
Novel Primers for Easy Detection of Yellow Fever Using Isothermal
PCR
Naufal Dimas Lingga Takbirrahman, Dea Syafira Alamsyah Sitompul
*
, Nicholas Gabriel Harefa
and Arief Budi Witarto
Department of Biology Cell and Molecular, Faculty of Military Medicine, The Republic of Indonesia Defense University,
IPSC Sentul, Bogor, 16810, Indonesia
Keywords: Yellow Fever Virus, Isothermal PCR, NS2a, NS5
Abstract: Yellow Fever is a disease caused by the Yellow Fever Virus (YFV), common in tropical areas in Africa and
South America. Hundreds of Indonesian soldiers are sent on peacekeeping missions to Congo -an endemic
region for YFV- every year. Therefore, an accurate technique is needed to screen those who possibly get
infections. Isothermal PCR, unlike conventional PCR, uses a single temperature with 4 to 6 combinations of
primers that can be performed with simple equipment such as a heat block and detected with a visual eye
instead of agarose electrophoresis. Thus, it is suitable for this challenge. YFV strain 17D genome was used
and particularly NS2a and NS5 genes were chosen as template for the design of primers. To check for
specificity, obtained primers were checked for sequence similarity using BLASTN. Literature studies showed
that various genes, including E and NS5, have been used for the target using conventional PCR. Meanwhile,
for isothermal PCR, only the NS1 gene and upstream non-coding gene have been used. From the NS2a gene
with length of 672 bp, 2 sets of primers were obtained. At the same time, NS5, with a longer length of 2,714
bp, gave 4 sets of primers. BLASTN results showed that all primers were specific to the YFV genome.
Original primers have been successfully designed for isothermal PCR of yellow fever. Primers designed are
the first step and also the most critical original works in nucleic acid testing. Therefore, towards self-sufficient
diagnostic reagents, the results are significant.
1 INTRODUCTION
1.1 Yellow Fever Virus
Yellow Fever is a disease caused by the Yellow Fever
Virus (YFV), which is common in tropical areas in
Africa and South America. The disease primarily
affects humans and nonhuman primates and is
transmitted through the bite of an infected mosquito.
Forty-seven countries in Africa, Central and South
America are either endemic for Yellow Fever Virus
(YFV) or have endemic areas. A modelling study
based on African data sources estimated the incidence
of Yellow Fever during 2013 was 84,000-170,000
severe cases and 29,000-60,000 deaths (Gardner et al,
2010).
Every year, hundreds of Indonesian soldiers are
sent on a peacekeeping mission to Congo - an
endemic region for YFV - who trained before
departure and returned home first to IPSC, where The
Republic of Indonesia Defense University is located.
Therefore, screening for possible YFV infection
among returning soldiers with an easy yet accurate
technique is necessary. Isothermal PCR, unlike
conventional PCR, uses single temperature and with
4 to 6 combinations of primers can be performed with
simple equipment such as a heat block, and can be
detected with a visual eye instead of using agarose
electrophoresis. Thus, it is suitable for this challenge
(Gardner et al, 2010).
1.2 Yellow Fever Virus Complete
Genome
Yellow Fever Virus is a single-stranded RNA virus
consisting of 10 constituent genes, namely 3
structural genes (C, prM, and E) and 7 nonstructural
genes (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and
NS5). A third of the YFV genome is encoded by 3
structural proteins, while two-thirds comprises 7
nonstructural proteins. One of the most targeted
Takbirrahman, N. D. L., Sitompul, D. S. A., Harefa, N. G. and Witarto, A. B.
Novel Primers for Easy Detection of Yellow Fever Using Isothermal PCR.
DOI: 10.5220/0013671600003873
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 1st International Conference on Medical Science and Health (ICOMESH 2023), pages 299-302
ISBN: 978-989-758-740-5
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
299

genomes for many researchers is Yellow Fever Virus-
17D, which is also very effective as a vaccine to form
antibodies against YFV itself. The genome was seen
as the most similar to the original YFV genome, so
that genome is used as the target sequence for this
test. Although further research is still needed on it,
and we must keep up to date with the latest
information about the virus (Nunes et al, 2015).
Among the 10 genes in the entire YFV genome,
NS2a is a small hydrophobic protein, one of the
nonstructural proteins of YFV and plays a role in its
virulence factors. Because this gene is essential for
the virus, we made it one of the target genes in the test
that we carried out. In addition, NS5 is another
protein in the YFV genome that has been the target of
molecular screening research that can be used as a
primer for PCR tests. It is undoubtedly in line with
our test's purpose, and is why NS5 is also one of the
target genes in this paper (Voßmann et al, 2015 and
Rezende et al, 2019).
1.3 Advantages of The LAMP Method
The LAMP (Loop-mediated isothermal amplification
of DNA) method was designed in 1998 by a Japanese
company. This technique increases the amount of
DNA amplified to one billion copies in less than an
hour and is very specific. Isothermal amplification
can be performed without sophisticated laboratory
equipment. Another innovative aspect of LAMP is its
high specificity due to the use of multiple primers
(from four to six), which can distinguish up to eight
specific sites on the DNA template (Keikha, 2018,
Wong et al, 2018, and Soroka et al, 2021).
Primary LAMP design can be done using online
software such as Primer Explorer
(https://primerexplorer.jp/e/), LAMP Designer
Optigene (www.optigene.co.uk/lamp-designer), or
Premier Biosoft
(http://www.premierbiosoft.com/isothermal/lamp.ht
ml), maybe using the neb lamp
(https://lamp.neb.com/#!/) can be done either which
was used in this experiment (Wong et al, 2018 and
Soroka et al, 2021).
The LAMP technique does not involve DNA
denaturation because, due to the strand, as mentioned
earlier, transfer activity of the Bst DNA polymerase,
the reaction can be carried out under isothermal
conditions. All LAMP steps were done at a stable
temperature of 60–65 °C, eliminating the need to use
a thermocycler to precisely adjust the thermal and
time profiles, which are required for commonly
known PCR techniques (Keikha, 2018, Wong et al,
2018, and Soroka et al, 2021).
The Loop-Mediated Isothermal Amplification
method has been used to diagnose various infectious
diseases and identify and differentiate pathogenic
microorganisms; for example, Mycobacterium
tuberculosis, Nocardia spp., Pseudomonas
fluorescens, Staphylococcus aureus, Helicobacter
pylori, Salmonella species, and several other bacteria
and medically necessary viruses (Keikha, 2018).
2 MATERIALS AND METHODS
This study uses the insilico method, which uses
software and computational techniques to test the
scientific data we get from the existing literature.
First, we look for diseases prone to occur in the
Garuda Contingent sent to Congo by looking for
literature showing the prevalence of viral illness in
that country, by looking at the possibility of this viral
disease occurrence in our country. In addition,
Yellow Fever Virus has the same intermediate host as
the dengue virus, namely the Aedes aegypti
mosquitoes. It is necessary to take precautions so our
country does not become a yellow fever virus
endemic area (Gardner et al, 2010).
After obtaining data on this viral disease in the
country, we determined the genome of the Yellow
Fever Virus, YFV strain 17D genome (Genbank entry
NC_002031), which will be the target of this research
through existing journals. We found that the genome
is most similar to the original viral genome, primarily
found in Africa, and the most used as the object of
research (Nunes et al, 2015).
Next, we searched for the target gene in the
genome by searching literature which states that the
NS2a gene has a significant role in the virulence
factor of the virus. In addition, we chose the NS5 gene
because it has become the target gene in molecular
screening literature using PCR (Voßmann et al, 2015
and Rezende et al, 2019).
Then, we searched for the genome sequence at
https://www.ncbi.nlm.nih.gov/ using the Genbank
feature. Following that procedure, we take the
genome into FASTA format, obtain the complete line
of the genome, and input the sequence into the
Microsoft Word application.
After that, using https://lamp.neb.com/#!/, we
entered the gene sequence and determined the
location of the base pair sequence, which we will
make into a primer. After completing the process, we
obtained 6 new primers and confirmed that the base
pair sequence of the reverse primer and backward
primer had been reversed and complemented, which
are in the viral genome sequence in Microsoft Word.
ICOMESH 2023 - INTERNATIONAL CONFERENCE ON MEDICAL SCIENCE AND HEALTH
300
Then, the specificity of those 6 primers was tested
for the viral genome at
https://blast.ncbi.nlm.nih.gov/Blast.cgi, and the
results were those 6 primers specific for the Yellow
Fever Virus genome.
3 RESULTS
Literature studies showed that various genes,
including E and NS5 (Nunes et al, 2011), have been
used for targets using conventional PCR. Meanwhile
for isothermal PCR, only NS1 gene (Nunes et al,
2015 and Kwallah et al, 2013) and upstream non-
coding gene (Escadafal et al, 2014) have been used.
From the NS2a gene with length of 672 bp, 2 sets of
primers were obtained. First set, start from nc 3,636
and total region length of 207 bp. While the second
set, from nc 3,956 and total region length of 207 bp.
While NS5 with a longer length of 2,714 bp gave 4
sets of primers. Each starts from nc 8,159; 8,714;
9,589; 9,776 and total region length of 218 bp, 222
bp, 172 bp, and 234 bp respectively. BLASTN results
showed that all primers were specific to the YFV
genome. At the end, we have 6 primers that are
eligible and have a high possible success rate to be
the primers for the purpose of this experiment which
has been mentioned before.
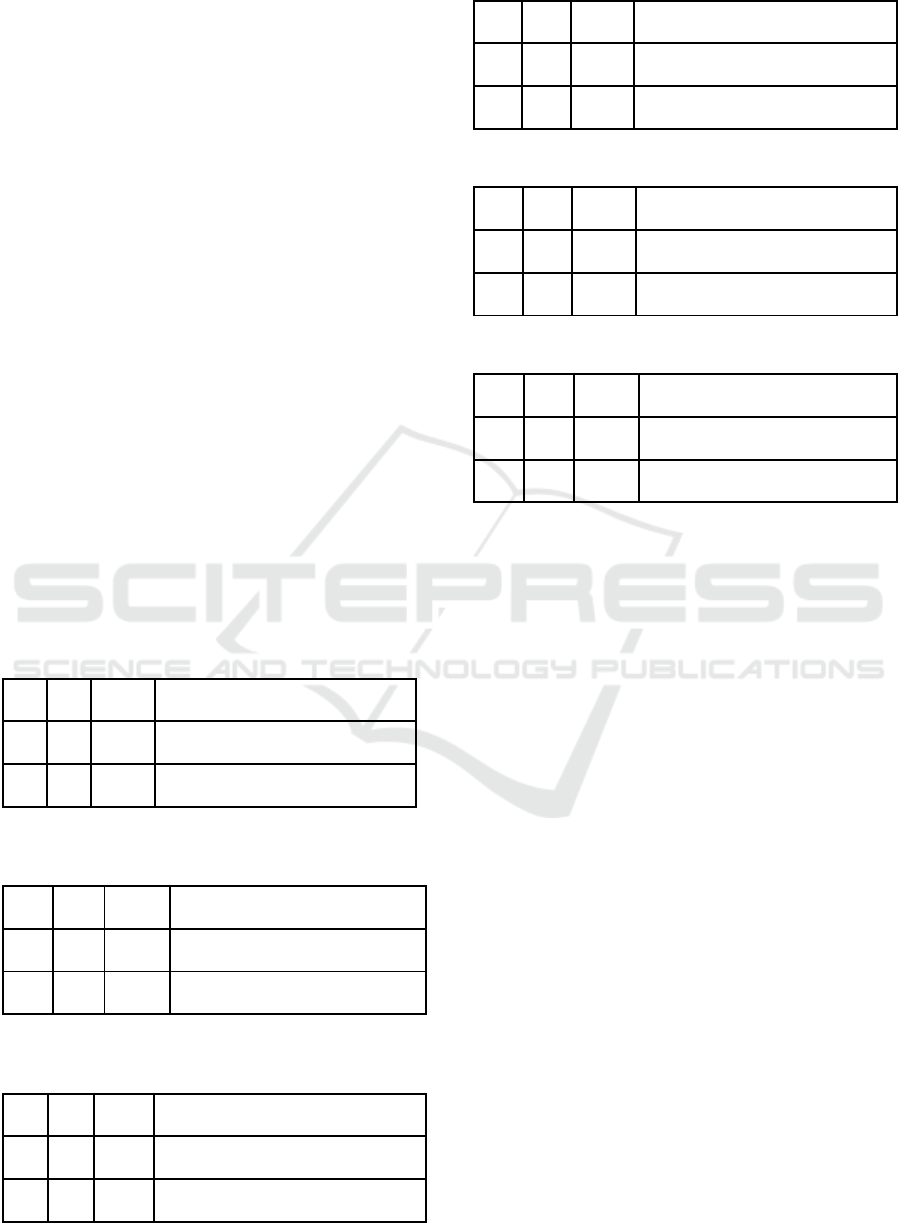
Table 1: Primer 1.
len Tm
F3 20 60.22
TCGGGCAAGTAACTCTCCTT
B3 18 60.55 CCACCATGGCTGCTCCTA
Table 2: Primer 2.
len Tm
F3 19 60.89 CCCCTCATGGCTCTGTTGA
B3 19 59.24 GGTTGCCAGAAATGCACAC
Table 3: Primer 3.
len T
m
F3 20 59.70 GTTGACACCAGAGCAAAGGA
B3 20 59.17 CCAGAACTTTGGGTCTTGGA
Table 4: Primer 4.
len Tm
F3 20 59.68 GGGGTTGACAACTTCTGTGT
B3 19 60.52 GGACGCCTCATTCTCCTCA
Table 5: Primer 5.
len Tm
F3 20 60.58 TCCCACCACTTCCATGAACT
B3 18 59.33 CCATGAGGTGGGAACAGC
Table 6: Primer 6.
len Tm
F3 19 59.92 CACTGAGCACGGATGTGAC
B3 20 59.10 CTCCCAATCATTCCACCCTT
4 DISCUSSION
For screening purposes, primers design is the first
step and is crucial for making a diagnostic tool that
can be used in the field. In its application, other
reagents such as enzymes and other chemicals may
use existing ones. However, with different primers,
the purpose or function of the diagnostic tool will also
be different. Isothermal PCR is not a common
diagnostic tool, especially in the military medical
environment in Indonesia. It is very suitable for the
conditions of the military environment in Indonesia,
so it can easily apply this diagnostic test (Keikha,
2018, Wong et al, 2018, and Soroka et al, 2021).
Primers design, as the first step, has been
completed. Indeed, it will continue with the
manufacture of prototypes, in vitro testing, until it is
proven that screening using isothermal PCR is
possible by attaching data obtained directly from all
the tests. Suppose all series of tests have been carried
out, and it is found that the primers we obtained
computationally can be used as primers in this
diagnostic tool. In that case, a new faster, easier, and
cheaper way to detect the Yellow Fever Virus in
Indonesia will be obtained (Nunes et al, 2015).
Indeed, it has a good impact on military medicine
in Indonesia, because it proves that Indonesia can
already produce its own diagnostic tool for a virus
Novel Primers for Easy Detection of Yellow Fever Using Isothermal PCR
301

that can quickly spread in Indonesia because of our
soldiers who go directly to its endemic areas. This is
a big step that can be used as an opening door for the
advancement of military medicine, especially in the
molecular field, so that we can become a producer
independently, not just a consumer who depends on
other countries.
5 CONCLUSIONS
Yellow Fever is a disease caused by the Yellow Fever
Virus, which is common in tropical areas in Africa
and South America. By easy yet accurate technique,
there is a need to screen for possible YFV infection
among returning soldiers from the YFV endemic
countries such as Congo. Original primers have been
successfully designed for isothermal PCR of yellow
fever. Isothermal PCR/LAMP is an easy and
inexpensive technology that is rarely used, but it can
be used to screen for Yellow Fever disease by
detecting the base pairs in the viral DNA genome.
Primers designed are the first step and also the most
critical original works in nucleic acid testing. From
this work, 6 gene primers were obtained that could be
used as primers for the planned screening. Therefore,
towards self-sufficient diagnostic reagents, the results
are significant.
REFERENCES
Gardner, CL., Ryman, KD., 2010. Yellow fever: a
Reemerging threat. Clin Lab Med, 30(1), 237-60.
Nunes, MRT., Vianez, JL., Nunes, KNB., da Silva, SP.,
Lima, CPS., Guzman, H., Martins, LC., Carvalho, VL.,
Tesh, RB., Vasconcelos, PFC., 2015. Analysis of a
Reverse Transcription Loop-mediated Isothermal
Amplification (RT-LAMP) for yellow fever diagnostic.
Journal of Virological Methods, 226, 40-51.
Voßmann, S., Wieseler, J., Kerber, R., Kümmerer, BM.,
2015. A basic cluster in the N terminus of yellow fever
virus NS2A contributes to infectious particle
production. J Virol, 89(9), 4951-65.
Rezende, IM., Alves, PA., Arruda, MS., Gonçalves, AP.,
Oliveira, GFG., Pereira, LS., Dutra, MRT., Campi-
Azevedo, AC., Valim, V., Tourinho. R., Oliveira, JG.,
Calzavara, CE., Said, RFDC., Kroon, EG., Martins-
Filho, OA., Teixeira-Carvalho, A., Drumond, BP.,
2019. Yellow Fever Virus Genotyping Tool and
Investigation of Suspected Adverse Events Following
Yellow Fever Vaccination. Vaccines (Basel), 7(4), 206.
Keikha, M., 2018. LAMP Method as One of the Best
Candidates for Replacing with PCR Method. Malays J
Med Sci, 25(1), 121-123.
Wong, YP., Othman, S., Lau, YL., Radu, S., Chee, HY.,
2018. Loop-mediated isothermal amplification
(LAMP): a versatile technique for detection of
microorganisms. J Appl Microbiol, 124(3), 626-43.
Soroka, M., Wasowicz, B., Rymaszewska, A., 2021. Loop
Mediated Isothermal Amplification (LAMP): The
Better Sibling of PCR? Cells, 10(8), 1931.
Nunes, MR., Palacios, G., Nunes, KN., Casseb, SM.,
Martins, LC., Quaresma, JA., Savji, N., Lipkin, WI.,
2011. Vasconcelos PF. Evaluation of two molecular
methods for the detection of Yellow fever virus
genome. J Virol Methods, 174(1-2), 29-34.
Kwallah, Ao., Inoue, S., Muigai, AW., Kubo, T., Sang, R.,
Morita, K., Mwau, M., 2013. A real-time reverse
transcription loop-mediated isothermal amplification
assay for the rapid detection of yellow fever virus. J
Virol Methods, 193(1), 23-7.
Escadafal, C., Faye, O., Sall, AA., Faye, O., Weidmann, M.,
Strohmeier, O., von Stetten, F., Drexler, J., Eberhard,
M., Niedrig, M., Patel, P., 2014. Rapid molecular
assays for the detection of yellow fever virus in low-
resource settings. PLoS Negl Trop Dis, 8(3), e2730.
ICOMESH 2023 - INTERNATIONAL CONFERENCE ON MEDICAL SCIENCE AND HEALTH
302
