
Visualizing, Analyzing and Constructing L-System from Arborized 3D
Model Using a Web Application
Nick van Nielen, Fons Verbeek
a
and Lu Cao
b
Leiden Institute of Advanced Computer Science, Leiden University, Leiden, The Netherlands
Keywords:
L-System, Arborized 3D Biological Model, Web Application.
Abstract:
In biology, arborized structures are well represented and typically complex for visualization and analysis.
In order to have a profound understanding of the topology of arborized 3D biological model, higher level
abstraction is needed. We aim at constructing an abstraction of arborized 3D biological model to an L-system
that provides a generalized formalization in a grammar to represent complex structures. The focus of this paper
is to combine 3D visualization, analysis and L-system abstraction into a single web application. We designed a
front-end user interface and a back-end. In the front-end, we used A-Frame and defined algorithms to generate
and visualize L-systems. In the back-end, we utilized the Vascular Modelling Toolkit’s (VMTK) centerline
analysis methods to extract important features from the arborized 3D models, which can be applied to L-system
generation. In addition, two 3D biological models: lactiferous duct and artery are used as two case studies to
verify the functionality of this web application. In conclusion, our web application is able to visualize, analyse
and create L-system abstractions of arborized 3D models. This in turn provides workflow-improving benefits,
easy accessibility and extensibility.
1 INTRODUCTION
Analysis and visualization of 3D models are unequiv-
ocally important in the biological studies. They have
been used to study physiological aspects, such as sim-
ulating the flow and particle transport of airways (van
Ertbruggen et al., 2005). This 3D model simulation
made it possible to obtain particle deposition patterns
and flow characteristics that could not be obtained
experimentally. 3D models have also been used to
analyse the complex anatomy of the canal network
in cortical bone (Cooper et al., 2003) to get a better
understanding of the mechanical properties. Further-
more, 3D models are used to analyze the phenotypical
changes of the lactiferous duct of newborn mice under
the exposure of endocrine disruptors (Cao, 2014). In
particular, these articles discuss the usage of 3D mod-
els with a tree-like branching structure (or arborized
3D models). These types of structures can be found
in plants, but also in animals, such as lactiferous duct
or vascular system. Typically, arborized structures are
complex due to their high branching order and visi-
bly indistinguishable individual branches. To realize
a
https://orcid.org/0000-0003-2445-8158
b
https://orcid.org/0000-0002-1847-068X
appropriate visualization and interpretation of these
complex structures, model abstraction with minimal
loss of features is needed. Theoretical computer sci-
ence methods can be used to extract important features
and represent these features in an abstracted model. A
well-known system that preserves most of the anatom-
ical features of arborized structures is Lindenmayer
systems (L-systems) (Prusinkiewicz and Lindenmayer,
1990). We will use L-systems in our research to de-
scribe and build abstractions of arborized 3D models.
Many different applications already exist to visual-
ize and analyse (arborized) biological 3D models. A
popular platform is 3D Slicer - a platform for medical
imaging which provides image registration, interac-
tive visualization, model-based analysis, and more
advanced functionality (Kikinis et al., 2014). This
can be used together with tools such as the Vascu-
lar Modelling Toolkit (VMTK) to analyse vascular-
like or arborized structures (Izzo et al., 2018). When
it comes to the abstraction of models, specifically
for generating L-systems, L-py can be used for com-
plex grammar (Boudon et al., 2010). More general
3D model visualization, processing and editing tools
can also be found, for example Blender (Foundation,
1998) and MeshLab (Cignoni et al., 2008). Mesh-
LabJS (of ISTI CNR, 2016) is a web-based version of
van Nielen, N., Verbeek, F. and Cao, L.
Visualizing, Analyzing and Constructing L-System from Arborized 3D Model Using a Web Application.
DOI: 10.5220/0012272100003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 1, pages 257-264
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
257

MeshLab that uses the JavaScript library Three.js (C,
2010) as a rendering engine. (Sawicki and Chaber,
2013) has shown that web applications can be a well-
structured and convenient way of visualizing 3D mod-
els. While having clear limitations in handling com-
plex models due to computational and spatial com-
plexities, web applications can improve accessibility
and portability from different devices and browsers.
Furthermore, web applications do not have to be down-
loaded or maintained by the end-user and the 3D mod-
els can be stored on and loaded from a database, im-
proving the ease of use and allowing for the possibility
to share models between multiple users.
Both system-based and web-based applications can
realize parts of the process needed to visualize, anal-
yse and create abstractions of arborized 3D models.
However, not many applications exist that combine
these parts which are typically related, especially in a
web-based format. To the best of our knowledge, there
does not seem to exist such a web-based application
that is capable of visualizing, analyzing and creating
L-system abstractions of arborized 3D models.
In this paper, A-frame was chosen to construct
the web-based application because of its rendering
performance, cross-compatibility and HTML Entity-
Component system. A-frame is a JavaScript web
framework for creating (complex) 3D scenes with ad-
ditional VR functionality. It is built on top of the
rendering engine Three.js (Mozilla, 2015). Concern-
ing L-system generation, the JavaScript library lin-
denmayer.js was selected to interpret and generate L-
system grammar in an object-oriented fashion (Brewe,
2016). In addition, the waterfall development method
was adopted to ensure good quality, performance, pro-
gramming and design (Balaji and Murugaiyan, 2012).
2 METHOD
To visualize and analyse 3D models as well as generate
L-systems, we need to design the desired functional-
ity. We will apply design patterns, discuss the design
choices and sketch the functionality of our web ap-
plication using A-Frame in section 2.1. Furthermore,
in section 2.2 we discuss how the model analysis is
implemented with VMTK, python and the user inter-
face of the web application. Lastly, we reason how we
combine Three.js and Lindenmayer.js to generate and
visualize L-systems in section 2.3.
2.1 Design
For the design of web-based applications, a modelling
method is needed to cover all the requirements. This
can help visualize and document the components and
corresponding functionality. UML diagrams have been
shown to be a powerful modelling method for covering
all required components of web applications (Koch
and Kraus, 2002). The UML diagrams of the front-
end and back-end layout of our web application are
available in https://surfdrive.surf.nl/files/index.php/s/
8weiJINAJTbplVs.
2.1.1 Front-End and Back-End
For 3D model visualization and manipulation, a
front-end is needed for the user input, display and
storing between static web page states. All these
functionalities are available on a single page called
index.html
. Furthermore,
index.html
is the view
node of the Model-View-Controller (MVC) program-
ming paradigm (Krasner et al., 1988), which is used
to display the user interface and is a part of the inter-
face component. Besides the view node, the states be-
tween static pages are stored in the cookie object data
structure altered by the model node in
Settings.js
and updated in the view node. Modifications to
this data structure are made by the controller node
UserInterface.js
, which reacts to the user input in
the view. Some user input does not change the model
and is directly updated in the view by the controller.
For the 3D model rendering and visualization, two
components are used which contain A-Frame with
Three.js rendering. The first component is the main A-
Frame. In this component, the main object or L-system
view is stored and displayed using the A-Frame web
framework inside
aframe.html
. The second compo-
nent is the preview A-Frames. As the name suggests,
this is where all the preview nodes of the L-system,
model and A-Frame settings are shown. The L-system
generation as well as the 3D model loading from I/O
are initiated by the controller using the preview event
handler.
Tasks in the user interface, that require compu-
tations on the server or I/O operations, are realized
in back-end. The back-end uses python to run the
development server
devServer.py
using flask, a mi-
cro web application framework for simple scalabil-
ity (Grinberg, 2018). For the deployment server, Gu-
nicorn (Gunicorn-Python, 2017) and Nginx (Reese,
2008) are used with flask as an HTTP interface and
web server with multi-threading.
2.2 Model Analysis
In the procedure of model analysis, a simplification of
the arborized 3D model is needed while preserving the
important topological features. The method of center-
line extraction (Verbeek and Cao, 2020) was applied
BIOIMAGING 2024 - 11th International Conference on Bioimaging
258

to preserve a one-dimensional representation of the
original skeletal structure in 3D. Many types of center-
line extraction methods were considered, but the 3D
geometric analysis tool VMTK(Izzo et al., 2018) was
chosen. Using the well-defined back-end structure,
VMTK tool that runs on python are deployed in the
request handler node. During the analysis, endpoints
on the model are selected in the front-end view node.
The endpoints and 3D model name are then send with
an analysis request to the request handler by the con-
troller node. In the request handler, three steps take
place: verification, instance creation and the analysis.
For verification, the request parameters are verified
by checking the 3D model filename and type. If ei-
ther the filename or file type are incorrect, the model
cannot be found in the objects folder and no analysis
will be possible. Thus, an error will occur. For the
second step, an analysis instance is created on a new
thread to prevent request blocking and fatal errors from
occurring on the development or deployment thread.
The analysis process and its structure can be explained
by the diagram in Figure 1. Endpoints and the 3D
model are used to compute the model centerline. Sub-
sequently, it is used to calculate the centerline mesh
points. For the final data extractions, the centerline
is used to calculate branch length, tortuously, torsion,
curvature and bifurcation angles.
Figure 1: Diagram of the analysis process of an arborized
3D model.
2.3 L-System
In section 2.1, the L-system algorithm node was intro-
duced in the preview A-Frame of the front-end design.
This section discusses the details of the algorithm, how
L-system models are generated and how parameters
extracted in the analysis are interpreted by the algo-
rithm.
2.3.1 L-System String Generation Algorithm
To understand how Algorithm 1 works, a formal defi-
nition of the L-system and a set of L-system variables
are needed. The L-system can be defined by the triplet
G = (V, ω, P). Here:
•
V consists of the variables I, B and a constant
D. I is a branch segment, B is a branch segment
candidate. D is a dead branch, which is a branch
segment candidate that will not expand any further.
• ω
is the axiom. For example, the axiom could be
III[/&B][/&B]. The number of I variables can be
chosen.
•
P is the set of production rules, these are defined
within the algorithm. For each variable, a new set
of symbols and variables is returned.
Variables I and B share the following parameters:
•
current iteration
∈ N
≥0
keeps track of the current
iteration in the algorithm.
•
centerline index
∈ Z
determines the centerline,
with a unique centerline ID.
Parameters unique to I are:
• Radius ∈ R
≥0
of the cylinder geometry.
• Length ∈ R
≥0
of the cylinder geometry.
Parameters unique to B are:
• Lifetime ∈ Z
≥0
of the variable.
•
Maximum centerline length
∈ N
≥0
determines the
maximum lifetime of both I and B. If the centerline
length exceeds this number, B will turn into a dead
branch D.
Furthermore, L-system parameters, italicized in
Algorithm 1, are defined as follows:
•
stemLength
∈ N
≥0
defines the number of branch
segments in the stem. This determines the number
of I variables in the axiom.
•
radialIncrement
∈ R
determines how much the
thickness of each I grows with each iteration.
•
upwardsAngle, upwardsAngleSpread are the mean
and the standard deviation of the normal distri-
bution
∈ [−180, 180]
of upwards growing branch
segments I.
•
lineLength, lineLengthStd
∈ R
≥0
are part of the
cylinder geometry length distribution of I.
•
nb axes prob is the probability a bifurcation will
occur each time a new segment is built.
•
iterations, iterationsStd
∈ N
≥0
sets the maximum
centerline length of B.
•
bifurcationAngle, bifurcationAngleSpread are the
mean and the standard deviation of the bifurcation
angle with a normal distribution ∈ [−180, 180].
•
maxOrder
∈ N
≥0
determines how spread out the
branches will be from the stem. A higher max-
Order will ensure a longer lifetime for B variables
at lower branch orders.
Visualizing, Analyzing and Constructing L-System from Arborized 3D Model Using a Web Application
259

From these definitions it can be concluded that
Algorithm 1 builds the scaffolding of the L-system
with B variables and replaces B variables with I vari-
ables. I Variables are constant in the sense that they do
not change to other symbols, only the segment radius
changes with each iteration. The lifetime of B ensures
that not many bifurcations occur at the start of the gen-
eration, which reduces intersections between segments
in later generation stages. Besides their lifetime, B
variables have a certain chance to create a bifurcation
or turn into a dead branch with each iteration. As
the variable and parameter definitions suggest, among
other things, the rates at which bifurcations occur, life-
times, number of iterations and radius increments can
be preset and adjusted by the user.
After generating the L-system object string,
an interpreter is designed to create a 3D model
corresponding to this object string using cylinder
geometries. Before the geometries are pushed, the
L-system model is checked for intersecting segments
and pruning them if needed.
2.3.2
Parameter Extraction from Model Analysis
Except the number of bifurcations, the centerline
points, branch and centerline curvature, tortuosity and
torsion, all parameters extracted in section 2.2 are used
in the L-system algorithm as shown in table 1. First,
the Bifurcation probability is set to
100%
that means
bifurcation is always happening. Furthermore, max.
order has not been included in the analysis, so it is set
to 0. Next, the mean number of iterations is set, and it
is multiplied by two because it takes the algorithm at
least two iterations to build a segment. The mean itera-
tions cannot exceed 50, as the L-system model would
grow too large. Subsequently, the upwards angle is
imported, which is not allowed to be larger than 30
degrees or smaller than -30 degrees based on empirical
experience. After that, the bifurcation angle mean is
divided by two for each segment in the bifurcation.
After importing the parameters and saving them on the
client side, the L-system generation can commence.
3 RESULTS
3.1 L-System Generation from
Arborized 3D Models
3.1.1 Lactiferous Duct
For the first case study, the section images of the lac-
tiferous duct are acquired and a stack of aligned im-
Algorithm 1: Pseudocode for L-system.
1:
Initialize stem with stemLength branch segments
and the first bifurcation
2: for number of iterations do
3: for each I do
4: Increment radius by radialIncrement
5: return I
6: end for
7: for each B do
8:
if max centerline length
<
current iteration
then
9: return D
10: end if
11:
if time is shorter than maximum duration
of B then
12:
Increment time by 1 and iteration by 1
13: ˆ ←
angle upwardsAngle with stan-
dard deviation upwardsAngleSpread
14:
I
←
length lineLength with standard
deviation lineLengthStd, radius radius,
store iteration and centerline in-
dex
15: return ˆIB
16: else
17:
Set time to 0, increment iteration and
order by 1
18:
if random chance of branching
>
nb axes prob or bifurcation has dead branch then
19:
/
←
angle with random integer be-
tween -60 and 60
20: return [/B]
21: else
22: branches ← new array
23: for each branch in bifurcation do
24:
set max centerline length to it-
erations with standard deviation iterationsStd
25:
set centerline index to random
integer
26:
/
←
angle with random integer
between -60 and 60
27:
&
←
angle bifurcationAngle
with standard deviation bifurcationAngleSpread
and iteration correction
28: push [/&B] to branches
29: end for
30: return branches
31: end if
32: end if
33: end for
34: end for
ages is used as input for the optimized reconstruction
pipeline (Cao and Verbeek, 2012; Cao and Verbeek,
2014) of the 3D model as shown in figure 2. The tree-
BIOIMAGING 2024 - 11th International Conference on Bioimaging
260

Table 1: A list of all available L-system parameters used in
algorithm 1.
Parameter name (units)
Segment radius (pixels)
Radial increment (pixels)
Bifurcation probability
Radial segments
Iterations
Iterations std.
Max. order
Segment branch length (pixels)
Segment branch length std. (pixels)
Upwards angle (degrees)
Upwards angle std. (degrees)
Bifurcation angle (degrees)
Bifurcation angle std. (degrees)
like structure, starting at the thicker bottom stem and
splitting to branches that vary in thickness, contains
few bifurcations and consequentially centerlines. For
this reason, it was used to test and verify the analysis
and the L-system parameter extraction methods dur-
ing the development of the web application. In this
section, the results of the final analysis and L-system
generation from the extracted parameters are found.
For the preparation of the analysis, endpoints are
manually selected. In figure 2a, the selected endpoints
are shown in red. The stem at the bottom was picked as
the source point of the centerline structure. Endpoints
are chosen based on distinct branches and where they
approximately end in the mesh structure. Using this
method, 13 endpoints were selected. After selecting
the endpoints, the centerline in figure 2b was extracted.
Table 2: Attributes extracted from the analysis of the model
mesh in figure 2. Only the important attributes used for the
L-system generation are shown in this table. Thus, attributes
such as curvature, torsion and tortuosity are omitted.
Attribute name (units) Mean Std.
Branch length (pixels) 63.96 39.20
Point radius (pixels) 14.46 4.13
Centerline length (pixels) 582.56 123.60
Stem length (# iterations) 4.32
Bifurcations 11
Centerlines 13
Branches 24
Iterations 4.79 1.93
Bifurcation Angle (degrees) 62.81 19.45
The data collected from the analysis both from the
model and the centerline can be seen in table 2. A few
observations can be made from the table. Firstly, from
the 13 endpoints selected in figure 2a, 13 centerlines
(a) (b)
Figure 2: Model mesh of a lactiferous duct from newborn
mice. (a) shows the model mesh and the endpoint selections
used for the centerline creation (in red). (b) is the computed
centerline model illustrated inside of the original mesh.
have been created, meaning there are no incorrectly
analyzed centerlines. Secondly, the number of bifur-
cations does not entirely match the number of bifurca-
tions which can be found in figure 2b, because there
is also one trifurcation mistaken for a bifurcation. An-
other observation is that the total number of iterations,
which is in terms of how many mean branch lengths it
takes to build one mean centerline, is approximately
9 ± 2
. Finally, most of the varying data has a relatively
high standard deviation, but the branch length, exclud-
ing the stem length, has the highest standard deviation.
Figure 2b also displays this large variety in branch
lengths between bifurcations.
The parameters from table 2 were then used to ran-
domly generate L-systems. First, two L-systems were
selected from many similarly generated systems using
the given parameters, which can be found in figure
3. During this process, some L-systems were created
with colliding branches due to randomness of the algo-
rithm. These results were left out by using the prune
intersections option, as the original model does not
contain intersections. Furthermore, models with simi-
lar data extracted from the original mesh were selected
for the best representation. The three models were all
generated with ten iterations, because building each
segment needs at least two iterations. Additionally,
they vary in branch lengths, number of centerlines and
number of branches. The branch lengths range from
frequent short branches in figure 3a to frequent long
branches or a mixture in figure 3b.
Lastly, Figure 4 shows two L-systems with 13 cen-
terlines and varying iterations, iteration standard devi-
ation and no branch length standard deviation. These
Visualizing, Analyzing and Constructing L-System from Arborized 3D Model Using a Web Application
261
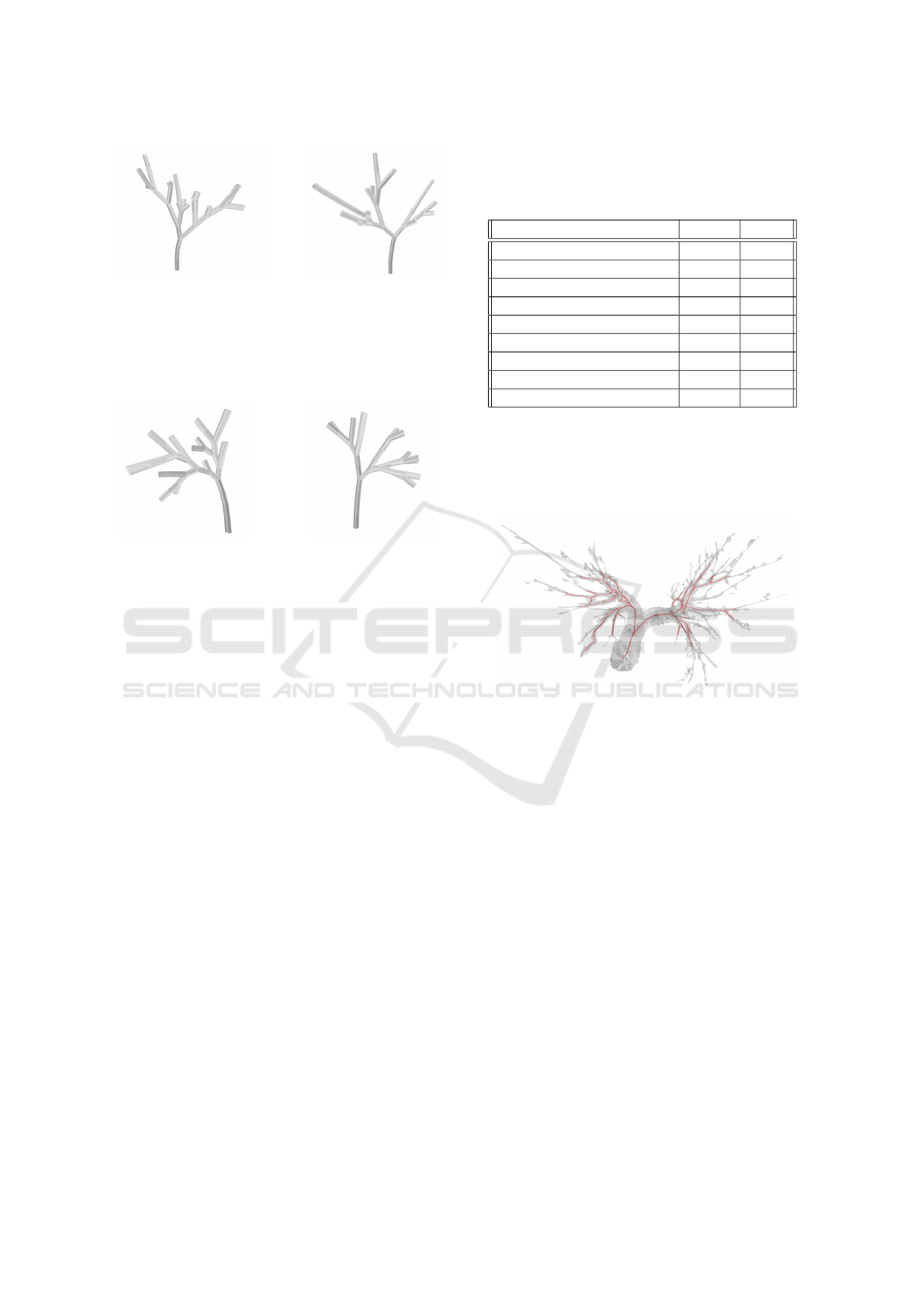
(a) (b)
Figure 3: A wide variety of randomly generated L-systems
based on the data from table 2 and with the number of iter-
ations set to 10. The upwards curve of the stem was set to
the standard value of 5 with a standard deviation of 2. (a) is
an L-system with 16 centerlines and 43 branches. (b) is an
L-system with 13 centerlines and 37 branches.
(a) (b)
Figure 4: Example of L-systems, based on the data from ta-
ble 2. The upwards curve of the stem was set to the standard
value of 5 with a standard deviation of 2. (a) is an L-system
with 38 branches, 10 iterations and no line std. (b) is an
L-system with 35 branches, 10 iterations, no iteration std.,
no branch length std. and 66% branch probability.
models were again selected from a pool of randomly
generated L-systems. Figure 4a shows a model where
the branch length standard deviation has been removed,
which removes extra long branches since all the branch
segment lengths are equal. The iteration standard devi-
ation has been reduced by one iteration. Consequently,
the variety in centerline lengths reduces. In Figure 4b,
the variety in centerline lengths has been completely
removed and the branch probability has been reduced
to retain a diverse amount of centerlines and bifurca-
tions. Due to the constant branch lengths, the overall
shape of the L-systems in figure 4 is more compact
compared to figure 3.
3.1.2 Artery
For the second case study, an artery 3D model was
used. The source of the model is derived from
www.sketchfab.com. This model has a more com-
plex tree-like structure starting at the much thicker bot-
tom stem and quickly decreasing in thickness. Some
branches are hardly connected to each other with
thin segments. Furthermore, the structure has a high
branching order and therefore contains many bifur-
cations and centerlines. Thus, it is used to test the
analysis and the L-system parameter extraction meth-
Table 3: Attributes extracted from the analysis of the artery
model. In the first column, the attribute name and units are
shown. The second and third column show the mean and the
standard deviation if applicable.
Attribute name (units) Mean Std.
Branch length (pixels) 17.25 12.70
Point radius (pixels) 6.39 4.48
Centerline length (pixels) 150.04 20.86
Stem Length (# iterations) 2
Bifurcations 32
Centerlines 40
Branches 72
Iterations 6.82 1.20
Bifurcation angle (degrees) 44.40 37.68
ods on a more complex structure. For the analysis, 40
endpoints were chosen. Figure 5 shows the resulting
centerline model in the original mesh. We observed
that it contains many long branches compared to the
lactiferous duct model.
Figure 5: The original artery model with computed center-
line.
The data collected both from the model and the
centerline, can be found in table 3. Once again, some
observations can be made from this data. First, the
number of centerlines is inline with the number of end-
points. Second, the number of bifurcations is correct
since there are no trifurcations in this centerline model.
Next, the total number of iterations is approximately
9 ± 2
. Furthermore, most of the data has a relatively
high standard deviation just like in table 2, even though
there is more data available. Relatively, the bifurca-
tion angle standard deviation contains the highest and
the point radius the second highest standard deviation.
Contrary to the bifurcation angle standard deviation,
the relatively large branch length and point radius stan-
dard deviations can be observed by the alternation
between long and short branches together with the
thick inner branches close to the stem and thin outer
branches further away from the stem.
For the L-system generation from the attributes
acquired in table 3, We observed an abundance of col-
liding branches and centerlines, indicating that with
BIOIMAGING 2024 - 11th International Conference on Bioimaging
262

the given 14 iterations, too many branches were gen-
erated. Figure 6 shows L-systems without the branch
length standard deviation. In Figure 6a, the iteration
standard deviation was reduced, which is demonstrated
by the reduced number of outliers and radial develop-
ment. Figure 6b has no iteration and branch length
standard deviations but a branch probability. This lead
to equal centerline lengths and no more cut-off center-
lines compared to the systems generated before.
(a) (b)
Figure 6: Two randomly generated L-systems based on the
data from table 3. For all of the systems, the number of
iterations was set to 12. The upwards curve of the stem was
set to the standard value of 5 with a standard deviation of
2. (a) is an L-system with 40 centerlines, 85 branches, 2.39
segment radius, no branch length std. and one iteration std.
(b) is an L-system with 38 centerlines, 86 branches, 2.39
segment radius, no branch length std, no iteration std. and
76% branch probability.
Lastly, what can be observed from all the L-
systems which have been generated from the lactif-
erous duct and artery models, is the increased number
of branches compared to the number of centerlines.
The data in table 2 and 3 show that the original mod-
els require less branches to obtain the same number
of centerlines compared to what is produced by the
algorithm.
4 CONCLUSION AND
DISCUSSION
In this paper, we have focused on combining visual-
ization, analysis and L-system abstraction in a web-
environment. We generalized this functionality for
numerous arborized 3D biological structures. The vi-
sualizing capabilities were shown to be similar with
other visualization tools such as MeshLab (Cignoni
et al., 2008). On the other hand, the ability to visualize
3D models is limited by the user’s (web) resources,
which restricts the complexity of the models that can
be rendered.
Secondly, it has been illustrated that the function-
ality is sufficient to perform proper statistical analysis
on branch-like structures and extract useful L-system
parameters. An important condition for the analysis
method is a sufficient branch radius. However, no
specification was given on which radii satisfy these
conditions. Another important condition, stated in the
VMTK documentation, requires the 3D model mesh
to be close-ended.
Next, L-system generation from extracted parame-
ters was shown. Changing the upwards and bifurcation
angle standard deviation did not seem to result in much
of a difference compared to the related work it was
derived from (Verbeek and Cao, 2020). In contrast,
the iteration and especially the branch length standard
deviation did have a significant impact on the resulting
models. Removing the branch length standard devi-
ation made the overall shape look less chaotic and
more comparable to the centerline models in both ex-
amples. Depending on the type of abstraction, either
the original branch probability or the iteration standard
deviation can be utilized to add more variety to center-
line lengths. Thus, extracted parameters are viable to
generate L-systems, particularly branch lengths, radii,
angles and iteration standard deviation.
From these key results, we can conclude the
method described in this paper could be applied to
develop a web application that is in fact capable of
visualizing, analyzing and creating L-system abstrac-
tions of arborized 3D models. In addition, easy accessi-
bility and portability can provide workflow-improving
benefits and the applied web development methods
contribute to extendibility.
For the future work, concerning visualization,
larger and more complex models could be supported
with server side rendering (Sawicki and Chaber, 2013)
or by rendering only specific sections of the 3D model.
The benefit of this is that the client requires less re-
sources, but at the cost of more server resources and
the loss of real-time navigation and interaction. Be-
sides visualization, more (arborized) 3D model analy-
sis functionality and methods can be further extended.
This could include automated endpoint selection found
in Slicer (Kikinis et al., 2014), model pre-processing
methods for open-ended model analysis and error re-
duction (Izzo et al., 2018) or support for analysis of
multiple similar 3D models at once.
For L-system generation, additional statistical re-
search can be conducted on the distributions of the
parameters and how the data can be applied optimally,
particularly when dealing with relatively high standard
deviations. This also includes the right estimate of
iterations, which consistently failed during the testing
phase. Further experiments on L-system parameters
and resulting abstractions may give rise to models
which are more consistent and in accordance with the
requirements.
To conclude, VR features that are readily avail-
able within the A-Frame framework can be utilized.
Visualizing, Analyzing and Constructing L-System from Arborized 3D Model Using a Web Application
263

This includes VR interaction methods with the user
interface and new visualization capabilities (such as
a free-floating camera) that can change the way 3D
structures are analyzed and perceived.
REFERENCES
Balaji, S. and Murugaiyan, M. S. (2012). Waterfall vs. v-
model vs. agile: A comparative study on sdlc. Interna-
tional Journal of Information Technology and Business
Management, 2(1):26–30.
Boudon, F., Cokelaer, T., Pradal, C., and Godin, C. (2010).
L-py, an open l-systems framework in python. In De-
Jong, Theodore, Silva, D., and David, editors, 6th In-
ternational Workshop on Functional-Structural Plant
Models, pages 116–119, Davis, CA, United States.
Brewe, T. (2016). A l-system library using modern (es6)
javascript with focus on a concise syntax. http://https:
//github.com/nylki/lindenmayer.
C, R. (2010). Three.js 3d javascript library. http://github.
com/mrdoob/three.js.
Cao, L. (2014). Biological model representation and anal-
ysis. Phd thesis, Leiden Insitute of Advanced Com-
puter Science (LIACS) , Leiden University, Leiden, the
Netherlands. Available at https://hdl.handle.net/1887/
29754.
Cao, L. and Verbeek, F. J. (2012). Evaluation of algorithms
for point cloud surface reconstruction through the anal-
ysis of shape parameters. In Baskurt, A. M. and Sit-
nik, R., editors, Three-Dimensional Image Processing
(3DIP) and Applications II. SPIE.
Cao, L. and Verbeek, F. J. (2014). Nature inspired phenotype
analysis with 3d model representation optimization. In
Abraham, A., Kr
¨
omer, P., and Sn
´
a
ˇ
sel, V., editors, Inno-
vations in Bio-inspired Computing and Applications,
pages 165–174. Springer International Publishing.
Cignoni, P., Callieri, M., Corsini, M., Dellepiane, M., Ganov-
elli, F., and Ranzuglia, G. (2008). Meshlab: an open-
source mesh processing tool.
Cooper, D., Turinsky, A., Sensen, C., and Hallgr
´
ımsson,
B. (2003). Quantitative 3d analysis of the canal net-
work in cortical bone by micro-computed tomography.
The Anatomical Record Part B: The New Anatomist,
274B(1):169–179.
Foundation, B. (1998). Blender. org—home of the blender
project—free and open 3d creation software. http:
//blender.org.
Grinberg, M. (2018). Flask web development: developing
web applications with python. ” O’Reilly Media, Inc.”.
Gunicorn-Python, W. (2017). Http server for unix. URL:
http://gunicorn. org.
Izzo, R., Steinman, D., Manini, S., and Antiga, L. (2018).
The vascular modeling toolkit: A python library for
the analysis of tubular structures in medical images.
Journal of Open Source Software, 3:745.
Kikinis, R., Pieper, S. D., and Vosburgh, K. G. (2014). 3D
Slicer: A Platform for Subject-Specific Image Analysis,
Visualization, and Clinical Support, pages 277–289.
Springer New York, New York, NY.
Koch, N. and Kraus, A. (2002). The expressive power of
uml-based web engineering. In Second International
Workshop on Web-oriented Software Technology (IW-
WOST02), volume 16, pages 40–41. Citeseer.
Krasner, G. E., Pope, S. T., et al. (1988). A description of
the model-view-controller user interface paradigm in
the smalltalk-80 system. Journal of object oriented
programming, 1(3):26–49.
Mozilla (2015). A-frame, a web framework for building
virtual reality (vr) experiences. http://github.com/
aframevr/aframe.
of ISTI CNR, V. C. L. (2016). A javascript, client-side, mesh
processing tool. inspired by the well known meshlab.
https://github.com/cnr-isti-vclab/meshlabjs.
Prusinkiewicz, P. and Lindenmayer, A. (1990). The algo-
rithmic beauty of plants. Springer Science & Business
Media.
Reese, W. (2008). Nginx: the high-performance web server
and reverse proxy. Linux Journal, 2008(173):2.
Sawicki, B. and Chaber, B. (2013). Efficient visualization
of 3d models by web browser. Computing, 95(1):661–
673.
van Ertbruggen, C., Hirsch, C., and Paiva, M. (2005).
Anatomically based three-dimensional model of air-
ways to simulate flow and particle transport using com-
putational fluid dynamics. Journal of Applied Physiol-
ogy, 98(3):970–980. PMID: 15501925.
Verbeek, F. J. and Cao, L. (2020). L-systems from 3d-
imaging of phenotypes of arborized structures. Funda-
menta Informaticae, 175(1-4):327–345.
BIOIMAGING 2024 - 11th International Conference on Bioimaging
264
