
Modeling iPSC-Derived Endothelial Cell Transition in Tumor
Angiogenesis Using Petri Nets
Ad
´
ela
ˇ
Sterberov
´
a
1
, Andreea Dincu
1
, Stijn Oudshoorn
1
, Vincent van Duinen
2
and Lu Cao
1 a
1
Leiden Insisute of Advanced Computer Science, Leiden University, Leiden, The Netherlands
2
Leiden University Medical Center, Leiden, The Netherlands
Keywords:
Tumor Angiogenesis, Petri Nets, Endothelial Cell Transition, VEGF Gradient.
Abstract:
Tumor angiogenesis concerns the development of new blood vessels supplying the necessary nutrients for
the further development of existing tumor cells. The entire process is complex, involving the production and
consumption of chemicals, endothelial cell transitions as well as cell interactions, divisions, and migrations.
Microfluidic cell culture platform has been used to study angiogenesis of endothelial cells derived from human
induced pluripotent stem cells (iPSC-ECs) for a physiological relevant micro-environment. In this paper, we
elaborate on how to define a pipeline for simulating the transformation and process that an iPSC-derived
endothelial cell goes through in this biological scenario. We leverage the robustness and simplicity of Petri
nets for modeling the cell transformation and associated constraints. The environmental and spacial factors
are added using custom 2-dimensional grids. Although the pipeline does not capture the entire complexity of
tumor angiogenesis, we are able to capture the essence of endothelial cell transitions in tumor angiogenesis
using this conceptually simplified solution.
1 INTRODUCTION
Angiogenesis is the process of the development of
new blood vessels. Microvascular processes in angio-
genesis are found to play an important role in kidney
diseases, and are even described as ‘the base of the
iceberg’ for this category of disease (Querfeld et al.,
2020). The formation of blood vessels in tumors has
also been shown to influence metastasis and growth
rates (Folkman, 2002; Carmeliet and Jain, 2000; Toi
et al., 2002).
In recent years, human induced pluripotent stem
cells derived endothelial cells (iPSC-ECs) became in-
fluential for disease modeling, drug discovery and
regenerative therapy. Microfluidic cell culture plat-
forms have been introduced to study angiogenesis of
iPSC-ECs in a physiological relevant cellular micro-
environment with controlled perfusion and gradients
(van Duinen et al., 2019). In this paper, we concen-
trated on modeling angiogenesis induced by VEGF
which is released by the hypoxic tumor cells in a mi-
crofluidic environment.
a
https://orcid.org/0000-0002-1847-068X
2 RELATED WORK
There are a number of mathematical and computa-
tional models developed to study different aspects of
angiogenesis (Peirce, 2008). A discrete mathemati-
cal model is developed for the dynamics of vascular
endothelial cells in angiogenic morphogenesis (Mat-
suya et al., 2016) It incorporates cell-mixing behavior
and temporal length generating behavior of the blood
vessel. A new mathematical model is designed to re-
produce the tumour-induced vascular networks under-
going stages of growth, regression and regrowth (Vi-
lanova et al., 2017). The model is able to capture cap-
illaries at full scale and the dynamics of vessel net-
works at long time scales. A mathematical formalism
is developed to simulate the early stages of angiogen-
esis based on a 3D in vitro model (Bookholt et al.,
2016). The model takes into account the dynamic in-
teraction and interchange of different phenotypes of
endothelial cells and several proteins playing a role in
the interaction. A hybrid model is developed to re-
alize silico experiments for tumor growth and angio-
genesis (Phillips et al., 2020). This model treats each
cell as an agent, incorporates phenotypic transitions
of each tumor and endothelial cell and allows VEGF
and nutrient fields to impact the dynamics.
Šterberová, A., Dincu, A., Oudshoorn, S., van Duinen, V. and Cao, L.
Modeling iPSC-Derived Endothelial Cell Transition in Tumor Angiogenesis Using Petri Nets.
DOI: 10.5220/0012268800003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 1, pages 337-346
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
337

These mathematical models are highly powerful
but they are not standardized enough for compari-
son. It also further hampers the reproducibility. We
are looking for a unified and versatile framework for
modeling biological systems. Petri nets is a graphi-
cal and mathematical formalism for the modeling and
analysis of concurrent, asynchronous, distributed sys-
tems (Chaouiya, 2007). It is shown to be a promising
mathematical tool to describe and study the relation-
ships and interactions between various parts of a bio-
logical systems e.g. metabolic pathways, organelles,
cells, and organisms (Carvalho et al., 2018; Valen-
tim et al., 2022; Liu et al., 2017). Furthermore, mul-
tiple variants of the initial formalism were created
(e.g. stochastic, timed, hybrid, coloured) to enable
analysing dynamical properties of complex processes,
from either a qualitative or a quantitative point of view
(Chaouiya et al., 2008).
In this paper, we utilized Petri nets to simulate the
transitions that iPSC-ECs undergo in the tumor angio-
genesis process in a microfluidic cell culture platform.
A complex pipeline is designed to model the diffusion
and consumption of the VEGF as well as the migra-
tion of the cells towards the VEGF gradient.
3 BIOLOGICAL DETAILS
A tumor induces the growth in quiescent arteries
when the tumor cells are depleted of nutrients, such
as oxygen. This growth induction is thus triggered in
hypoxic cells, causing them to excrete VEGF. VEGF
diffuses to the environment around the tumor and
causes the phalanx. Endothelial cells, that make up
the artery, start turning into tip cells by the influence
of VEGF. In addition, the VEGF acts as a chemoat-
tractant to tip cells. These cells inhibit neighboring
cells from turning into tip cells through notch sig-
nalling (Sainson and Harris, 2008). These neighbor-
ing cells turn into stalk cells instead. Stalk cells pro-
liferate and, meanwhile, follow the tip cell that is near
them. The tip cells inhibit the stalk cells from transi-
tioning back into phalanx cells within a certain dis-
tance. Therefore, the stalk cells can turn into pha-
lanx cells to stabilise the growing artery structure. In
Figure 1, an example of this stage in angiogenesis is
shown.
New physiologically relevant in vitro screening
was recently developed (van Duinen et al., 2019)
using iPSC-ECs in a microfluidic cell culture plat-
form, to model angiogenesis towards a VEGF gradi-
ent. This could be seen as a simplification of vessel
growth near tumors, where the hypoxic tumor cells
are replaced by a VEGF gradient. In this paper, the
Figure 1: A visualization of the sprouting process. Red cells
are phalanx cells. Brown cells are tip cells and light brown
cells are stalk cells.
vessel formation is modeled in silico. The aim is to
create a model which includes transitions, movement,
differentiation and VEGF consumption of the cell. In
addition, the formation of a new blood vessel is also
induced. In this process, there are several cell types
with different roles. The methods that were used to
simulate these are further described in the following
sections.
4 METHOD
We developed a hybrid model that includes a central
Petri net and two grid matrices. The central Petri net
realizes the cell type transitions. The two grid matri-
ces deal in part with spatially connected features, such
as the cell positioning and movement, and the growth
vector (i.e. VEGF) concentrations.
4.1 Endothelial Cell Transitions Model
The endothelial cell type transitions are modeled us-
ing a timed hybrid Petri net in order to incorpo-
rate division/growing time, distance to the closest tip
cell and the VEGF concentration for each cell. The
scheme that we use is originally from (Phillips et al.,
2020) as illustrated in Figure 2. Because this scheme
is designed to mimic the movement of the tip cell ac-
cording to the gradient of VEGF which fits our mi-
crofluidic environment the best. In addition, the pa-
per provides a detailed baseline parameters to work
with. It helps to bring biologically relevant proper-
ties to our model and makes our model more realistic.
The places in our model represent all the possible cell
types that an endothelial cell can adopt, namely: pha-
lanx cell P, stalk cell S, and tip cell T.
An additional place is added to represent the divi-
sion of stalk cells (SS). As the effect of this transition
is the separation of one cell into two smaller cells, two
different tokens have to be generated, adjacent to each
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
338

Figure 2: Schematic illustration of endothelial cell tran-
sitions. d
tip
: the distance from the current cell to the
nearest tip cell. d
SP
: Minimum distance from tip cell for
S → P transition. d
PS
: Maximum distance from tip cell for
P → S transition. d
min
: Minimum distance from tip cell
for P → T transition. [VEGF]: VEGF concentration. α
v
:
VEGF threshold for P → T transition. τ: the amount of
time that a cell spends in a given place. τ
sp
: a predefined
time interval for a stalk cell to divide. τ
gs
: growing time of
a stalk cell. τ
grow
: mandatory growing time.
other (positions (i, j) and (i − 1, j)). The introduction
of another token in the system has to be captured also
in the positioning grid, by filling an additional posi-
tion ((i − 1, j)). A stalk cell can divide after a prede-
fined time interval (τ
sp
), with each being half of its
original size and volume. For the newly divided stalk
cells to be considered again as integral stalk cells, they
have to go through a growing period to increase their
volume. This mandatory growing time is defined as
a pre-set threshold, τ
grow
. Even if we have uniformed
the volume of the cell to a constant, the mechanisms
behind the growing phase are preserved.
The transitions in our Petri net correspond to pos-
sible changes in phenotype that a cell with a given
type can reach.
Each transition has to be conditioned in different
manners, as shown in Figure 2. The division of a stalk
cell (S → SS) and the transition of divided parts back
to an integral stalk cell (SS → S) are conditioned by
the passing of specific time intervals, τ
sp
and τ
grow
re-
spectively. Additionally, the S → SS transition is con-
strained by the space available in the positioning grid.
Both the transition from phalanx cell P to stalk cell S
(P → S) and the reversed one (S → P) are conditioned
by the distance of a given cell to the nearest tip cell
d
tip
. For transitioning a phalanx cell P to a stalk cell
S, we have to ensure that the distance from a given
cell to a tip cell T is less than a pre-defined threshold
d
PS
, while when transitioning back from a stalk cell S
to a phalanx cell P, we need to have a tip cell T further
than a pre-defined threshold d
SP
. Finally, the transi-
tion from phalanx cell P to tip cell T (P → T ) is avail-
able only when there is no other tip cell in a certain
immediate vicinity (specified by d
min
) and the VEGF
at the location of the cell exceeds a certain value α
v
.
The distance to the nearest tip cell d
tip
is calculated
Table 1: Table with the fixed parameters in our simulation.
Parameter Meaning Value
α
v
VEGF threshold for P → T
transition
0.1
γ Fixed consumption rate 10h
−1
d
SP
Minimum distance from tip
cell for S → P transition
1.55
d
PS
Maximum distance from tip
cell for P → S transition
1.55
d
min
Minimum distance from tip
cell for P → T transition
10
R Endothelial cell radius 1
S delay Stalk cell may divide after
this delay
4
SS delay Stalk cell growth time 3
based on the positioning grid, while the VEGF con-
centration is retrieved from the VEGF grid. The fixed
thresholds used in the implementation can be found in
Table 1. These parameters are derived from baseline
set of model parameter values in (Phillips et al., 2020)
after unifying both tumor and endothelial cell radii to
1 so as to make our model biologically sound.
In order to prevent transitions in conflict situation
during simulation. The order of firing the transitions
is set based on biological reasoning. We first fire the
S → SS transition, as there is no notion in our biologi-
cal framework that inhibits a cell from dividing when
the conditions for division are met. The same rea-
soning applies to the SS → S transition (which we set
as the second transition to be fired) as when the cell
reaches the volume of a full stalk cell, it should be
immediately assigned to that phenotype. As the pos-
sible divisions of the stalk cells are already conducted,
the transition of the remaining stalk cells S to phalanx
cells P can be freely handled. The last two transitions
P → T and P → S are executed in this order. Even
if generally these two transitions are not concurrent,
in case such a situation might occur (based on the set
values of d
min
and d
PS
), we would like to prioritize the
transition to a tip cell T when the VEGF condition is
met. By defining a clear ordering, any concurrency
problems are mitigated.
The final component of Petri nets that we need to
discuss is the token. In our model, a token represents
a specific cell, encapsulating various characteristics of
the defined cell. The token’s internal representation is
{x, y, d
tip
, VEGF, τ}. x and y define the position of
the cell in the positioning matrix. d
tip
specifies the
distance from the current cell to the nearest tip cell
(as computed using the positioning matrix). VEGF
indicates the VEGF concentration retrieved from the
VEGF grid at the cell position (x, y). τ stores the
amount of time that a cell spends in a given place.
Modeling iPSC-Derived Endothelial Cell Transition in Tumor Angiogenesis Using Petri Nets
339
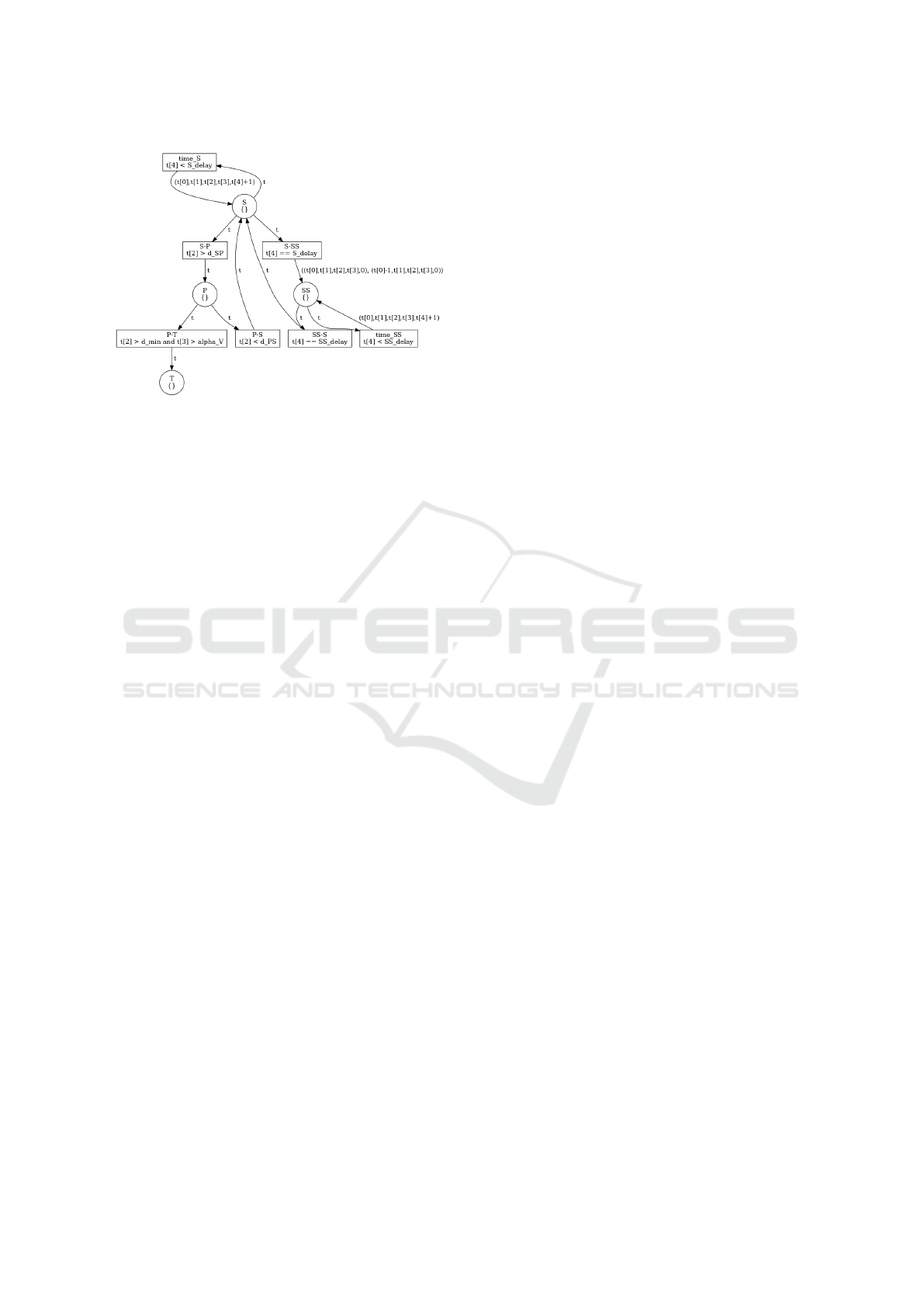
Figure 3: Schematic illustration of the Petri net.
To create the Petri net, we used the Python
library SNAKES (Pommereau, 2015), which pro-
vides flexibility in terms of modeling options or
extending the features. The source code is avail-
able at https://github.com/LuLIACS/angiogenesis-
modeling. The net scheme can be seen in Figure 3.
We can observe the places for each endothelial cell
phenotype (marked by circles). There are two types
of transition in the net. One type is a transition be-
tween different phenotypes. The other type of transi-
tion is a representation of time. The time is valid only
for two places, S and SS. In all transitions, t[i] refers to
specific information preserved in the token at position
i.
4.2 Positioning and VEGF Grid
As previously introduced, there is a need in our model
to determine the distance between two cells. To de-
fine a proper distance metric, one first needs to set
the space in which the cells are placed. For conve-
nience, we chose to model this environment as a 2-
dimensional space, with the distance metric being the
Euclidean distance. In this manner, the integrity of
the biological concepts is preserved, and the extension
to the 3-dimensional space is fairly straightforward.
This kind of spatial representation can be interpreted
as a cross section of the original environment.
The grid was implemented as a 2-dimensional NumPy
(Harris et al., 2020) matrix, with configurable size
(H,W ). Each position in the matrix can hold a cell
or zero value when it is empty. This introduces the
limitation of all cells having the same volume, thus
the same growth factor consumption rate γ. The cell
is implemented as an object, initialized with the cell
type (phalanx, stalk, or tip). Subsequently, the func-
tions relevant to the cells are implemented on this ma-
trix, such as cell movement (changing positions of
cells), cell transition (changing cell types), calcula-
tion of the distance to the nearest tip cell, or division
of stalk cells. All of these functions are connected to
the Petri net and enabled for firing. In order to simu-
late the formation of new blood vessels, the matrix is
initialized with all positions empty, except the bottom
row, filled with phalanx cells P. This row is meant
to depict a section of the micro-vessel culture in the
microfluidic well-plate.
The VEGF grid is similar to the positioning grid
and must match its shape. The VEGF grid was
also implemented as a 2-dimensional NumPy matrix,
which retains the concentration of VEGF present at
each position. The VEGF grid is initialized with only
the first row holding non-zero concentrations. It sim-
ulates the existence of some tumor cells that emit the
VEGF signal. At each time step, a diffusion model is
applied. The diffusion model should define the way
in which the VEGF vector diffuses in our system.
We implemented two types of diffusion. The first
one is regular. The part of the VEGF that is diffused
from the central position to a neighbor position with a
lower value of VEGF is given by diffusion factor f
d
,
which is set to 0.1 in our implementation. Therefore,
the value of VEGF in the center position and VEGF
in the neighborhood position can be given by
V EGF
center
= VEGF
center
− f
d
·V EGF
center
, (1)
V EGF
neighbor
= V EGF
neighbor
+ f
d
·V EGF
center
. (2)
But they are too regular and pleasant for a natural phe-
nomenon. Therefore, we implemented a second type
of diffusion with some level of randomness. In this
type of diffusion for each position in the grid, we ran-
domly decide which part of the VEGF amount should
diffuse in which direction (Hill, 2017). We again look
at each element in the VEGF matrix, and from this
center element, a random part of the VEGF amount is
diffused into each neighborhood position.
Additionally, the cells continuously consume part
of the VEGF according to their consumption rate γ.
In this manner, the VEGF grid has to be continuously
updated.
4.3 Cell Movement
Regarding the cell movement, we adopt a simplified
model. The movement is conditioned to happen only
upwards. After a cell transition to a tip cell, if the
tip cell is surrounded by stalk cells, the tip cell is al-
lowed to update its position. An additional type of
movement is the one when a stalk cell S divides. The
old cell has to be replaced by two new tokens which
represent the division into two smaller cells. In order
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
340
to create space for the two tokens, all of the cells po-
sitioned in the previous rows are simply shifted one
row above, if there is space. In case there is no space,
the cell division does not take place, and the move-
ment does not happen. It particularly helps us to have
a natural end of the simulation, instead of allowing
cells to continuously multiply and push others outside
the grid. This design choice can be easily adapted as
needed.
4.4 Time Integration
As illustrated in Figure 2, certain transitions of the
cell type are time constrained. Moreover, the cell
movement and the diffusion of the VEGF have to be
discretized in order to be integrated into the model. A
natural way of doing this was to use time as our sam-
pling factor and update both the cell positions and the
VEGF concentrations once per time step. Thus, the
time factor is a necessary piece in all components of
our model. We introduced a custom way of model-
ing the time in order to better match the biological
scenario. In our approach, each cell (token) has an
internal individual timer τ which is reset to 0 when
the token enters a new place. This timer increases
for each cell independently until it reaches the time
constraint necessary for the connected transitions (i.e.
for the S → SS, the condition is τ > τ
sp
). When the
condition is met, the transition can be fired, and the
token has its timer reset. The timer increase can be
easily integrated into the Petri net using a loop transi-
tion so as to increment the τ variable inside the token
representation, until a set threshold (i.e. τ
sp
). This
loop transition should be fired at the start of each time
step. In this manner, the biological model is not com-
promised when implementing the time aspect, as any
number of cells are able to transition at the same time
step if needed.
5 EXPERIMENTS AND RESULTS
5.1 Diffusion Simulations
In the first experiment, the two diffusion types are
compared with the same matrix initialization. The re-
sults of the simulations are shown in Figures 5 and 6
for normal and random diffusion, respectively.
In the first simulation with the normal diffusion,
tip cells form on two sides of the grid at the same
time as shown in (a) of Figure 5. When these tip cells
are 1 square away from the top of the grid, enough
distance has accumulated and the two new tip cells in
the center form, both in time step 51.
(a)
(b)
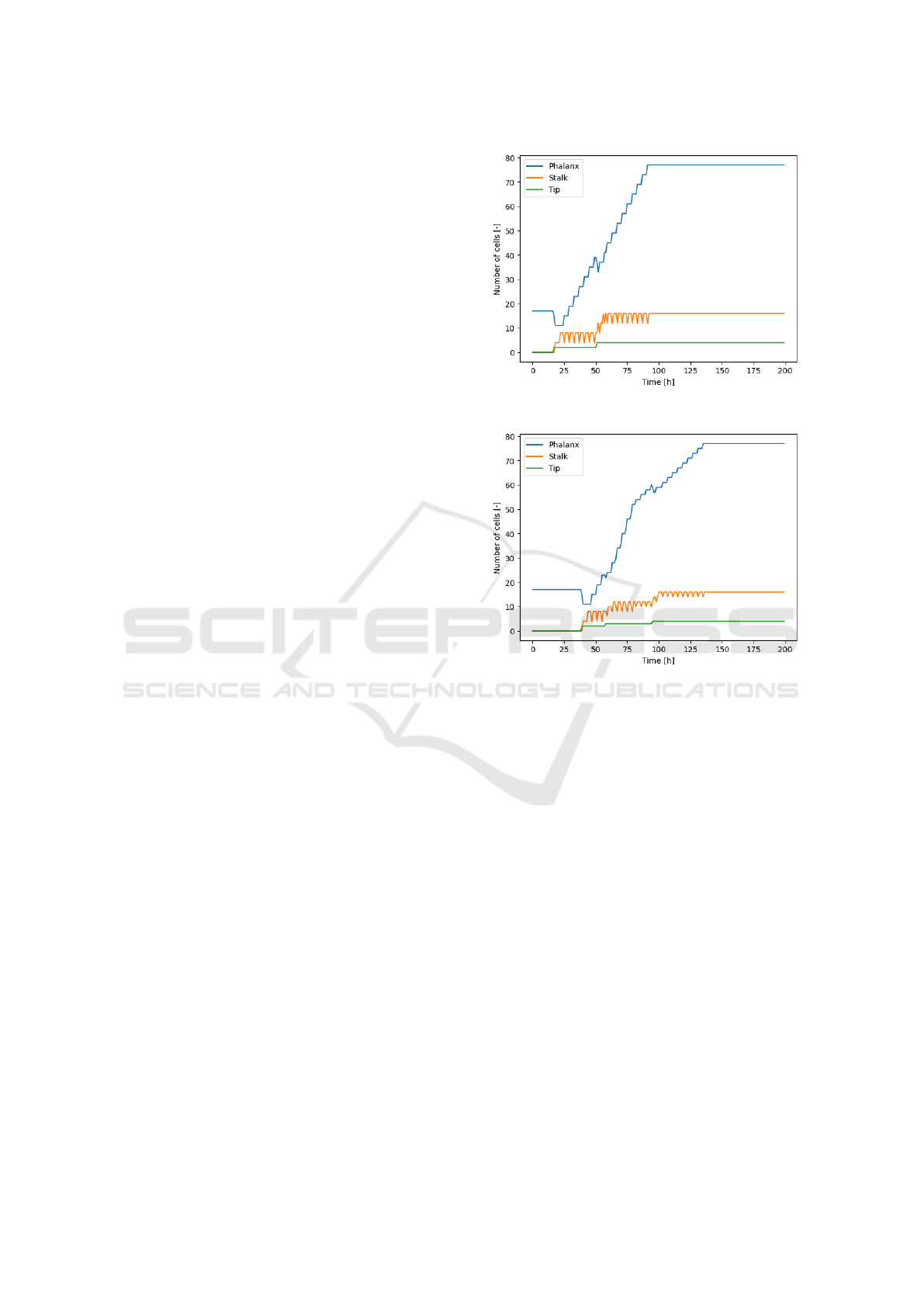
Figure 4: The number of cells shown per type for the normal
and random diffusion experiments.
The number of cells throughout the simulation is
depicted for each cell type in Figure 4(a). In this
graph the moments where the number of tip cells in-
creases, followed by the constant addition of phalanx
cells are visible.
The simulation with random diffusion, as shown
in Figure 6, also has two phalanx cells that transfer to
tip cells but on one side of the matrix at first, which
happens 22 time steps later than with the normal dif-
fusion.
The count of the different cell types for this exper-
iment is shown in Figure 4(b). The third transition to
a tip cell happens on the left side at time step 58. The
two initial tip cells reach the top at 80, so the increase
in phalanx cells turns less steep. The fourth and fi-
nal transition takes place as the third tip cell reaches
the top after which the increase in phalanx cells is
still less steep than during the growth of the first two
branches.
The corresponding VEGF matrices for the nor-
mal and random diffusion are shown in Figures 7 and
Modeling iPSC-Derived Endothelial Cell Transition in Tumor Angiogenesis Using Petri Nets
341

(a) Cell positioning - Iteration 17
(b) Cell positioning - Iteration 92
Figure 5: The cell matrix for the first time step where tip
cells are formed (left) and where the final movement of the
tip cells is seen (right). The time step is shown below the
figures.
8. The states on the left show the concentrations at
the time of the first transition to a tip cell, while the
ones on the right are from the moment the last tip cell
reaches the top of the grid. A structured gradient is
observed and the matrix turns more symmetrical as
time passes in the simulation. After 115 steps the ma-
trix holds no VEGF anymore.
As random diffusion is not deterministic, differ-
ent states could be reached with the same initial state.
Note that the spreading patterns of the concentrations
do look similar at the same phase despite using dif-
ferent diffusion methods. The key difference is the
asymmetry that is observed, which follows from the
branching that started at one side. We quantified
the random diffusion, by running this experiment ten
times and averaging as shown in Table 2.
The average number of branches is observed to be
the same as in the normal diffusion simulation, which
suggests that random diffusion can be used to approx-
imate the normal diffusion.
5.2 VEGF Amount Experiments
In this set of experiments, we examined the influence
of the initial value of VEGF that is inserted into our
(a) Cell positioning - Iteration 39
(b) Cell positioning - Iteration 136
Figure 6: The cell matrix during the random diffusion ex-
periment when the first tip cells form (left) and when the
final movement is made by a tip cell (right). The number
below indicates the time step during the event.
Table 2: The average time step at which the described
events took place, and the average number of branches are
shown with their standard deviation.
Measurement µ ± σ
First tip cell 33.6 ± 6.7
First grown branch 74.6 ± 6.7
Number of branches 4.0 ± 0.7
Last grown branch 153.7 ± 23.7
environment. From the biological point of view, we
can assume that the environment closer to the tumor
contains a higher level of VEGF concentration than
the environment more distant from the tumor. There-
fore, a small amount of VEGF inserted into the model
may be seen as equivalent to the angiogenesis affected
by the tumor in the distance.
We performed these experiments with the same
initial settings; only the initial VEGF value differed.
We used the grid size 11 × 15 so that the cells have
enough space to grow. Parameter d
tip
was set to
10. And then, we performed 200 iterations with each
value of VEGF inserted into the grid. And all experi-
ments were conducted for both types of diffusion.
The results from the experiments are noted in
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
342
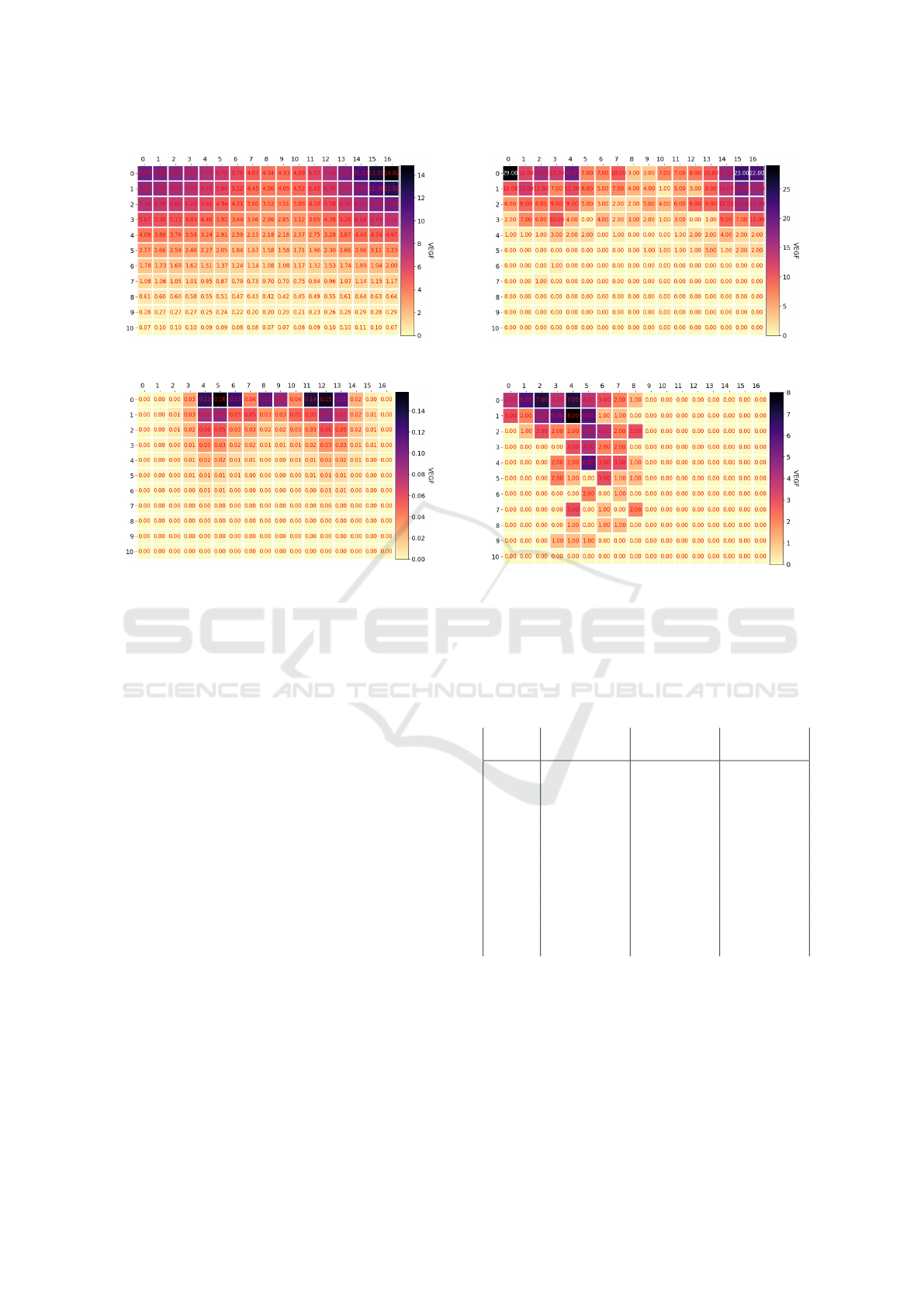
(a) VEGF grid - Iteration 17
(b) VEGF grid - Iteration 92
Figure 7: The VEGF concentrations for the normal dif-
fusion experiment, where the left frame shows the matrix
when the first tip cell has formed and the right shows the
matrix when the last tip cell reaches the top of the matrix.
3, where we summed up the essential findings. 3
shows the results from the experiments with random
diffusion and regular diffusion. The first column
(”VEGF”) shows the amount of initial VEGF. The
second column (”First transition”) indicates at which
iteration the first transition of the cell phenotype oc-
curred. The third column (”First branch”) tells at
which iteration the first branch was fully grown. The
last column (”Number of branches”) shows the num-
ber of branches that was grown in total. If the value is
noted by ’-’, the situation didn’t occur at all. Regard-
ing to regular diffusion, we observe that the minimum
amount of VEGF from the phalanx cell to the first tip
cell is 300. Otherwise, the amount of VEGF, that dif-
fuses to the positions of the initial phalanx cells, is
trivial. Because it is consumed in each iteration until
there is no more VEGF left. We can also observe a
relation between the amount of VEGF and iteration
where the first transition occurs. After exceeding the
limit amount necessary for the first transition, the iter-
ations in which the first transition took place decrease.
Furthermore, the iterations in which the first branch
was fully grown decreased. On the other hand, with
random diffusion, we can not observe such relations.
The iterations in which the first transition took place
(a) VEGF grid - Iteration 17
(b) VEGF grid - Iteration 92
Figure 8: The VEGF concentrations in the diffusion exper-
iments at the same time steps as in the normal diffusion in
Figure 7.
Table 3: Table of results from experiments with different
initial VEGF values, with random diffusion and regular dif-
fusion.
VEGF FT FB # branches
rand regu rand regu rand regu
50 27 - 68 - 3 -
100 30 - 72 - 4 -
150 36 - 78 - 3 -
200 53 - 94 - 3 -
300 43 19 84 60 3 1
400 32 15 74 56 3 3
500 26 13 68 54 4 3
600 33 12 73 54 3 3
700 27 11 68 52 4 3
800 37 10 78 52 5 3
900 35 10 76 52 3 4
1000 33 9 74 50 3 3
FT: First transition; FB: First branch; # branches:
number of branches
are around 27-53. But no pattern can be observed.
In addition, no pattern can be observed in a num-
ber of branches related to the amount of VEGF. For
example, the values are again random around 3-5.
With regular diffusion, it seems that the number of
branches is increasing with the amount of VEGF, but
it does not hold in all cases.
Modeling iPSC-Derived Endothelial Cell Transition in Tumor Angiogenesis Using Petri Nets
343
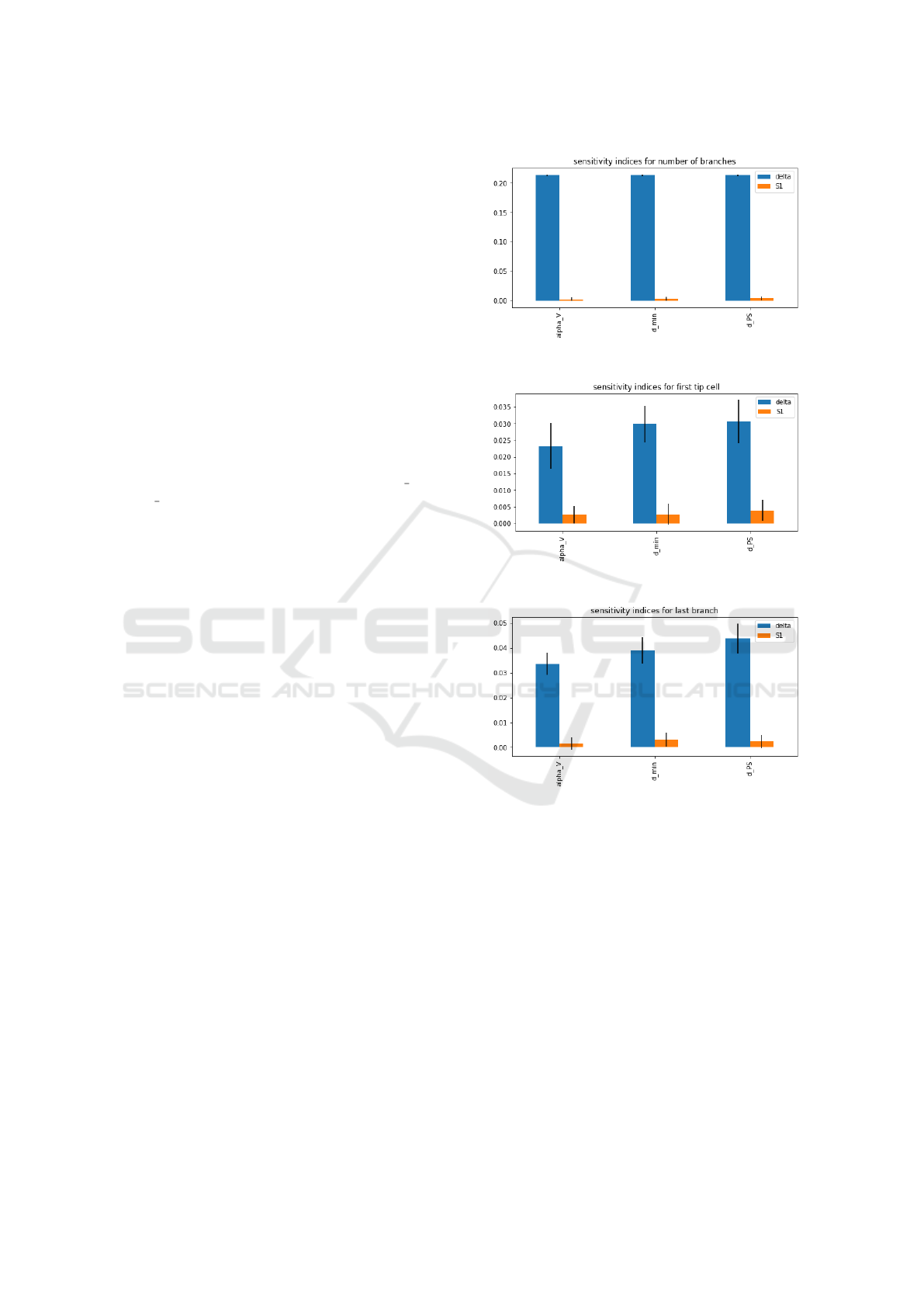
Table 4: Parameter range for sensitivity analysis.
Parameter range
α
v
[0, 3]
d
SP
[0.5, 2.5]
d
min
[10, 15]
In general, we can observe that the number of
branches created is higher using random diffusion.
Moreover, the first transition occurs sooner with ran-
dom diffusion. Therefore, we can conclude that using
random diffusion gives the model higher chances to
transition and grow new branches.
5.3 Sensitivity Analysis
Some of the parameters 1 that we used in our model
such as α
v
, γ, d
SP
and d
PS
are carefully estimated
in (Phillips et al., 2020). Some of them are fine-
tuned to fit our model scale such as d
min
, R, S delay
and ss delay. Nevertheless, we don’t know whether
iPSC-ECs behave the same during angiogenesis as
endothelial cells derived from adult mice or human.
Therefore, the parameters should be able to adjust and
we are interested to learn how sensitive is the model
to the choice of parameters. We selected three pa-
rameters, which have space to adjust in our model,
to conduct a sensitivity analysis. We set a range of
each parameter so that the cell transition is still hap-
pening within 200 iteration and the branch still grows
in a reasonable manner. A reasonable manner means
that there is only one tip cell on top of a branch while
growing and there is no single column branch that can
grow without a tip cell. With all the factors consid-
ered, we set the range for the three parameters as fol-
lowing:
We selected three features to be the output of the
model. They are the time step for the first tip cell, the
time step for the last grown branch and the total num-
ber of branches. Delta Moment-Independent Measure
(Borgonovo, 2007) is used for sensitive analysis. It is
a global sensitive indicator which looks at the influ-
ence of input uncertainty on the entire output distribu-
tion. In addition, the method returns first-order sobol
indices which is a variance-based sensitivity (Sobol’
and Kucherenko, 2009). We set number of samples
equal to 1024 per parameter. The sensitive indices are
shown in Figure 9. We observed that, apart from the
sensitive indices for number of branches, the rest of
the indices are all very low and close to 0. That means
the parameters do not contribute much to the uncer-
tainty of the output of the model. That is probably
because our analysis model is relatively small with a
grid size 11 × 15. It does not give much variance in
the output features. In the future, a larger grid size
(a) sensitivity indices for number of branches
(b) sensitivity indices for first tip cell
(c) sensitivity indices for last branch
Figure 9: Results of sensitivity analysis.
should be considered for sensitivity analysis.
5.4 Scalability Experiment
The final experiment that we conducted was designed
to test the scalability of our current implementation.
To attain this goal, we ran our simulation for a log-
arithmically increasing grid size, with the height and
weight varying in the interval [10, 10 000]. The cur-
rent implementation has an exponential increase of
running time, when the grid size is logarithmically in-
creased. Functions, such as the ones computing the
distance to the closest tip cell, executing the cell divi-
sion in the positioning grid and both VEGF diffusion
mechanisms, are computationally heavy. In the future
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
344

work, we will look into the possibility of improving
these functions with the use of multi-processing or by
vectorizing the computations.
6 CONCLUSION
In this paper, we modeled the transformations that
iPSC-ECs undergo in the process of tumor angiogen-
esis in a microfluidics environment. The angiogen-
esis is guided by a gradient of VEGF. The model’s
behaviour was tested in various scenarios. Moreover,
sensitivity analysis and scalability analysis are con-
ducted to evaluate its performance.
The diffusion models were varied to find the most
realistic one for model simulation. The diffusion
approximation through random particle movement
reached results that were very similar to those with
normal diffusion. It suggests that random diffusion
can be used to approximate the VEGF diffusion. The
non-symmetric formation of branches is also more
comparable to the chaotic branching seen in the in
vitro models. However, both approaches have their
drawbacks. Where the normal diffusion is too perfect
to be realistic, the random diffusion may result in too
large ‘jumps’ of particles to be realistic. In addition,
we found that the initial amount of VEGF does not in-
fluence the speed of angiogenesis when using random
diffusion. But it is an essential parameter for regular
diffusion, where, with too low values of initial VEGF,
the angiogenesis does not start at all.
In this work, we used a rather simplified model
for cell movement, which only allows cells to move
upward. Side-way movement of tip cells should be
included in the future to make the model more real-
istic. Furthermore, the direction of motion should be
decided by the concentration of VEGF, while preserv-
ing contact with stalk cells.
Another limitation is the scale of the model with
a grid size 11x15. It is relatively small to indicate the
contribution of parameters to the uncertainty of the
output of the model. A larger grid size is preferred
for the sensitivity analysis. However, in scalability
analysis, we observed that the current implementation
has an exponential increase of running time when the
grid size grows logarithmically. In the future work,
functions with heavy computations should be further
investigated and the limitation on scalability of the
model should be solved using parallel computing.
REFERENCES
Bookholt, F. D., Monsuur, H. N., Gibbs, S., and Vermolen,
F. J. (2016). Mathematical modelling of angiogene-
sis using continuous cell-based models. Biomechanics
and Modeling in Mechanobiology, 15(6):1577–1600.
Borgonovo, E. (2007). A new uncertainty importance
measure. Reliability Engineering & System Safety,
92(6):771–784.
Carmeliet, P. and Jain, R. (2000). Carmeliet, p & jain, rk.
angiogenesis in cancer and other disease. nature, 407:
249-257. Nature, 407:249–57.
Carvalho, R. V., Verbeek, F. J., and Coelho, C. J. (2018).
Bio-modeling Using Petri Nets: A Computational Ap-
proach, pages 3–26. Springer International Publish-
ing, Cham.
Chaouiya, C. (2007). Petri net modelling of biological net-
works. Briefings in Bioinformatics, 8(4):210–219.
Chaouiya, C., Remy, E., and Thieffry, D. (2008). Petri net
modelling of biological regulatory networks. Journal
of Discrete Algorithms, 6(2):165–177. Selected pa-
pers from CompBioNets 2004.
Folkman, J. (2002). Role of angiogenesis in tumor growth
and metastasis. Seminars in Oncology, 29(6, Supple-
ment 16):15–18.
Harris, C. R., Millman, K. J., van der Walt, S. J., Gommers,
R., Virtanen, P., Cournapeau, D., Wieser, E., Taylor,
J., Berg, S., Smith, N. J., Kern, R., Picus, M., Hoyer,
S., van Kerkwijk, M. H., Brett, M., Haldane, A., del
R
´
ıo, J. F., Wiebe, M., Peterson, P., G
´
erard-Marchant,
P., Sheppard, K., Reddy, T., Weckesser, W., Abbasi,
H., Gohlke, C., and Oliphant, T. E. (2020). Array pro-
gramming with NumPy. Nature, 585(7825):357–362.
Hill, C. (2017). A very simple 2-d diffusion model.
Liu, F., Heiner, M., and Gilbert, D. (2017). Coloured Petri
nets for multilevel, multiscale and multidimensional
modelling of biological systems. Briefings in Bioin-
formatics, 20(3):877–886.
Matsuya, K., Yura, F., Mada, J., Kurihara, H., and Toki-
hiro, T. (2016). A discrete mathematical model for
angiogenesis. SIAM Journal on Applied Mathematics,
76(6):2243–2259.
Peirce, S. (2008). Computational and mathematical model-
ing of angiogenesis. Microcirculation (New York, N.Y.
: 1994), 15:739–51.
Phillips, C. M., Lima, E. A. B. F., Woodall, R. T., Brock, A.,
and Yankeelov, T. E. (2020). A hybrid model of tumor
growth and angiogenesis: In silico experiments. Plos
One, 15(4).
Pommereau, F. (2015). SNAKES: A flexible high-level
petri nets library (tool paper). In Application and The-
ory of Petri Nets and Concurrency, pages 254–265.
Springer International Publishing.
Querfeld, U., Mak, R., and Pries, A. (2020). Microvascular
disease in chronic kidney disease: The base of the ice-
berg in cardiovascular comorbidity. Clinical Science,
134:1333–1356.
Sainson, R. and Harris, A. (2008). Regulation of angiogen-
esis by homotypic and heterotypic notch signalling in
Modeling iPSC-Derived Endothelial Cell Transition in Tumor Angiogenesis Using Petri Nets
345

endothelial cells and pericytes: From basic research to
potential therapies. Angiogenesis, 11:41–51.
Sobol’, I. and Kucherenko, S. (2009). Derivative based
global sensitivity measures and their link with global
sensitivity indices. Mathematics and Computers in
Simulation, 79(10):3009–3017.
Toi, M., Bando, H., Ogawa, T., Muta, M., Hornig, C., and
Weich, H. (2002). Significance of vascular endothelial
growth factor (vegf)/soluble vegf receptor-1 relation-
ship in breast cancer. International Journal of Cancer
- INT J CANCER, 98:14–18.
Valentim, R. A. M., Caldeira-Silva, G. J. P., da Silva, R. D.,
Albuquerque, G. A., de Andrade, I. G. M., Sales-
Moioli, A. I. L., Pinto, T. K. d. B., Miranda, A. E.,
Galv
˜
ao-Lima, L. J., Cruz, A. S., Barros, D. M. S., and
Rodrigues, A. G. C. D. R. (2022). Stochastic petri net
model describing the relationship between reported
maternal and congenital syphilis cases in brazil. BMC
Medical Informatics and Decision Making, 22(1).
van Duinen, V., Stam, W., Borgdorff, V., Reijerkerk, A.,
Orlova, V., Vulto, P., Hankemeier, T., and van Zon-
neveld, A. J. (2019). Standardized and scalable as-
say to study perfused 3d angiogenic sprouting of ipsc-
derived endothelial cells in vitro. Journal of Visualized
Experiments, (153).
Vilanova, G., Colominas, I., and Gomez, H. (2017). A
mathematical model of tumour angiogenesis: growth,
regression and regrowth. Journal of The Royal Society
Interface, 14(126):20160918.
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
346
