
Detection of Non-Coding RNA (Hsa_circ_0003416) in Pulmonary
Arterial Hypertension Patients’ Plasma by qRT-PCR Analysis
Fajri Marindra Siregar
1,2 a
, Dicka Wahyu Setiasari
3b
, Anggoro Budi Hartopo
4,* c
,
Lucia Kris Dinarti
4d
and Sofia Mubarika Haryana
5e
1
Doctorate Program, Faculty of Medicine, Public Health and Nursing, Gadjah Mada University, Yogyakarta, Indonesia
2
Department of Biochemistry, Faculty of Medicine, Universitas Riau, Pekanbaru, Indonesia
3
Faculty of Medicine, Public Health and Nursing, Gadjah Mada University, Yogyakarta, Indonesia
4
Department of Cardiology and Vascular Medicine, Faculty.of Medicine, Public Health and Nursing,
Universitas Gadjah Mada–Dr. Sardjito Hospital, Yogyakarta, Indonesia
5
Department of Histology and Cell Biology, Faculty of Medicine, Public Health and Nursing,
Gadjah Mada University, Yogyakarta, Indonesia
Keywords: Circular RNA, hsa_circ_0003416, Pulmonary Arterial Hypertension, Non-Coding RNA, qRT-PCR.
Abstract: Circular RNA refers to a type of non-coding RNA molecule and possesses a circular conformation. Circular
RNA has greater stability compared to linear RNA due to its resistance to RNAse activity. hsa_circ_0003416
is a circular RNA reported to have potential as a biomarker in patients with pulmonary arterial hypertension
(PAH). This study aims to detect hsa_circ_0003416 using qRT-PCR in the plasma of PAH patients. The
sample was taken from RSUP Dr. Sardjito Yogyakarta. The miRNeasy Serum/Plasma Advanced kit was
used to isolate total RNA. Following that, cDNA was generated with an Applied Biosystems thermal cycler
and the ExcelRT
TM
Reverse Transcription Kit II reagent, and qPCR was performed with an Applied
Biosystems
TM
7500 Real-Time PCR equipment and the SensiFAST
TM
SYBR® Lo-ROX Kit reagent.
Specific divergent primers were utilized. Melting curve analysis and visualization of the amplified products
were performed using gel electrophoresis. The qRT-PCR technique achieved a single amplification, resulting
in a melting curve value observed at 81.8
o
C. Examination by visualizing the gel electrophoresis results
showed a single band that matched the target size, specifically 124 base pairs. In conclusion, the qRT-PCR
analysis successfully identified the presence of hsa_circ_0003416 in the plasma of PAH patients.
1 INTRODUCTION
Circular RNA or circRNA is a unique form of RNA
molecule that does not code for proteins (non-coding
RNA) (Greene et al., 2017). It is conserved across
different species and exhibits a strong preference for
specific tissues and cells (Z. Yu et al., 2021).
Additionally, circRNA can be identified in the
peripheral blood (Wen et al., 2021). Circular RNA
has enhanced stability compared to other RNA types
due to its closed-loop structure, resulting in an
extended half-life (Sharma et al., 2021). The
attributes above make circRNA a promising
a
https://orcid.org/0000-0002-1239-7338
b
https://orcid.org/0009-0007-4646-5216
c
https://orcid.org/0000-0002-6373-1033
d
https://orcid.org/0000-0002-4011-2184
e
https://orcid.org/0000-0001-7205-652X
candidate for future use as noninvasive and liquid
biopsy biomarkers.
Circular RNAs can modulate gene expression
both during and after transcription by interacting with
microRNAs (miRNAs) or RNA-binding proteins
(C.-Y. Yu & Kuo, 2019). They play a part in a diverse
array of biological processes. Newly available data
uncovers complex connections between several
forms of RNA, including protein-coding messenger
RNAs and non-coding RNAs (Sharma et al., 2021).
The presence of this circRNA-miRNA-mRNA
regulatory pathway is associated with the
pathogenesis of numerous diseases, including
Siregar, F. M., Setiasari, D. W., Hartopo, A. B., Dinarti, L. K. and Haryana, S. M.
Detection of Non-Coding RNA (Hsa_circ_0003416) in Pulmonary Arterial Hypertension Patients’ Plasma by qRT-PCR Analysis.
DOI: 10.5220/0013672000003873
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 1st International Conference on Medical Science and Health (ICOMESH 2023), pages 335-339
ISBN: 978-989-758-740-5
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
335

pulmonary arterial hypertension (PAH). (Tang et al.,
2021; Wu et al., 2022).
Dysregulated circRNA expression may lead to
the development of PAH. A study found that
hsa_circ_0003416 was markedly reduced in children
with PAH associated with congenital heart diseases
(CHD) and has potential as a biomarker (Huang et al.,
2022). An in-silico study has discovered that
hsa_circ_0003416 is a circRNA molecule that is 124
base pairs in length. It originates from exon three of
the thymosin beta 4 X-linked gene (TMSB4X)
(Huang et al., 2022). It plays a crucial function in
regulating the process of angiogenesis (Huang et al.,
2022). There is a scarcity of studies on the expression
of hsa_circ_0003416 in PAH associated with CHD.
Furthermore, the utilization of qRT-PCR for the
analysis of circular RNA remains unfamiliar,
particularly in Indonesia. This study aims to detect
hsa_circ_0003416 using qRT-PCR approaches from
the plasma of PAH patients.
2 MATERIALS AND METHODS
The study was conducted in the Integrated Research
Laboratory of the Faculty of Medicine, Public
Health, and Nursing at Universitas Gadjah Mada
(UGM). The specimens utilized were plasma samples
obtained from patients diagnosed with PAH
associated with CHD, who had an examination at
RSUP Dr. Sardjito Yogyakarta and were
subsequently enrolled in the Congenital HeARt
Disease in adult and Pulmonary Hypertension
(COHARD-PH) registry (Dinarti et al., 2020).
Medical And Health Research Ethics Committee
(MHREC) Faculty of Medicine, Public Health and
Nursing UGM approved this study. (number
KE/FK/0429/EC/2023)
2.1 RNA Isolation
The RNA isolation process utilised the miRNeasy
Serum/Plasma Advanced kit from Qiagen [217204].
The sample volume utilized was 200 µl. Next, the
isolation technique is executed in accordance with
the manufacturer's protocol. The isolation results
were further analyzed using a nanodrop to ascertain
the concentration and purity of RNA.
2.2 Synthesis of cDNA
The cDNA synthesis was performed using the
ExcelRT™ Reverse Transcription Kit II from
Smobio [RP1400], following the manufacturer's
instructions. The RNA templates utilized were 50 ng.
The cDNA synthesis method was conducted using an
Applied Biosystems thermal cycler, the protocol is as
follows: incubation step at 25
o
C (10 minutes),
reverse-transcription step at 42
o
C (50 minutes), and
inactivation of reaction 85
o
C (5 minutes).
2.3 qPCR hsa_circ_0003416
The SensiFAST™ SYBR® Lo-ROX Kit [BIO-
94005] was employed in accordance with the
manufacturer's instructions, utilising the Applied
Biosystems™ 7500 Real-Time PCR instrument. The
cDNA product was diluted in a ratio of 1:5 before
being used as a template. The primers used are
Forward divergent
CCCCTTTCACACATCAAAGAAC and Reverse
divergent ATTTAAACTTGATCCAACATGC
(Huang et al., 2022). The primer concentration used
was 400nM with a total reaction volume of 20 µl. The
qPCR protocol is as follows: polymerase activation at
95
o
C (2 minutes), 40 cycles of denaturation step at
95
o
C (5 seconds), and annealing/extension step at
60
o
C (30 seconds). The amplicons were subjected to
melting curve analysis and visualization using gel
electrophoresis when they were completed.
3 RESULTS
A study was done using the protocol described in the
methods section. The findings from the
hsa_circ_0003416 analysis presented in Figure 1.
According to the qRT-PCR technique employed, a
single amplification was achieved, resulting in a
melting curve value observed at a temperature of
81.8
o
C.
Subsequently, gel electrophoresis was conducted
to verify the amplification product. The visualization
results with gel electrophoresis are shown in Figure
2. There is a visible band within the sample that is
about 124 base pairs in length.
4 DISCUSSIONS
A qRT-PCR study was performed to amplify the
hsa_circ_0003416 target in the plasma of patients
diagnosed with PAH associated with CHD at Dr.
RSUP. Sardjito Yogyakarta. Circular RNA is a
circular-shaped non-coding RNA. Unlike linear
RNA, the analysis of circRNA requires the utilization
of random primers during the cDNA synthesis step
(Panda
et al., 2017). The subsequent phase involves
ICOMESH 2023 - INTERNATIONAL CONFERENCE ON MEDICAL SCIENCE AND HEALTH
336
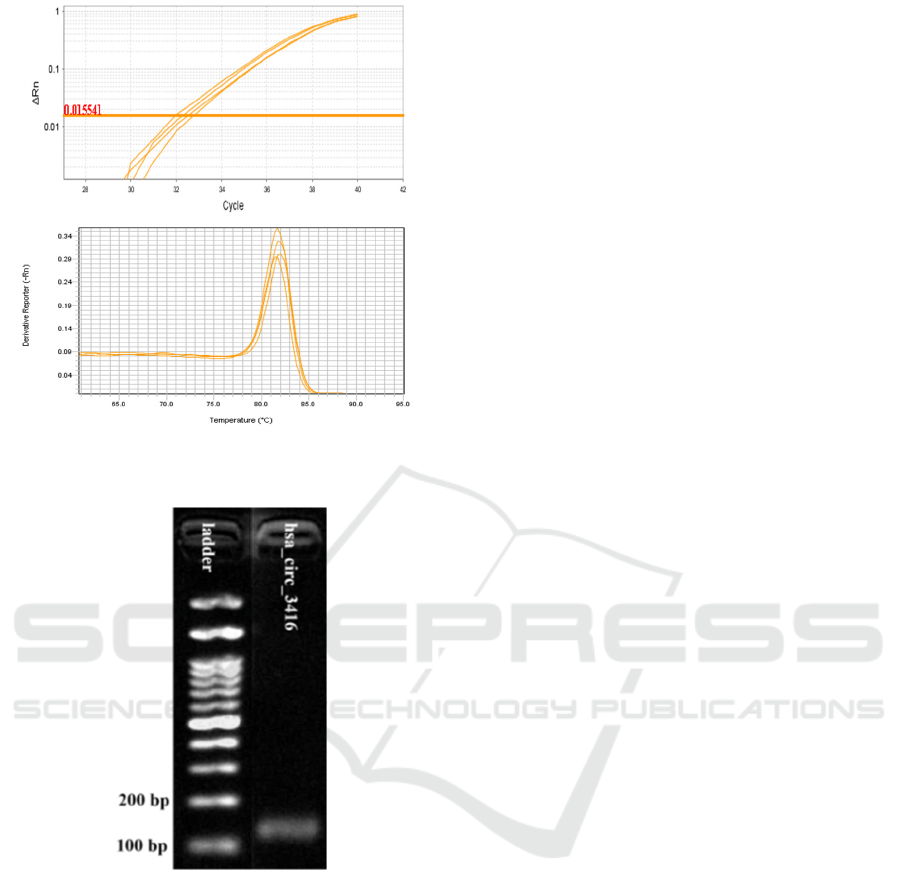
Figure 1: qPCR amplification (top) and melting curves
(bottom) of hsa_circ_0003416.
Figure 2: Visualization with 2% gel electrophoresis.
the utilization of a unique primer known as a
divergent primer. Divergent primers are designed to
amplify a specific portion of circRNA, known as the
backsplice junction, by working in the opposite
manner as shown in Figure 3 (Dieterich &
Papantonis, 2018). Amplification will take place if
the cDNA template utilized includes the desired
circRNA.
The approach we employ, while able to identify
the target circRNA, has limitations, specifically that
the target circRNA is detected at a relatively high
cycle threshold value (CT Value) of approximately
32. These results suggest a low abundance of
circRNA targets in the cDNA template. Therefore,
additional steps, such as the circRNA purification
procedure, must be considered. The RNase R
treatment might enhance the presence of circular
RNAs in an RNA sample. This strategy can
effectively eliminate linear RNAs (Xiao & Wilusz,
2019).
A single peak in the melting curve analysis
signifies the existence of a distinct qPCR product.
Subsequently, the qPCR product can be observed by
gel electrophoresis. Essentially, the amplicons
generated by qPCR will migrate across the medium
and come to a halt at a specific location based on their
size. A band will be formed at the location where
these amplicons accumulate, which will be visible
(Rana & Joshi, 2023). Thus, the size of the amplicon
generated by the qPCR reaction can be determined
using gel electrophoresis. The investigation revealed
that the length of hsa_circ_0003416 was 124 base
pairs. The acquired gel electrophoresis results
showed a solitary band within the 100-150 bp range,
confirming the findings. Nevertheless, a sequencing
analysis is required to validate this result (Dieterich
& Papantonis, 2018).
This study aims to verify the qRT-PCR method
for assessing the expression of the hsa_circ_0003416
gene in patients with PAH at RSUP Dr. Sardjito
Yogyakarta. It is a preliminary research report.
Subsequently, all research samples will undergo
examination in order to acquire expression patterns.
Patients with PAH may have an aberrant regulation
of hsa_circ_0003416 expression, which could play a
role in the development of the disease.
Detection of Non-Coding RNA (Hsa_circ_0003416) in Pulmonary Arterial Hypertension Patients’ Plasma by qRT-PCR Analysis
337
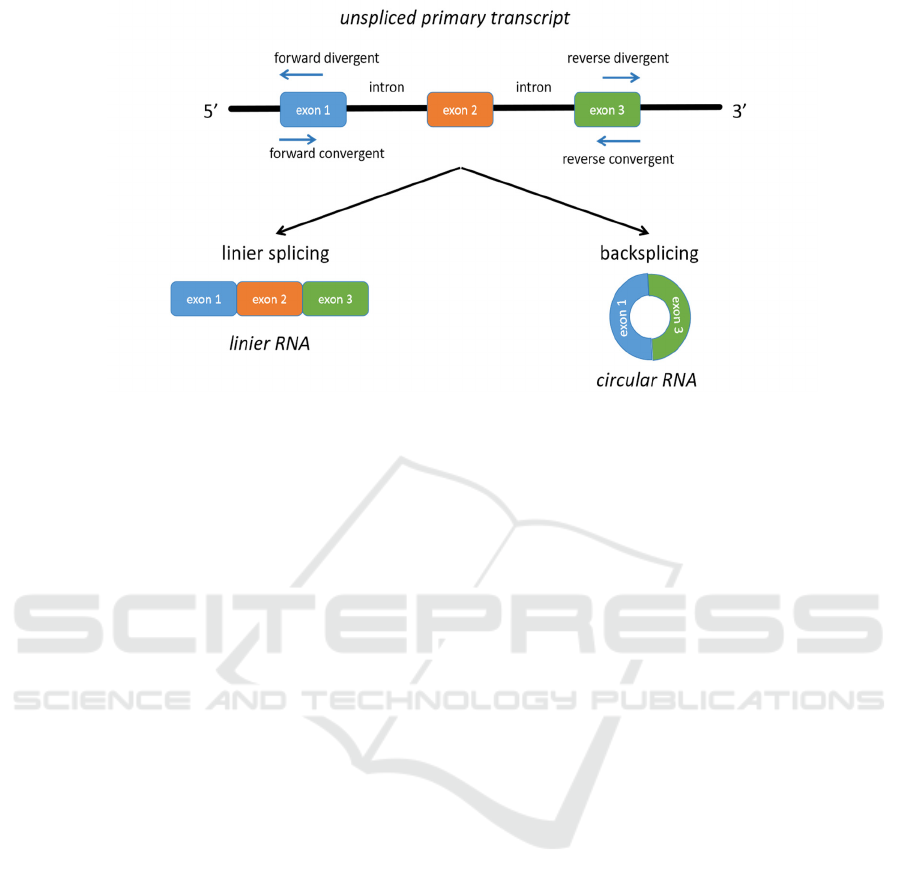
Figure 3: The linear RNA that is made by linear splicing is shown on the left, and the circRNA that is made by back splicing
is shown on the right. These two types of primaries (convergent and divergent) work in different directions
5 CONCLUSIONS
The qRT-PCR approach, as described in the protocol,
is capable of detecting hsa_circ_0003416 in the
plasma of patients diagnosed with PAH associated
with CHD at RSUP Dr. Sardjito Yogyakarta.
Additional research, utilizing a sufficient number of
samples, is required to ascertain the expression
pattern of hsa_circ_0003416.
ACKNOWLEDGEMENTS
This work was supported by Hibah Penelitian Dana
Masyarakat (Damas) FKKMK UGM 2023. We also
thank the PAH team from FKKMK UGM and RSUP
Dr. Sardjito Yogyakarta for their invaluable support
in supplying the samples.
REFERENCES
Dieterich, C., & Papantonis, A. (2018). Circular RNAs
methods and protocols. Methods in Molecular Biology,
1724. https://doi.org/10.1007/978-1-4939-7562-4
Dinarti, L. K., Hartopo, A. B., Kusuma, A. D., Satwiko, M.
G., Hadwiono, M. R., Pradana, A. D., & Anggrahini,
D. W. (2020). The COngenital HeARt Disease in adult
and Pulmonary Hypertension (COHARD-PH) registry:
a descriptive study from single-center hospital registry
of adult congenital heart disease and pulmonary
hypertension in Indonesia. BMC Cardiovascular
Disorders, 20(1), 163. https://doi.org/10.1186/s12872-
020-01434-z
Greene, J., Baird, A.-M., Brady, L., Lim, M., Gray, S. G.,
McDermott, R., & Finn, S. P. (2017). Circular RNAs:
biogenesis, function and role in human diseases.
Frontiers in Molecular Biosciences, 4, 38.
https://doi.org/10.3389/fmolb.2017.00038
Huang, Y., Su, D., Ye, B., Huang, Y., Qin, S., Chen, C.,
Zhao, Y., & Pang, Y. (2022). Expression and clinical
significance of circular RNA hsa_circ_0003416 in
pediatric pulmonary arterial hypertension associated
with congenital heart disease. Journal of Clinical
Laboratory Analysis, 36(4), e24273.
https://doi.org/10.1002/jcla.24273
Panda, A. C., Abdelmohsen, K., & Gorospe, M. (2017).
RT-qPCR detection of senescence-associated circular
RNAs. Methods in Molecular Biology, 1534, 79–87.
https://doi.org/10.1007/978-1-4939-6670-7_7
Rana, B., & Joshi, G. K. (2023). Electrophoresis: Basic
principle, types, and applications. In Basic
biotechniques for bioprocess and bioentrepreneurship
(pp. 183–193). Academic Press.
https://doi.org/10.1016/B978-0-12-816109-8.00011-8
Sharma, A. R., Bhattacharya, M., Bhakta, S., Saha, A., Lee,
S.-S., & Chakraborty, C. (2021). Recent research
progress on circular RNAs: Biogenesis, properties,
functions, and therapeutic potential. Molecular
Therapy Nucleic Acids, 25, 355–371.
https://doi.org/10.1016/j.omtn.2021.05.022
Tang, Y., Bao, J., Hu, J., Liu, L., & Xu, D.-Y. (2021).
Circular RNA in cardiovascular disease: Expression,
mechanisms and clinical prospects. Journal of Cellular
and Molecular Medicine, 25(4), 1817–1824.
https://doi.org/10.1111/jcmm.16203
Wen, G., Zhou, T., & Gu, W. (2021). The potential of using
blood circular RNA as liquid biopsy biomarker for
ICOMESH 2023 - INTERNATIONAL CONFERENCE ON MEDICAL SCIENCE AND HEALTH
338

human diseases. Protein & Cell, 12(12), 911–946.
https://doi.org/10.1007/s13238-020-00799-3
Wu, M., Xun, M., & Chen, Y. (2022). Circular RNAs:
regulators of vascular smooth muscle cells in
cardiovascular diseases. Journal of Molecular
Medicine. https://doi.org/10.1007/s00109-022-02186-
3
Xiao, M.-S., & Wilusz, J. E. (2019). An improved method
for circular RNA purification using RNase R that
efficiently removes linear RNAs containing G-
quadruplexes or structured 3’ ends. Nucleic Acids
Research, 47(16), 8755–8769.
https://doi.org/10.1093/nar/gkz576
Yu, C.-Y., & Kuo, H.-C. (2019). The emerging roles and
functions of circular RNAs and their generation.
Journal of Biomedical Science, 26(1), 29.
https://doi.org/10.1186/s12929-019-0523-z
Yu, Z., Huang, Q., Zhang, Q., Wu, H., & Zhong, Z. (2021).
CircRNAs open a new era in the study of
cardiovascular disease (Review). International Journal
of Molecular Medicine, 47(1), 49–64.
https://doi.org/10.3892/ijmm.2020.4792
Detection of Non-Coding RNA (Hsa_circ_0003416) in Pulmonary Arterial Hypertension Patients’ Plasma by qRT-PCR Analysis
339
