
Teeth Localization and Lesion Segmentation in CBCT Images
Using SpatialConfiguration-Net and U-Net
Arnela Hadzic
1
, Barbara Kirnbauer
2
, Darko
ˇ
Stern
3
and Martin Urschler
1 a
1
Institute for Medical Informatics, Statistics and Documentation, Medical University of Graz, Graz, Austria
2
Department of Dental Medicine and Oral Health, Medical University of Graz, Graz, Austria
3
Institute of Computer Graphics and Vision, Graz University of Technology, Graz, Austria
Keywords:
Teeth Localization, Lesion Segmentation, SpatialConfiguration-Net, U-Net, CBCT, Class Imbalance.
Abstract:
The localization of teeth and segmentation of periapical lesions in cone-beam computed tomography (CBCT)
images are crucial tasks for clinical diagnosis and treatment planning, which are often time-consuming and re-
quire a high level of expertise. However, automating these tasks is challenging due to variations in shape, size,
and orientation of lesions, as well as similar topologies among teeth. Moreover, the small volumes occupied
by lesions in CBCT images pose a class imbalance problem that needs to be addressed. In this study, we pro-
pose a deep learning-based method utilizing two convolutional neural networks: the SpatialConfiguration-Net
(SCN) and a modified version of the U-Net. The SCN accurately predicts the coordinates of all teeth present in
an image, enabling precise cropping of teeth volumes that are then fed into the U-Net which detects lesions via
segmentation. To address class imbalance, we compare the performance of three reweighting loss functions.
After evaluation on 144 CBCT images, our method achieves a 97.3% accuracy for teeth localization, along
with a promising sensitivity and specificity of 0.97 and 0.88, respectively, for subsequent lesion detection.
1 INTRODUCTION
Cone-beam computed tomography (CBCT) is a
highly effective medical imaging technique used to
generate a 3D image of the oral and maxillofacial re-
gion, with various applications in dentistry (Khana-
gar et al., 2021; Umer and Habib, 2022). However,
the analysis and documentation of CBCT images typ-
ically require a significant amount of time and exper-
tise from professionals. While automated methods
have been proposed to localize and segment anatomi-
cal structures in general medical images, this remains
challenging for dental structures due to the presence
of similar topologies and appearances among teeth,
unclear boundaries, artifacts, or variations in shapes,
appearance and size of lesions. Moreover, in dental
images lesions occupy a significantly smaller volume
compared to the background and they only affect a
small percentage of teeth in patients, resulting in class
imbalance which poses another major challenge.
Early approaches for localizing anatomical struc-
tures in medical images were based on statistical
models of shape and appearance (Cootes et al., 1994),
which were later improved by incorporating random
forest-based machine learning models (Donner et al.,
a
https://orcid.org/0000-0001-5792-3971
2013; Unterpirker et al., 2015; Urschler et al., 2018).
Recently, deep learning-based methods have outper-
formed traditional machine learning localization ap-
proaches in terms of accuracy and efficiency. (Zhang
et al., 2017) proposed two deep convolutional neu-
ral networks (CNNs) for detecting anatomical land-
marks in brain MR volumes, where the first net-
work captures inherent relationships between local
image patches and target landmarks, while the sec-
ond network predicts landmark coordinates directly
from the input image. (Jain et al., 2014) found that
regressing heatmaps rather than coordinates improves
the overall performance of landmark detection and
also simplifies the analysis of network’s predictions.
Building on this idea, (Payer et al., 2019) proposed
the SpatialConfiguration-Net, which combines local
appearance responses with the spatial configuration
of landmarks in an end-to-end manner and achieved
state-of-the-art performance on a variety of medical
datasets. Regarding teeth detection in CBCT im-
ages, several studies that use CNNs have been pub-
lished. (Chung et al., 2020) adopted a faster R-
CNN framework to localize individual tooth regions
inside volume of interest regions, which were previ-
ously extracted and realigned using a 2D pose regres-
sion CNN. More recently, (Du et al., 2022) employed
a classification network to extract tooth regions and
290
Hadzic, A., Kirnbauer, B., Štern, D. and Urschler, M.
Teeth Localization and Lesion Segmentation in CBCT Images Using SpatialConfiguration-Net and U-Net.
DOI: 10.5220/0012305200003660
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 19th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2024) - Volume 3: VISAPP, pages
290-297
ISBN: 978-989-758-679-8; ISSN: 2184-4321
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

trained a YOLOv3 network to detect teeth bounding
boxes within these regions.
Early approaches for segmenting structures in
medical images often relied on active contours or sta-
tistical shape models (Heimann and Meinzer, 2009;
Gan et al., 2017). Their limitation of requiring pre-
defined handcrafted features was recently overcome
by deep learning-based methods. To date, several re-
search studies have been proposed for the automated
detection of periapical lesions in dental images using
deep learning. Most of these studies focus on peri-
apical or panoramic radiographs (Ekert et al., 2019;
Endres et al., 2020; Krois et al., 2021; Pauwels et al.,
2021), which exhibit lower accuracy compared to us-
ing CBCT scans (Antony et al., 2020), potentially
leading to missed or hidden lesions. To the best of our
knowledge, only a limited number of research studies
have addressed the automated detection or segmen-
tation of periapical lesions in CBCT images. (Lee
et al., 2020) employed a CNN architecture based on
the GoogLeNet Inception v3 model and trained it on
cropped 2D slices from CBCT images. They achieved
a sensitivity value of 0.94 for periapical lesion detec-
tion. (Setzer et al., 2020) used a U-Net-based archi-
tecture to segment periapical lesions in limited field-
of-view CBCT data. Their model achieved a sensi-
tivity of 0.93 and a specificity of 0.88, with an aver-
age Dice score of 0.52 for all positive examples and
0.67 for true positive examples. However, their train-
ing and testing involved 2D scans from only 20 CBCT
volumes with 61 roots. In another study, (Zheng et al.,
2020) trained an anatomically constrained Dense U-
Net using 2D slices from 20 CBCT images. They in-
corporated oral-anatomical knowledge that periapical
lesions are located near the roots of teeth and achieved
a sensitivity value of 0.84. (Orhan et al., 2020) em-
ployed two separate U-Net-like CNN architectures for
teeth localization and periapical lesion segmentation
in CBCT images. The first network localized each
tooth, and the second network used the extracted teeth
with their context to detect lesions. However, the au-
thors did not provide details about the architecture or
the training/testing procedure of their networks. They
reported detecting 142 out of 153 lesions correctly,
resulting in a sensitivity of 92.8% for lesion detec-
tion, with only one misidentified tooth. They did not
provide an evaluation of negative examples, nor did
they clarify whether 3D volumes were used through-
out their method.
Although deep learning techniques have demon-
strated potential in automating the detection of peri-
apical lesions in CBCT images, current methods of-
ten rely on training procedures based solely on 2D
slices, which may result in the loss of valuable infor-
mation. To address this concern, it is important to
incorporate 3D volumes into the training process to
effectively utilize all available data. Additionally, the
issue of class imbalance should be taken into account,
since lesions occupy only small volumes in images,
and the majority of teeth are lesion-free.
In this work, we have developed a fully automated
deep learning method for the detection of teeth and
periapical lesions in 3D CBCT images in a multi-step
process. First, we use the 3D SpatialConfiguration-
Net to perform the teeth localization, i.e., to gen-
erate a 3D coordinate of each tooth in an image.
Then, we automatically crop each tooth in the im-
age based on the generated coordinates. Finally, we
train a 3D U-Net using the previously cropped vol-
umes to segment periapical lesions. To address the
commonly encountered class imbalance problem in
medical datasets, we use and compare three differ-
ent reweighting loss functions during both the training
and testing procedures. Utilization of state-of-the-art
(SOTA) 3D network architectures for respective tasks
as well as adaptation to address class imbalance prob-
lems contribute to the reliability and high accuracy of
our method. The objective of this study is to provide
a comprehensive description of the techniques used
to obtain the results that we recently published in a
clinical journal (Kirnbauer et al., 2022), and to shed
light on the significant issues of class imbalance and
preservation of 3D volumetric information.
2 METHOD
2.1 Data
Our method was trained and tested on a dataset that
consists of 144 3D CBCT images provided by the
University Clinic for Dentistry and Oral Health Graz.
Ethical approval was granted by the Medical Univer-
sity of Graz under review board number 33-048 ex
20/21. Out of the 144 images, 16 images visualize
both jaws, while the remaining 128 images visualize
either the upper or lower jaw. The images visualize
2128 teeth, and in most images at least one periapi-
cal lesion was found. In total, approximately 10% of
the teeth in the dataset were affected by a periapical
lesion, thus leading to class imbalance.
To obtain ground truth data, we first perform a
manual localization of each tooth location, thus cre-
ating a set of 32 coordinates for each image. If a
tooth is missing, we annotate it with the coordinate
(−1,−1,−1). Second, for the ground truth segmen-
tation of periapical lesions in an image, we use the
semi-automatic Total Variation (TV) framework pro-
Teeth Localization and Lesion Segmentation in CBCT Images Using SpatialConfiguration-Net and U-Net
291

posed in (Urschler et al., 2014). Each lesion segmen-
tation was then verified by an experienced dentist us-
ing the ITK-Snap software (Yushkevich et al., 2006)
and manually adjusted if necessary.
2.2 CNN Architecture
The developed method consists of two networks (see
Figs. 1 and 2): the SpatialConfiguration-Net (Payer
et al., 2019), and a modified version of the U-Net
(Ronneberger et al., 2015). The method includes an
additional cropping step in between, a design inspired
by (Payer et al., 2020).
2.2.1 Teeth Localization
As the first step towards the detection of periapical
lesions in 3D CBCT images, we perform the teeth lo-
calization using the SpatialConfiguration-Net (SCN).
SCN is a fully convolutional neural network that con-
sists of two main components: local appearance (LA)
and spatial configuration (SC). The LA component
generates locally accurate predictions, but they may
be ambiguous. To solve this issue, the SC com-
ponent incorporates the spatial relationship between
landmarks into the network.
The network is trained to produce heatmap images
of landmarks, where each heatmap image represents
the probability of a specific landmark being present at
a particular location in the image. The predicted co-
ordinate x
′
i
of a landmark L
i
, where i ∈ {1,. . . , 32}, is
defined as the coordinate where the predicted heatmap
h
i
(x;w,b) has its highest value.
For each image in our dataset, we create a tar-
get heatmap image g by merging Gaussian heatmaps
of its ground truth landmarks. A Gaussian heatmap
g
i
(x;σ
i
) of a ground truth landmark L
i
is defined by
g
i
(x;σ
i
) = exp
−
∥
x − ˜x
i
∥
2
2
2σ
2
i
!
, (1)
where x are image coordinates and ˜x
i
is the ground
truth coordinate of the landmark L
i
. The heatmap
peak widths are determined by the standard deviation
σ and depend on the distance between the image co-
ordinates and the ground truth coordinate. Higher val-
ues are assigned to voxels that are closer to the ground
truth coordinate ˜x
i
, while the values of voxels further
away from ˜x
i
decrease gradually.
To minimize differences between the predicted
h
i
(x;w,b) and the target heatmaps g
i
(x;σ
i
) for each
landmark L
i
, we minimize the objective function
min
w,b,σ
N−1
∑
i=0
∑
x
∥
h
i
(x;w,b) − g
i
(x;σ
i
)
∥
2
2
· M(x) + T, (2)
where
T = α
∥
σ
∥
2
2
+ λ
∥
w
∥
2
2
. (3)
We calculate the distance between the predicted
heatmaps and the target heatmaps using the L
2
mea-
sure, which is multiplied with a binary mask M(x).
When the ground truth annotation at location x is
annotated as missing, the value of x in M(x) is set
to zero. This way, the network ignores the predic-
tions for missing teeth. The heatmap peak widths
σ = (σ
0
,σ
1
,...,σ
N−1
)
T
, the network weights w, and
the bias b are learnable parameters of the network.
The factor α determines how strong the heatmap peak
widths σ are being penalized, while λ determines the
impact of the L
2
norm of the weights w.
The LA component consists of four levels, where
each level includes three convolutional layers and one
average pooling layer, except for the last level where
downsampling is not performed. The SC component
consists of one level with four 7x7x7 convolutional
layers, three of which have 64 outputs and one has
32 outputs. The inputs to the first convolutional layer
are the local appearance heatmaps H
LA
generated by
the LA component, downsampled by a factor of 4.
To generate the set of spatial configuration heatmaps
H
SC
, the 32 outputs of the last convolutional layer are
upsampled to the input resolution using tricubic in-
terpolation. All convolutional layers, except the ones
generating H
LA
and H
SC
, have a LeakyReLU acti-
vation function with a negative slope of 0.1. The
weights are initialized using the He initializer (He
et al., 2015). The layer generating H
LA
has a linear
activation function, while the layer generating H
SC
has a TanH activation function. Both layers initial-
ize the biases with 0 and the weights using a Gaus-
sian distribution with a standard deviation of 0.001.
A dropout rate of 0.3 is applied after the first convo-
lution layer in each level.
As shown in Fig. 1, the LA and SC components
generate separate heatmap images, which are then
multiplied voxel-wise to generate the final output of
the SCN. Using the coordinates predicted by the SCN,
all teeth present in the images are cropped to the size
of [64, 64, 64] and fed into the U-Net for lesion seg-
mentation.
2.2.2 Lesion Segmentation
For the segmentation of periapical lesions, we use a
modified version of the U-Net. Our adaptation con-
sists of 5 levels, where each convolutional layer has
a kernel size of [3,3,3] and 16 filter outputs. In the
contracting path, we use convolutional operations fol-
lowed by downsampling through average pooling. In
the expansive path, each upsampling layer performs
trilinear interpolation followed by two convolutional
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
292

Input
H
LA
H
LA
H
SC
⊙
H
Local Appearance
Spatial Configuration
Figure 1: Teeth localization using SpatialConfiguration-Net. The local appearance component generates a set of local appear-
ance heatmaps denoted as H
LA
, while the spatial configuration component produces a set of spatial configuration heatmaps
H
SC
. The final heatmap images H for teeth landmarks are obtained by performing voxel-wise multiplication of H
LA
and H
SC
.
The arrows represent different operations: green arrow – convolution, blue arrow – downsampling, yellow arrow – upsam-
pling, and orange arrow – voxel-wise addition.
operations. After each convolutional layer we apply a
dropout of 0.3, and use ’same’ padding to maintain the
same input and output size. All layers have a ReLU
activation function except the last one, where no acti-
vation function is used in order to obtain logits instead
of probabilities as the network’s output. Same as for
the SCN, the weights in the last layer are initialized
with the He initializer. In all other layers, the initial
weights are sampled from a truncated normal distri-
bution with a standard deviation of 0.001. The last
layer consists of a single output, generating an image
with predicted intensity values. The network’s output
is an image of size [64, 64,64], with voxel intensities
in the range (−∞,+∞). Finally, a threshold of 0 was
applied to the output in order to produce the predicted
binary segmentation map, where all voxel values be-
low the threshold are considered background, while
non-negative values represent a detected lesion.
To address the class imbalance problem, we com-
pare three objective functions, Focal Loss FL (Lin
et al., 2017), Focal Tversky Loss FTL (Abraham and
Khan, 2019), and Combo Loss CL (Taghanaki et al.,
2019) based on Dice Similarity Coefficient (DSC):
FL(p
t
) = −α
t
(1 − p
t
)
γ
log(p
t
), (4)
FTL(P,G) = (1 − TI(P, G))
1
γ
, (5)
and
CL(y, p) = δ · L
BBCE
(y, p) − (1 − δ) · DSC(y, p). (6)
In the above equations, Tversky Index (TI) and Bal-
anced Binary Cross-Entropy (BBCE) are defined by
TI(P,G) =
|PG| + ε
|PG| + β|P\G| + (1 − β)|G\P| + ε
(7)
and
L
BBCE
(y, p) = −α· y log(p) − (1 − α) · (1 − y)log(1 − p).
(8)
The parameters α and β in the above equations are
used to weight positive and negative examples. α can
be set as the inverse class frequency or considered as a
hyperparameter, while values of β larger than 0.5 give
more significance to false negative examples. To en-
hance the focus on misclassified predictions, γ should
be larger than 0 in the FL function and larger than 1
in the FTL function. The parameter δ regulates the
contribution of the BBCE to the CL function.
2.3 Data Augmentation
To prepare images for training the SCN for local-
ization, we resize all images from the original size
of [501, 501, 501] to a new size of [64,64,32], while
maintaining a fixed aspect ratio. As we do not need
highly precise locations of teeth for the lesion seg-
mentation task, the SCN can be trained on downsam-
pled images. Image intensities are then scaled to the
range [−1,1]. Additionally, images undergo transla-
tion, rotation, and scaling using random factors sam-
pled from uniform distributions ([−10, 10] for trans-
lation, [−0.1,0.1] for rotation, [0.9,1.1] for scaling).
Before training our modified U-Net for the lesion
segmentation task, original high-resolution images
are cropped along with their corresponding ground
truth segmentation maps for each tooth. This process
generates 32 cropped images from a single original
image, where the center of each cropped image corre-
sponds to the center coordinate of a particular tooth.
Teeth Localization and Lesion Segmentation in CBCT Images Using SpatialConfiguration-Net and U-Net
293

Input image
Output image
Predicted seg. map
Ground truth map
t = 0
U-Net
Figure 2: Periapical lesion segmentation using a modified U-Net architecture. A cropped image of a tooth along with its
corresponding ground truth map is used as input to the U-Net. The network is trained to output an image of intensity values,
where each value lies in the range (−∞, +∞). A threshold t is applied to the predicted intensity image in order to generate a
final binary segmentation map.
Each cropped image has dimensions of [64,64,64]
and a spacing of [0.4,0.4,0.4]. Since periapical os-
teolytic lesions are always located in the area of the
root tips, we translate cropped images by fixed fac-
tors to ensure visibility of the entire tooth root within
a cropped image. The translation factors of [0, 1, −9]
are applied to the cropped images with a tooth in
the upper jaw. The cropped images with a tooth in
the lower jaw are translated by factors [0,3,−9] and
rotated by 180 degrees. Moreover, all cropped im-
ages undergo random translation within the range of
[−1,−1], random rotation, and random scaling with-
ing the range of [−0.2, 0.2]. As a post-processing
step, we normalize cropped images to the range [0, 1]
and perform shift scale intensity transformation using
a random factor of 0.6. We perform label smoothing
by applying a Gaussian kernel with a standard devia-
tion of 0.1 to the ground truth segmentation maps.
2.4 Implementation Details
An Intel(R) CPU 920 with an NVIDIA GeForce GTX
TITAN X was used for the training and testing of the
model. The CNNs were running on Ubuntu 20.04
operating system with Python 3.7 and TensorFlow
1.15.0. To evaluate the performance of our model,
we used 4-fold cross validation (cv). Total training
time of the SCN for one cv fold took about 20 hours,
while one fold for training the U-Net took about 17
hours. The teeth localization network took around 15
seconds for inference on a CBCT volume, while the
segmentation network, including cropping, took ap-
proximately 2 to 3 minutes.
To minimize the loss function during training of
SCN, we use the Nesterov Accelerated Gradient with
a learning rate of 10e − 7 and momentum value of
0.99. We set the number of iterations to 15,000 as
we did not observe any substantial improvement after
that. Remaining hyper-parameters batch size, weight
decay, and sigma regularization term are determined
empirically and set to 1, 5e−5, and 100, respectively.
The U-Net is trained for 113,000 iterations using a
batch size of 8. As mentioned earlier, we address the
class imbalance issue by utilizing Focal Loss, Focal
Tversky Loss, and Combo Loss. We set the param-
eters of these loss functions as follows: Within our
dataset, positive voxels in each cropped image occupy
less than 10% of the volume. Therefore, we set the
weighting factors α and β to 0.9 in the corresponding
loss functions. The parameter γ controls the down-
weighting of easy examples. We set γ to 2 in the Focal
Loss and the Focal Tversky Loss function. In the case
of the Combo Loss function, we set the parameter δ
to 0.5, which determines the contribution of the Bal-
anced Binary Cross-Entropy. Furthermore, in order
to minimize the loss functions, we employ the Adam
optimizer with a learning rate of 1e − 4.
Both networks are trained and tested using the 4-
fold cross validation technique. For teeth localization,
our dataset of 144 3D CBCT images is divided into
four cv folds, each containing 36 images. For lesion
segmentation, we split a total of 2128 cropped images
into four cv folds. Since only 206 teeth in our dataset
are affected by a lesion, we distribute them uniformly
over all cv folds, resulting in 10% of teeth with lesions
and 90% teeth without lesions per fold.
2.5 Metrics
To evaluate the teeth localization performance, we
use the point-to-point error (PE) and accuracy met-
rics. For a landmark i, the point-to-point error PE
i
represents the Euclidean distance between the ground
truth landmark ˜x
i
and the predicted landmark x
′
i
. Ac-
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
294
curacy is defined as the percentage of correctly identi-
fied landmarks over all predicted landmarks (i.e., av-
erage precision). A predicted landmark x
′
i
is identified
correctly if the closest ground truth landmark is the
correct one and the Euclidean distance to the ground
truth landmark is less than a specified radius r.
To evaluate the lesion detection performance, we
use the metrics sensitivity (TP/(TP+FN)) and speci-
ficity (TN/(TN + FP)). Each predicted segmentation
is evaluated based on the Dice score, an overlap mea-
sure between the ground truth segmentation map X
and the predicted segmentation map Y , defined by
DSC = 2 ·
|X ∩Y |
|X| + |Y |
. (9)
Sensitivity represents the network’s ability to cor-
rectly detect teeth with lesions, whereas specificity
represents its ability to correctly detect teeth without
lesions. A predicted segmentation map for a tooth
with a lesion is classified as a True Positive (TP) if
the Dice score between the ground truth and predicted
segmentation map is larger than 0. Otherwise, the pre-
dicted segmentation map is classified as a False Neg-
ative (FN). A predicted segmentation map for a tooth
without a lesion is classified as a True Negative (TN)
if all voxels in the predicted segmentation map are 0.
Otherwise, the predicted segmentation map is classi-
fied as a False Positive (FP) prediction.
2.6 Delaunay Triangulation
For the training of the U-Net, each original image
is cropped to the size of [64,64,64] for each tooth,
where the center coordinate of a cropped image corre-
sponds to the center coordinate of the particular tooth
in the original image. Since teeth can have different
shapes, sizes, and orientations, the resulting cropped
images may contain multiple teeth, which compli-
cates automated tooth-based evaluation. To address
this issue, we use the definitions of the convex hull
and the Delaunay triangulation.
First, we annotate a cuboid around each tooth in
the original image using Planmeca Romexis
®
soft-
ware. This annotation ensures that each cuboid en-
closes only the area of a single tooth. The software
then automatically generates a set of 3D coordinates
representing the cuboid. We use these coordinates as
input for the ’Convex-Hull’ and ’Delaunay’ functions
from the SciPy 1.6.2 library. By using the ’Delau-
nay.find simplex’ function, we obtain the indices of
all the simplices that contain all cuboid voxels. Using
these indices, we are able to generate a segmentation
map for an annotated cuboid. Finally, during the eval-
uation of a specific tooth, we only consider the voxels
that belong to its corresponding annotated cuboid.
3 RESULTS AND DISCUSSION
The results of the teeth localization and lesion seg-
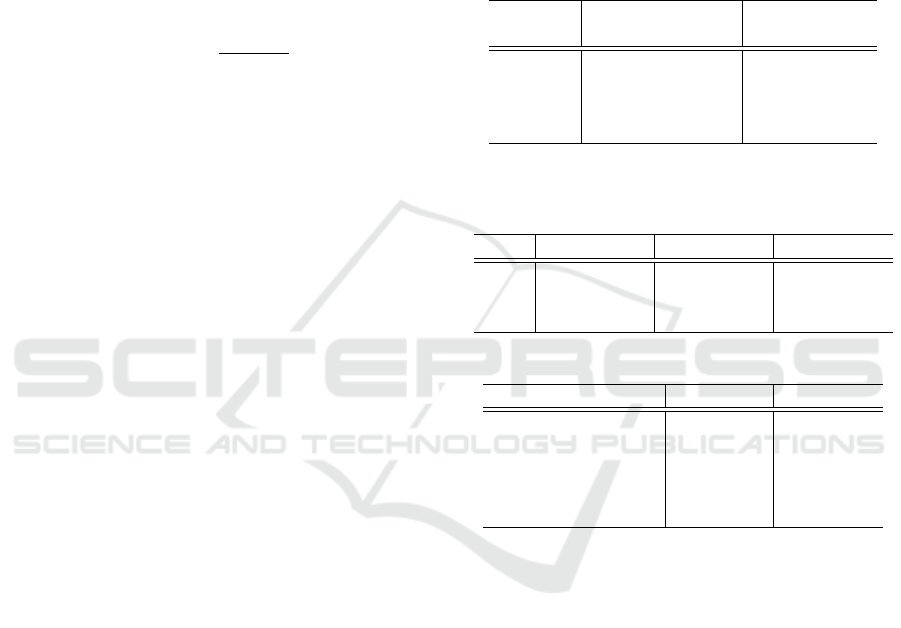
mentation tasks are shown in Table 1 and Table 2, re-
spectively. Moreover, we provide a comparison of the
results with SOTA methods for periapical lesion de-
tection in Table 3.
Table 1: Teeth localization results for 4-fold cross valida-
tion, showing point-to-point error with standard deviation,
as well as accuracy of the model for different radii r.
Radius r
PE in mm
Accuracy (%)
Mean ± SD
2 mm
72.6
3 mm 1.74 ± 1.44 89.1
4 mm 94.7
6 mm 97.3
Table 2: Lesion detection and segmentation results for 4-
fold cross validation, shown as sensitivity, specificity, and
Dice score obtained using different loss functions.
Loss Sensitivity Specificity DICE
FL 0.97 ± 0.03 0.88 ± 0.04 0.67 ± 0.03
FTL 0.92 ± 0.05 0.89 ± 0.08 0.70 ± 0.04
CL 0.85 ± 0.04 0.94 ± 0.02 0.70 ± 0.04
Table 3: Comparison with SOTA methods.
Method Sensitivity Specificity
(Lee et al., 2020) 0.94 -
(Setzer et al., 2020) 0.93 0.88
(Zheng et al., 2020) 0.84 -
(Orhan et al., 2020) 0.93 -
Ours 0.97 0.88
We evaluated the performance of the SCN on 4
folds and achieved an average accuracy of 97.3% in
teeth detection, with a mean point-to-point error of
1.74 ± 1.44 mm. In other words, for 97.3% of the
predicted teeth, the closest ground truth landmark was
the correct one and the distance between the predicted
and ground truth landmarks was less than 6 mm. De-
creasing the radius to 4, 3, and 2 mm resulted in av-
erage accuracy values of 94.7%, 89.1%, and 72.6%,
respectively. By observing individual predictions, we
could notice that the most incorrect predictions oc-
curred in images affected by artifacts or images with
misaligned teeth. This can be attributed to these cases
being rare in our dataset and thus deviating from the
learned data distribution. In future work, this could
be addressed by combining generative models with
CNNs to incorporate global shape/landmark configu-
rations into the training process. This way, the detec-
tion of out-of-distribution data could be improved.
Teeth Localization and Lesion Segmentation in CBCT Images Using SpatialConfiguration-Net and U-Net
295

The U-Net was trained using the Focal Loss (FL),
Focal Tversky Loss (FTL), and Combo Loss (CL)
functions. By adjusting the parameters of these loss
functions, we were able to reweight the hard and easy
class examples. Each setup was evaluated on 4 folds,
with uniformly distributed teeth with lesions. The
highest lesion detection rate of 0.97 was achieved
using FL, whereas FTL and CL achieved the detec-
tion rates of 0.92 and 0.88, respectively. The high-
est specificity value of 0.94 was achieved using CL,
while the values of 0.88 and 0.89 were achieved us-
ing FL and FTL, respectively. The highest Dice score
of 0.70 was achieved using the FTL and CL func-
tions. The setup trained with FL achieved a slightly
smaller value of 0.66 for the Dice score. By observ-
ing the predictions for individual lesions, we noticed
a larger number of false negative predicted voxels and
consequently lower Dice scores in images with very
small or very large lesions. Small differences between
predicted and ground truth segmentation maps have a
significant impact on the Dice score for images with
very small lesions, while a significant predicted por-
tion of a lesion can result in a low Dice score for
images with very large lesions. Since false negative
predictions are less tolerable than false positive pre-
dictions in the lesion detection task, we conclude that
the best performance of our method was achieved us-
ing the FL function with a sensitivity value of 0.97
and a specificity value of 0.88. In future work, we
plan to explore the use of stronger anatomical con-
straints via generative models to improve the crucial
teeth localization step.
When comparing our method with other SOTA
methods, our approach achieved the highest sensitiv-
ity value and the same specificity value as (Setzer
et al., 2020). However, it is important to note that
a direct comparison is not feasible due to the use of
different datasets in the evaluation of these methods.
Furthermore, all SOTA methods, except (Orhan et al.,
2020), utilized 2D slices rather than 3D volumes dur-
ing the training and testing procedures. Addition-
ally, the studies conducted in (Setzer et al., 2020) and
(Zheng et al., 2020) were limited to small respective
datasets. When considering the negative class, which
refers to teeth without periapical lesions, only (Setzer
et al., 2020) reported a specificity value. However,
their dataset is highly selective as it consists of only
20 CBCT volumes with limited field-of-view. More-
over, among these volumes, there are 29 roots with le-
sions and 32 roots without lesions, thus creating a bal-
anced distribution of roots with and without lesions,
which does not reflect clinical practice.
4 CONCLUSION
In this paper, we have presented a fully automated
two-step method for detecting periapical lesions in
CBCT images. In the first step, we utilize the 3D
SpatialConfiguration-Net (SCN) for teeth localiza-
tion. By using the teeth coordinates generated by the
SCN, we extract relevant subregions from the original
images, and use them to train the 3D U-Net for lesion
segmentation in the second step. In contrast to other
SOTA methods, our method incorporates 3D volumes
throughout all stages, ensuring no loss of valuable in-
formation. Additionally, to the best of our knowledge,
we are the first to address the class imbalance issue
associated with automatic periapical lesion detection,
which is commonly observed in clinical data. De-
spite the challenges posed by dental CBCT images,
our method achieved promising results in localizing
teeth and detecting periapical lesions in CBCT data.
REFERENCES
Abraham, N. and Khan, N. M. (2019). A novel focal
Tversky loss function with improved Attention U-
Net for lesion segmentation. In 2019 IEEE 16th In-
ternational Symposium on Biomedical Imaging (ISBI
2019), pages 683–687. IEEE.
Antony, D. P., Thomas, T., and Nivedhitha, M. (2020). Two-
dimensional periapical, panoramic radiography versus
three-dimensional cone-beam computed tomography
in the detection of periapical lesion after endodontic
treatment: A systematic review. Cureus, 12(4).
Chung, M., Lee, M., Hong, J., Park, S., Lee, J., Lee, J.,
Yang, I.-H., Lee, J., and Shin, Y.-G. (2020). Pose-
aware instance segmentation framework from cone
beam ct images for tooth segmentation. Computers
in Biology and Medicine, 120:103720.
Cootes, T. F., Hill, A., Taylor, C. J., and Haslam, J. (1994).
Use of active shape models for locating structures
in medical images. Image and Vision Computing,
12(6):355–365.
Donner, R., Menze, B. H., Bischof, H., and Langs, G.
(2013). Global localization of 3d anatomical struc-
tures by pre-filtered hough forests and discrete opti-
mization. Medical Image Analysis, 17(8):1304–1314.
Du, M., Wu, X., Ye, Y., Fang, S., Zhang, H., and Chen,
M. (2022). A combined approach for accurate and
accelerated teeth detection on cone beam ct images.
Diagnostics, 12(7):1679.
Ekert, T., Krois, J., Meinhold, L., Elhennawy, K., Emara,
R., Golla, T., and Schwendicke, F. (2019). Deep learn-
ing for the radiographic detection of apical lesions.
Journal of Endodontics, 45(7):917–922.
Endres, M. G., Hillen, F., Salloumis, M., Sedaghat, A. R.,
Niehues, S. M., Quatela, O., Hanken, H., Smeets, R.,
Beck-Broichsitter, B., Rendenbach, C., et al. (2020).
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
296

Development of a deep learning algorithm for peri-
apical disease detection in dental radiographs. Diag-
nostics, 10(6):430.
Gan, Y., Xia, Z., Xiong, J., Li, G., and Zhao, Q. (2017).
Tooth and Alveolar Bone Segmentation From Den-
tal Computed Tomography Images. IEEE Journal of
Biomedical and Health Informatics, 22(1):196–204.
He, K., Zhang, X., Ren, S., and Sun, J. (2015). Delving
deep into rectifiers: Surpassing human-level perfor-
mance on ImageNet classification. In Proceedings of
the IEEE International Conference on Computer Vi-
sion, pages 1026–1034.
Heimann, T. and Meinzer, H.-P. (2009). Statistical shape
models for 3D medical image segmentation: A review.
Medical Image Analysis, 13(4):543–563.
Jain, A., Tompson, J., Andriluka, M., Taylor, G. W., and
Bregler, C. (2014). Learning human pose estima-
tion features with convolutional networks. In 2nd In-
ternational Conference on Learning Representations
(ICLR), pages 1–11.
Khanagar, S. B., Al-Ehaideb, A., Vishwanathaiah, S., Ma-
ganur, P. C., Patil, S., Naik, S., Baeshen, H. A., and
Sarode, S. S. (2021). Scope and performance of ar-
tificial intelligence technology in orthodontic diagno-
sis, treatment planning, and clinical decision-making-
a systematic review. Journal of Dental Sciences,
16(1):482–492.
Kirnbauer, B., Hadzic, A., Jakse, N., Bischof, H., and Stern,
D. (2022). Automatic detection of periapical oste-
olytic lesions on cone-beam computed tomography
using deep convolutional neuronal networks. Journal
of Endodontics, 48(11):1434–1440.
Krois, J., Garcia Cantu, A., Chaurasia, A., Patil, R., Chaud-
hari, P. K., Gaudin, R., Gehrung, S., and Schwen-
dicke, F. (2021). Generalizability of deep learning
models for dental image analysis. Scientific Reports,
11(1):6102.
Lee, J.-H., Kim, D.-H., and Jeong, S.-N. (2020). Diagno-
sis of cystic lesions using panoramic and cone beam
computed tomographic images based on deep learn-
ing neural network. Oral Diseases, 26(1):152–158.
Lin, T., Goyal, P., Girshick, R. B., He, K., and Doll
´
ar, P.
(2017). Focal loss for dense object detection. In IEEE
International Conference on Computer Vision, pages
2999–3007. IEEE.
Orhan, K., Bayrakdar, I., Ezhov, M., Kravtsov, A., and
¨
Ozy
¨
urek, T. (2020). Evaluation of artificial intelli-
gence for detecting periapical pathosis on cone-beam
computed tomography scans. International Endodon-
tic Journal, 53(5):680–689.
Pauwels, R., Brasil, D. M., Yamasaki, M. C., Jacobs, R.,
Bosmans, H., Freitas, D. Q., and Haiter-Neto, F.
(2021). Artificial intelligence for detection of periapi-
cal lesions on intraoral radiographs: Comparison be-
tween convolutional neural networks and human ob-
servers. Oral Surgery, Oral Medicine, Oral Pathology
and Oral Radiology, 131(5):610–616.
Payer, C., Stern, D., Bischof, H., and Urschler, M. (2019).
Integrating spatial configuration into heatmap regres-
sion based CNNs for landmark localization. Medical
Image Analysis, 54:207–219.
Payer, C., Stern, D., Bischof, H., and Urschler, M. (2020).
Coarse to fine vertebrae localization and segmentation
with SpatialConfiguration-Net and U-Net. In VISI-
GRAPP (5: VISAPP), pages 124–133.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-
net: Convolutional networks for biomedical im-
age segmentation. In Medical Image Computing
and Computer-Assisted Intervention – MICCAI 2015,
pages 234–241. Springer International Publishing.
Setzer, F. C., Shi, K. J., Zhang, Z., Yan, H., Yoon, H., Mup-
parapu, M., and Li, J. (2020). Artificial intelligence
for the computer-aided detection of periapical lesions
in cone-beam computed tomographic images. Journal
of Endodontics, 46(7):987–993.
Taghanaki, S. A., Zheng, Y., Zhou, S. K., Georgescu, B.,
Sharma, P., Xu, D., Comaniciu, D., and Hamarneh, G.
(2019). Combo loss: Handling input and output im-
balance in multi-organ segmentation. Computerized
Medical Imaging and Graphics, 75:24–33.
Umer, F. and Habib, S. (2022). Critical analysis of artificial
intelligence in endodontics: a scoping review. Journal
of Endodontics, 48(2):152–160.
Unterpirker, W., Ebner, T.,
ˇ
Stern, D., and Urschler, M.
(2015). Automatic third molar localization from 3D
MRI using random regression forests. In 19th Interna-
tional Conference on Medical Image Understanding
and Analysis (MIUA), pages 195–200, Lincoln, UK.
Urschler, M., Ebner, T., and
ˇ
Stern, D. (2018). Integrating
geometric configuration and appearance information
into a unified framework for anatomical landmark lo-
calization. Medical Image Analysis, 43:23–36.
Urschler, M., Leitinger, G., and Pock, T. (2014). Interac-
tive 2D/3D image denoising and segmentation tool for
medical applications. In Proceedings MICCAI Work-
shop Interactive Medical Image Computation (IMIC).
Yushkevich, P. A., Piven, J., Cody Hazlett, H., Gim-
pel Smith, R., Ho, S., Gee, J. C., and Gerig, G.
(2006). User-guided 3D active contour segmentation
of anatomical structures: Significantly improved effi-
ciency and reliability. Neuroimage, 31(3):1116–1128.
Zhang, J., Liu, M., and Shen, D. (2017). Detecting
anatomical landmarks from limited medical imaging
data using two-stage task-oriented deep neural net-
works. IEEE Transactions on Image Processing,
26(10):4753–4764.
Zheng, Z., Yan, H., Setzer, F. C., Shi, K. J., Mupparapu,
M., and Li, J. (2020). Anatomically constrained deep
learning for automating dental cbct segmentation and
lesion detection. IEEE Transactions on Automation
Science and Engineering, 18(2):603–614.
Teeth Localization and Lesion Segmentation in CBCT Images Using SpatialConfiguration-Net and U-Net
297
