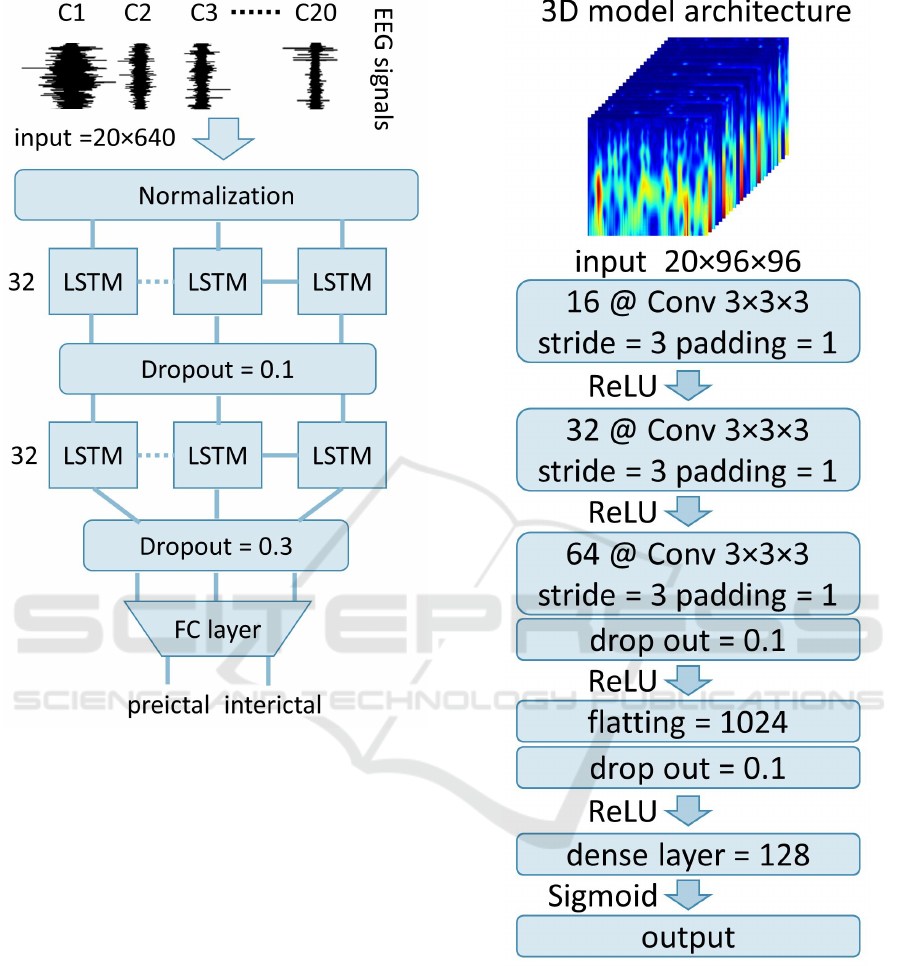
Figure 3: Representation of the final LSTM model architec-
ture. The model comprises two LSTM layers with 32 units
each and two drop-out layers with 0.1 and 0.3 rates, respec-
tively.
Accuracy = ((t p + tn)/(t p +tn + f n + f p)) (2)
Sensitivity = (t p/(t p + f n)) (3)
Speci f icity = (tn/(tn + f p)) (4)
In order to evaluate the performance gain of deep
learning models compared to traditional machine
learning classifiers, the best-performing architectures
were further benchmarked against an XGBoost clas-
sifier trained on a set of 53 EEG features (for details,
see (Shafiezadeh et al., 2023)).
Figure 4: Representation of the final 3D CNN model ar-
chitecture. The model comprises three convolutional layers
and two drop-out layers. The 2D CNN architecture is iden-
tical, with a kernel dimension of 3 × 3.
3 RESULTS
As shown in Figure 5, the LSTM model converged
to higher accuracy in a smaller number of training
epochs. The final performance of the 2D model was
significantly lower than the LSTM and 3D models,
suggesting that inter-channel correlations constitute
A Comparison of Recurrent and Convolutional Deep Learning Architectures for EEG Seizure Forecasting
587

Figure 5: Improvement of training accuracy during the fit-
ting of the three deep learning models (3D, LSTM, and 2D).
an important source of information that can be lever-
aged for the seizure prediction task.
In the testing phase, the LSTM model achieved the
highest specificity at 99.01%, while the 3D model ex-
hibited the highest accuracy and sensitivity, with val-
ues of 98.95% and 98.91%, respectively. ACC, SEN,
and SPE average values of each model in the testing
set, as well as standard deviations of each testing set’s
performance metric after implementation of five-fold,
are presented in Table 1. This table also illustrates the
results of statistical comparisons (one-way ANOVA)
between the three models after tuning and their best
results. The findings show significant differences be-
tween all performance metrics, and post-hoc Tukey
tests confirmed that the 2D architecture performed
worse than the LSTM and the 3D architectures (see
Table 2).
The LSTM model demonstrated significant im-
provements in performance metrics compared to the
2D model, with differences of 9.49%, 7.62%, and
12.46% in ACC, SEN, and SPE, respectively. Also,
the 3D model achieved substantial improvements
compared to the 2D model, respectively of 9.53%,
7.72%, and 11.57%. Although slight variations exist
in the performance metrics between the LSTM and
the 3D model, such differences were not statistically
significant. A graphical representation of all perfor-
mance metrics is given in Figure 6.
3.1 Comparison with XGBoost
Having identified the LSTM and 3D models as the
top-performing models according to all performance
metrics, we then proceeded with a comparison against
a standard machine learning model, specifically XG-
Boost, trained on a set of 53 ad hoc features extracted
from the EEG signal (for details, see (Shafiezadeh
et al., 2023)). The performance metrics of XGBoost
are shown in the gray column of Figure 6.
After applying cross-validation on the fea-
Figure 6: Comparison of the average performance metrics
of the testing set for classifying interictal and preictal phases
with the LSTM, 2D, 3D, and XGBoost models. Error bars
represent the standard deviation of each performance metric
after the implementation of five-fold cross-validation.
ture data set, the XGBoost model achieved ACC
(75.86%±3.37), SEN (75.90%±5.74), and SPE
(74.72%±4.61). The LSTM model significantly out-
performed XGBoost across all evaluation metrics:
23.05% in ACC, 22.91% in SEN, and 24.29% in SPE
(all p-values <0.01). The same results were found for
the 3D model, which significantly outperformed XG-
Boost with increases of 23.09% in ACC, 23.01% in
SEN, and 23.40% in SPE (all p-values <0.01). It is
also worth noting that deep learning models are asso-
ciated with substantially reducing the standard error
associated with each evaluation metric, suggesting a
more stable classification performance.
4 DISCUSSION
In this study, we leveraged raw signals and scalogram
images derived from multichannel EEG recordings of
epileptic patients to conduct a comparative analysis
between the LSTM, 2D, and 3D models in seizure
forecasting. Our investigation revealed that incorpo-
rating inter-channel correlations by feeding all chan-
nels to the LSTM model and the 3D CNN model
yielded a significant (almost 10%) improvement in
accuracy, increasing the classifier precision in distin-
guishing between interictal and preictal states.
A direct benchmarking against a more tradi-
tional XGBoost machine learning classifier further
confirmed that deep learning architectures allow to
achieve substantial performance gains across all eval-
uation metrics. At the same time, it should be noted
that our results are not directly comparable with other
recently published findings, since our analyses were
carried out using a novel clinical dataset recently col-
lected in a local hospital. However, as a qualita-
tive comparison, we might argue that our findings
BIOSIGNALS 2024 - 17th International Conference on Bio-inspired Systems and Signal Processing
588
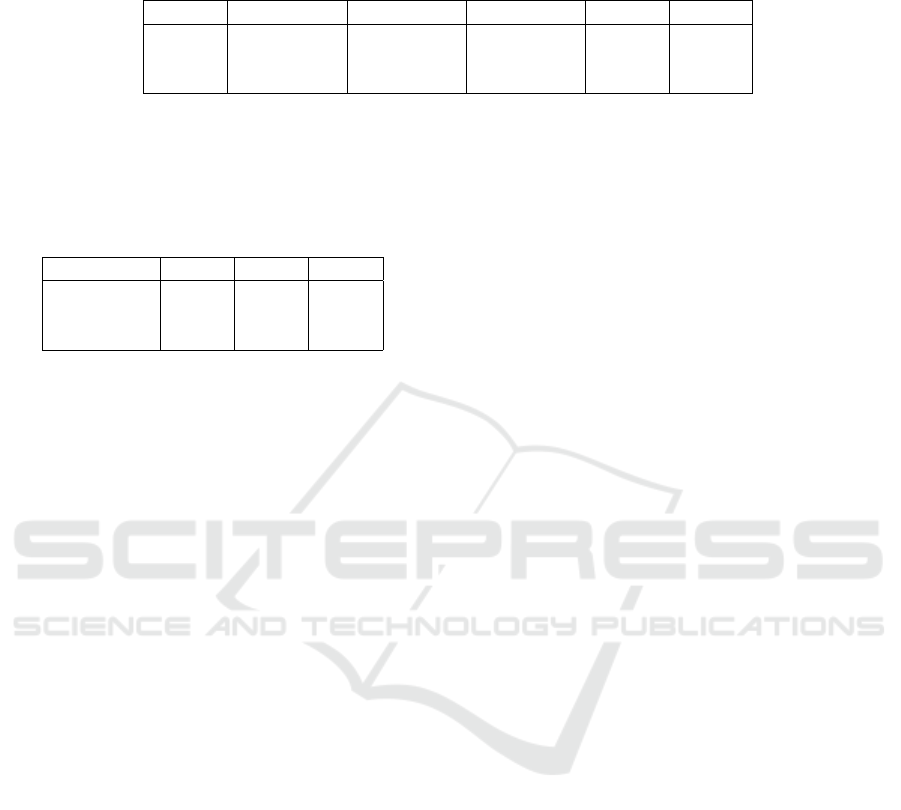
Table 1: Average accuracy (ACC), sensitivity (SEN), and specificity (SPE) of the testing set obtained by different architectures.
All performance metrics are illustrated by mean(%)±standard deviation after the implementation of five-fold cross-validation.
The last two columns report the results from the ANOVA.
Metrics LSTM 2D 3D F-value p-value
ACC 98.91±0.34 89.42±1.13 98.95±0.39 234.96 <0.01
SEN 98.81±0.56 91.19±3.79 98.91±0.26 15.97 <0.01
SPE 99.01±0.75 86.55±2.23 98.12±0.73 95.30 <0.01
Table 2: Results of the Tukey post-hoc test to compare per-
formance metrics of the LSTM, 2D, and 3D models. Each
row in the table corresponds to the p-value calculated for
the comparison between two target models in terms of ac-
curacy (ACC), sensitivity (SEN), and specificity (SPE) of
the testing set.
Models ACC SEN SPE
LSTM - 2D <0.01 <0.05 <0.01
LSTM - 3D 0.996 0.998 0.660
3D - 2D <0.01 <0.01 <0.01
are well-aligned, if not superior, with the accuracy
and sensitivity levels reported in the recent literature:
for example, LSTMs achieved an accuracy of 85.1%
and a sensitivity of 86.8% in a similar seizure predic-
tion task (Varnosfaderani et al., 2021), and 3D CNNs
have been recently shown able to reach an accuracy of
80.50% (Wang et al., 2021) and sensitivity of 85.7%
(Ozcan and Erturk, 2019) in seizure prediction.
5 CONCLUSIONS
While several studies have recently demonstrated that
deep learning models can achieve high accuracy in
seizure prediction tasks by analyzing EEG record-
ings from individual channels, our study suggests that
feeding multiple channels simultaneously, as it hap-
pens in the LSTM and 3D CNN architectures, allows
to exploit inter-channel correlations to improve fore-
casting performance. Furthermore, our results show
that deep learning methods significantly outperform
traditional machine learning approaches, supporting
the recent trend of applying neural network models to
the automated analysis of biological signals.
For future work, it would be interesting to investi-
gate channel selection techniques (Jana and Mukher-
jee, 2021) to further establish whether some channels
are more important than others in the seizure predic-
tion task. Especially for focal epilepsy, this would
likely depend on the localization of the seizure source
(e.g., temporal vs. occipital). It would thus be cru-
cial to collect EEG recordings from a larger cohort of
patients in order to make it possible to train source-
specific models and/or exploit a more heterogeneous
training set to extract generalizable features that can
be used across different patients’ profiles. A related
research direction would be to move from random-
ized cross-validation setups to more robust evaluation
schemes, such as those based on leave-one-patient-
out cross-validation (Shafiezadeh et al., 2023). How-
ever, this would also require to significantly scale-up
the available training datasets, which should allow to
discover more robust predictive features.
Last but not least, it would be important to de-
sign and implement interpretability methods that can
be used to better understand the output produced by
deep learning models. Indeed, in medical settings it
is of paramount importance to deploy explainable AI
methods to improve the reliability of automatic deci-
sion systems (Gunning et al., 2019).
REFERENCES
Abbasi, B. and Goldenholz, D. M. (2019). Machine learn-
ing applications in epilepsy. Epilepsia, 60(10):2037–
2047.
Akiba, T., Sano, S., Yanase, T., Ohta, T., and Koyama, M.
(2019). Optuna: A next-generation hyperparameter
optimization framework. In Proceedings of the 25rd
ACM SIGKDD International Conference on Knowl-
edge Discovery and Data Mining.
Allen, P., Fish, D., and Smith, S. (1992). Very high-
frequency rhythmic activity during EEG suppression
in frontal lobe epilepsy. Electroencephalography and
clinical neurophysiology, 82(2):155–159.
Arroyo, S. and Uematsu, S. (1992). High-frequency EEG
activity at the start of seizures. Journal of Clinical
Neurophysiology, 9(3):441–448.
Assi, E. B., Nguyen, D. K., Rihana, S., and Sawan, M.
(2017). Towards accurate prediction of epileptic
seizures: A review. Biomedical Signal Processing and
Control, 34:144–157.
Bostanov, V. (2004). Bci competition 2003-data sets ib and
iib: feature extraction from event-related brain poten-
tials with the continuous wavelet transform and the t-
value scalogram. IEEE Transactions on Biomedical
engineering, 51(6):1057–1061.
Calesella, F., Testolin, A., De Filippo De Grazia, M., and
Zorzi, M. (2021). A comparison of feature extraction
methods for prediction of neuropsychological scores
from functional connectivity data of stroke patients.
Brain Informatics, 8(1):1–13.
A Comparison of Recurrent and Convolutional Deep Learning Architectures for EEG Seizure Forecasting
589

Falamarzi, Y., Palizdan, N., Huang, Y. F., and Lee, T. S.
(2014). Estimating evapotranspiration from tempera-
ture and wind speed data using artificial and wavelet
neural networks (wnns). Agricultural Water Manage-
ment, 140:26–36.
Gunning, D., Stefik, M., Choi, J., Miller, T., Stumpf, S.,
and Yang, G.-Z. (2019). Xai—explainable artificial
intelligence. Science robotics, 4(37):eaay7120.
Hochreiter, S. and Schmidhuber, J. (1997). Long short-term
memory. Neural computation, 9(8):1735–1780.
Hussein, R., Lee, S., Ward, R., and McKeown, M. J.
(2021). Semi-dilated convolutional neural networks
for epileptic seizure prediction. Neural Networks,
139:212–222.
Jana, R. and Mukherjee, I. (2021). Deep learning based ef-
ficient epileptic seizure prediction with EEG channel
optimization. Biomedical Signal Processing and Con-
trol, 68:102767.
Ji, S., Xu, W., Yang, M., and Yu, K. (2012). 3d convolu-
tional neural networks for human action recognition.
IEEE transactions on pattern analysis and machine
intelligence, 35(1):221–231.
Kareem, A. and Kijewski, T. (2002). Time-frequency anal-
ysis of wind effects on structures. Journal of Wind
Engineering and Industrial Aerodynamics, 90(12-
15):1435–1452.
LeCun, Y., Bengio, Y., et al. (1995). Convolutional net-
works for images, speech, and time series. The
handbook of brain theory and neural networks,
3361(10):1995.
Liu, Y., Jain, A., Eng, C., Way, D. H., Lee, K., Bui, P.,
Kanada, K., de Oliveira Marinho, G., Gallegos, J.,
Gabriele, S., et al. (2020). A deep learning system
for differential diagnosis of skin diseases. Nature
medicine, 26(6):900–908.
Niknazar, H., Maghooli, K., and Nasrabadi, A. M. (2015).
Epileptic seizure prediction using statistical behavior
of local extrema and fuzzy logic system. international
journal of computer applications, 113(2).
Ozcan, A. R. and Erturk, S. (2019). Seizure prediction
in scalp EEG using 3d convolutional neural networks
with an image-based approach. IEEE Transactions
on Neural Systems and Rehabilitation Engineering,
27(11):2284–2293.
Ozdemir, M. A., Cura, O. K., and Akan, A. (2021). Epilep-
tic EEG classification by using time-frequency images
for deep learning. International journal of neural sys-
tems, 31(08):2150026.
Peng, Z., Chu, F., and He, Y. (2002). Vibration signal
analysis and feature extraction based on reassigned
wavelet scalogram. Journal of Sound and Vibration,
253(5):1087–1100.
San-Segundo, R., Gil-Martin, M., D’Haro-Enr
´
ıquez, L. F.,
and Pardo, J. M. (2019). Classification of epileptic
EEG recordings using signal transforms and convo-
lutional neural networks. Computers in biology and
medicine, 109:148–158.
Shafiezadeh, S., Duma, G. M., Mento, G., Danieli, A., An-
toniazzi, L., Del Popolo Cristaldi, F., Bonanni, P., and
Testolin, A. (2023). Methodological issues in evaluat-
ing machine learning models for EEG seizure predic-
tion: Good cross-validation accuracy does not guaran-
tee generalization to new patients. Applied Sciences,
13(7):4262.
Thangavel, P., Thomas, J., Peh, W. Y., Jing, J., Yuvaraj, R.,
Cash, S. S., Chaudhari, R., Karia, S., Rathakrishnan,
R., Saini, V., et al. (2021). Time–frequency decompo-
sition of scalp electroencephalograms improves deep
learning-based epilepsy diagnosis. International jour-
nal of neural systems, 31(08):2150032.
Tsiouris, K. M., Pezoulas, V. C., Zervakis, M., Konitsio-
tis, S., Koutsouris, D. D., and Fotiadis, D. I. (2018).
A long short-term memory deep learning network for
the prediction of epileptic seizures using EEG signals.
Computers in biology and medicine, 99:24–37.
T
¨
urk,
¨
O. and
¨
Ozerdem, M. S. (2019). Epilepsy detection by
using scalogram based convolutional neural network
from EEG signals. Brain sciences, 9(5):115.
Usman, S. M., Khalid, S., and Aslam, M. H. (2020). Epilep-
tic seizures prediction using deep learning techniques.
Ieee Access, 8:39998–40007.
Van Mierlo, P., Vorderw
¨
ulbecke, B. J., Staljanssens,
W., Seeck, M., and Vulli
´
emoz, S. (2020). Ictal
EEG source localization in focal epilepsy: Review
and future perspectives. Clinical Neurophysiology,
131(11):2600–2616.
Varlı, M. and Yılmaz, H. (2023). Multiple classification
of EEG signals and epileptic seizure diagnosis with
combined deep learning. Journal of Computational
Science, 67:101943.
Varnosfaderani, S. M., Rahman, R., Sarhan, N. J.,
Kuhlmann, L., Asano, E., Luat, A., and Alhawari,
M. (2021). A two-layer lstm deep learning model for
epileptic seizure prediction. In 2021 IEEE 3rd Inter-
national Conference on Artificial Intelligence Circuits
and Systems (AICAS), pages 1–4. IEEE.
Wang, Z., Yang, J., and Sawan, M. (2021). A novel multi-
scale dilated 3d cnn for epileptic seizure prediction.
pages 1–4.
World Health Organization (2023). Epilepsy, https://www.
who.int/en/news-room/fact-sheets/detail/epilepsy.
Yildiz, A., Zan, H., and Said, S. (2021). Classification and
analysis of epileptic EEG recordings using convolu-
tional neural network and class activation mapping.
Biomedical signal processing and control, 68:102720.
BIOSIGNALS 2024 - 17th International Conference on Bio-inspired Systems and Signal Processing
590
