
In Silico Virtual Screening Studies Using Molecular Docking of
Isoflavonoid Compounds as Potential Antimalarials on the
Plasmodium Falciparum Dihydroorotate Dehydrogenase
(PfDHODH) Enzyme
Semuel Sandy
*
Research Center for Public Health and Nutrition, National Research and Innovation Agency, Kawasan Kerja Bersama
(KKB) BRIN Jayapura, Jl. Isele Kampung Waena, Distrik Heram, Jayapura Selatan, 99358, Papua, Indonesia
Keywords: Isoflavonoids, Malaria, Enzyme, Drug, Docking Simulations.
Abstract: Malaria remains a health problem in Indonesia, leading to an increase in morbidity and mortality. The use of
antimalarial drugs has been reported to result in cases of resistance in several Plasmodium spp. Therefore,
there is a need for the discovery and development of new drugs to address the broader impact of drug
resistance. The aim of this study is to screen flavonoid compounds from plants that have the potential to be
developed as antimalarial agents. Virtual screening method is employed using flavonoid compounds obtained
from secondary metabolites database server (http://pscdb.appsbio.utalca.cl/viewIndex/index.php). The
chemical structures of the flavonoid compounds are then prepared and optimized. The macromolecule used
in the docking simulation is the Plasmodium falciparum dihydroorotate dehydrogenase (PfDHODH) enzyme,
which plays a role in the synthesis of pyrimidine and purine in the parasite's DNA. The protein structure
preparation and optimization are performed with 3D protonation using the AMBER10HT force field in the
MOE 2015 application. Docking simulations are carried out using the London scoring system (dG) and the
results are visualized in two dimensions using the MOE 2015 application. The goal of this study is to identify
potential flavonoid compounds from plants that can be further developed as raw materials for antimalarial
drugs. The screening results of molecular docking of isoflavonoid compounds against the PfDHODH enzyme
yielded seven compounds: Retonone, Retononone, Degueline, 12a-hydroxyrotanenone, Toxicarol, Tephrosin,
and Cristacarpin. These isoflavonoid compounds have lower docking scores than the native ligand 2EN603.
1 INTRODUCTION
Globally, an estimated 1.7 billion malaria cases and
10.6 million malaria deaths were averted in the period
2000–2020. Most of the cases (82%) and deaths
(95%) averted were in the WHO African Region,
followed by the WHO South-East Asia Region (cases
10% and deaths 2%) (WHO, 2021). Malaria is a
serious infectious disease caused by the Plasmodium
species and is endemic in more than 90 countries.
Malaria is caused by protozoan parasites of the genus
Plasmodium, including P. falciparum, P. vivax, P.
ovale, P. malariae, and P. knowlesi, and it is
exclusively transmitted through the bite of female
Anopheles mosquitoes. The most deadly form of
malaria is caused by Plasmodium falciparum. If not
treated within 24 hours, P. falciparum malaria can
progress to severe disease, often resulting in death.
Around half of the world's population is at risk of
malaria infection, with the highest burden in tropical
regions such as Africa, Asia, and Latin America
(World Health Organization, 2023).
Malaria remains a common global infectious
disease that causes significant morbidity and
mortality. Despite the availability of several approved
drugs for its treatment, drug resistance has
jeopardized many of them, making the development
of new drugs for the treatment and prevention of
malaria crucial. The completion of the Plasmodium
falciparum genome and the growing understanding of
parasite biology have triggered the search for new
drug targets. However, only a few targets have been
chemically validated in vivo. The pyrimidine
biosynthesis pathway represents one of the best
examples of successful identification of anti-malarial
drug targets (Phillips and Rathod, 2010).
Sandy, S.
In Silico Virtual Screening Studies Using Molecular Docking of Isoflavonoid Compounds as Potential Antimalarials on the Plasmodium Falciparum Dihydroorotate Dehydrogenase (PfDHODH)
Enzyme.
DOI: 10.5220/0013667000003873
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 1st International Conference on Medical Science and Health (ICOMESH 2023), pages 131-142
ISBN: 978-989-758-740-5
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
131

Purine and pyrimidine bases are essential for the
synthesis of RNA and DNA. If a cell cannot
synthesize its own RNA or DNA, it will die (Löffler
et al., 2005). In human cells, pyrimidine bases can be
accessed through salvage pathways or de novo
synthesis. If de novo synthesis is inhibited, the cell
will rely on salvage pathways, and the cell will not
die. However, because Plasmodium species lack the
pyrimidine salvage pathway, inhibiting de novo
synthesis in the parasite cells leads to their death,
making them vulnerable to the inhibition of
dihydroorotate dehydrogenase (DHODH) (Vyas et
al., 2016). Plasmodium falciparum dihydroorotate
dehydrogenase (PfDHODH) catalyzes the fourth
reaction of de novo pyrimidine biosynthesis in the
parasite and is an important target for malaria
treatment (Vyas et al., 2016). PfDHODH is a Class 2
enzyme consisting of 569 amino acids. Inhibition of
PfDHODH disrupts the pyrimidine biosynthesis
pathway in the parasite, leading to its death, making
it a promising target for developing anti-malarial
drugs(Hoelz et al., 2018).
Plants produce a wide variety of organic
compounds, which are collectively known as Plant
Secondary Compounds (PSC). These compounds,
distributed differently among limited taxonomic
groups within the plant kingdom, contribute
significantly to specific colors, flavors, and odors, as
well as provide defense properties. One of the most
important applications of plant secondary compounds
for humans is their use as traditional medicines and
pharmaceuticals. (Wink, 2003).
One promising chemical compound as a candidate
for antimalarial treatment is the isoflavonoid.
Molecular docking screening analysis is conducted to
select isoflavonoid compounds that have similar
interactions with the inhibitor compound of the
PfDHODH enzyme, in this case, the native ligand
E2N603. The compound E2N603 has inhibitory
abilities at the active site with specific amino acids on
the PfDHODH enzyme (Vyas and Ghate, 2011).
Newer classes of antimalarial agents target
molecular-based enzymes in nucleoside biosynthesis
pathways, such as PfDHODH for pyrimidine
biosynthesis inhibition, purine nucleoside
phosphorylase for purine nucleoside biosynthesis
inhibition, adenosine deaminase for nucleoside
synthesis inhibition, and dihydrofolate reductase for
folate biosynthesis in the parasite (Leartsakulpanich
et al., 2002; Belén Cassera et al., 2011; Frame et al.,
2015).
The goal of the docking study is to identify new
hits from the database of natural compound
flavonoids that have the potential to be potent and
selective antimalarial agents against the PfDHODH
enzyme.
2 MATERIAL AND METHODS
2.1 Protein Preparation for Molecular
Docking
Molecular docking simulations for drug discovery
were conducted using the crystal structure of
Plasmodium falciparum dihydroorotate
dehydrogenase (DHODH) co-crystallized with 3-
Hydroxy-1-methyl-5-((3 (trifluoromethyl)phenoxy)
methyl)-1H-pyrazole-4-carboxylic acid (ID: 6I4B)
obtained from X-ray diffraction with a resolution of
1.98Å, R-free = 0.226, and R-work = 0.192. The
crystal structure of the DHODH enzyme was
downloaded from the RCSB Protein Data Bank
(https://www.rcsb.org/structure/6I4B). Ligands,
heteroatoms, and water molecules were separated
from the PfDHODH enzyme structure prior to the
molecular docking simulations (Shivanika et al.,
2020). The PfDHODH enzyme's structure was
optimized and subjected to 3D protonation using the
AMBER10HT force field with the assistance of the
MOE 2015 application (Abhimanyu, Srivastava and
Jain, 2022).
2.2 Ligands Preparation for Molecular
Docking
Chemical compounds of isoflavones from various
plants were downloaded from the Plant Secondary
Compounds database
(http://pscdb.appsbio.utalca.cl/viewIndex/index.php)
in sdf format. A total of 122 groups of isoflavone
chemical compounds were screened using the
Lipinski rule of five, resulting in 103 chemical
compounds (Lipinski, 2000, 2004; Valdés-Jiménez et
al., 2021).
These compounds were then prepared by
adding hydrogen atoms and force field AMBER10HT
charges, followed by energy minimization of the test
compounds using the MOE 2015 application.
2.3 Molecular Docking Simulation
Molecular docking was performed by identifying the
binding site of the PfDHODH enzyme using MOE
2015 software. The binding site was automatically
identified and saved as a target for the docking
simulations of the native ligand E2N603 as the
reference and the docking simulations of the test
ligands. The docking process was conducted with
ICOMESH 2023 - INTERNATIONAL CONFERENCE ON MEDICAL SCIENCE AND HEALTH
132
flexible ligands and a rigid enzyme PfDHODH
receptor. The obtained docking results were scored
using the London scoring system (dG) (Rachman and
Mutalib, 2008; Sarwar, 2013)
3 RESULT AND DISCUSSION
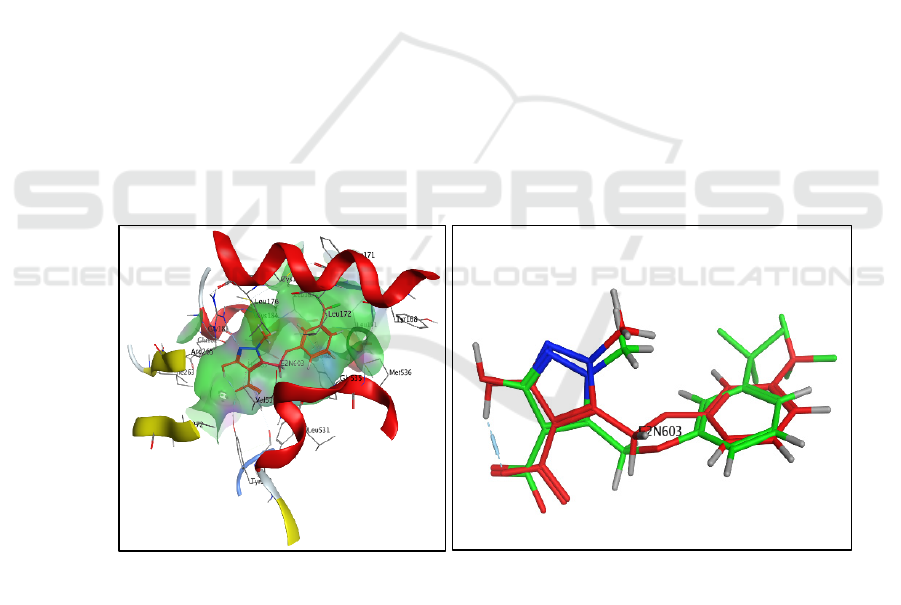
The results of the binding site analysis for the
E2N603 compound can be seen in Figure 1a. The
binding site of the PfDHODH enzyme is dominated
by a hydrophobic zone, with only a small portion of
the hydrogen bonding and polar zones found.
Molecular docking validation is performed to ensure
the validity of the docking scores. One commonly
used validation method is to perform redocking on the
active site of the compound with known
conformation and orientation, usually obtained from
co-crystal structures. A program capable of
reproducing poses with a Root Mean Square
Deviation (RMSD) value below the pre-selected
threshold (usually 1.5 or 2 Å, depending on the ligand
size) is considered successful. The selection of poses
is then followed by assessment and ranking to
determine which scoring function provides the most
accurate ranking of poses based on their RMSD
values (Hevener et al., 2010)
In the redocking
simulation using the reference ligand E2N603 on the
PfDHODH enzyme, a root mean square deviation
(RMSD) value of 0.9 A was obtained, indicating that
the docking simulation process conducted is valid.
The redocking simulation of the native ligand
E2N603 resulted in a docking score of -7.87 kcal/mol.
The hydrogen bond interactions between the
PfDHODH enzyme complex and the E2N603 ligand
involve the amino acid residues ARG265, HIS185,
and TYR258. Additionally, ionic interactions are
formed with HIS185, HIS185, and Pi-Interactions
with PHE188 and VAL532 (Table 2). The 2-
dimensional visualization of the interaction between
the PfDHODH enzyme complex and the ligand
E2N603 can be seen in Figure 2(a) and Figure 2(b).
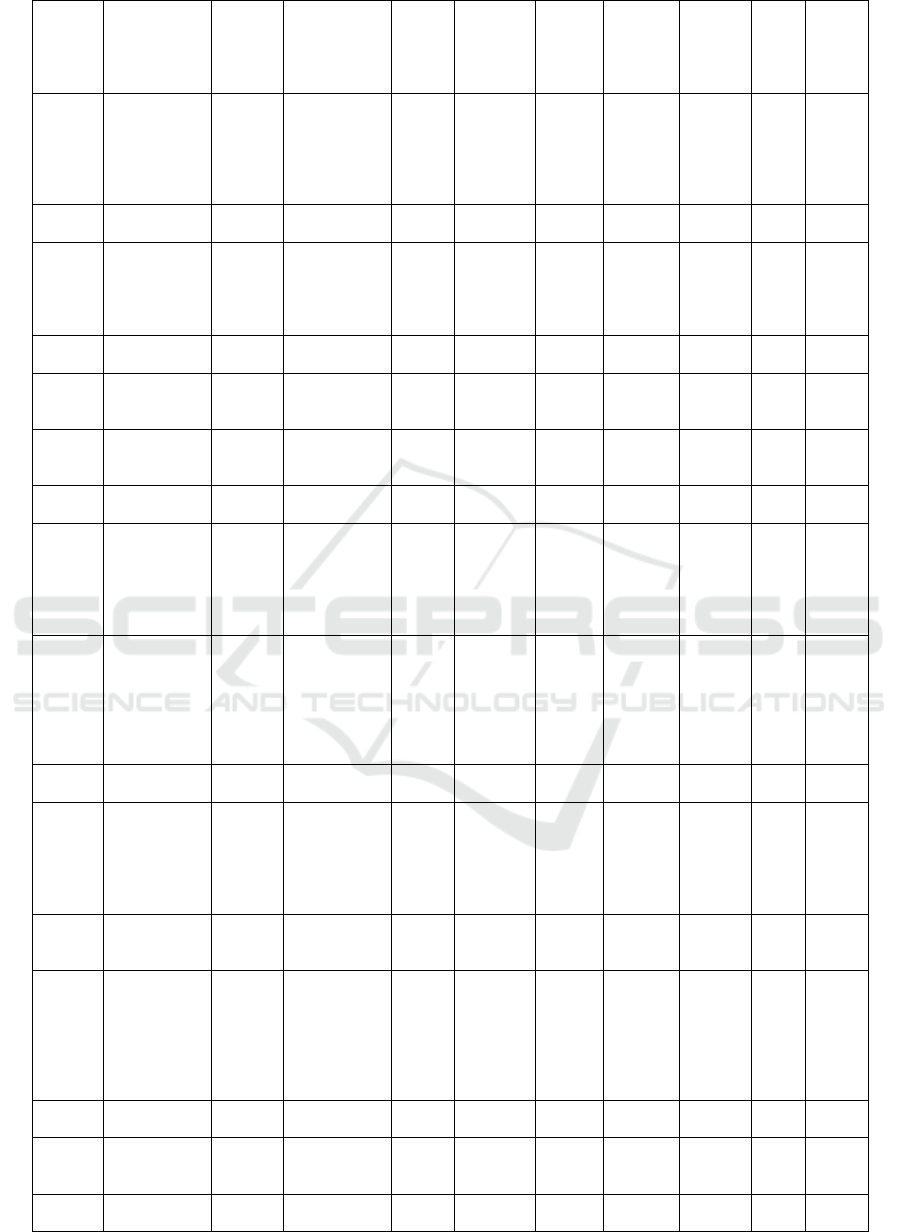
A total of 122 ligands were obtained from the
database, and after screening using the Lipinski's rule
of five, 103 isoflavonoid compounds were selected
(Hebbar et al., 2022).
These isoflavonoid compounds
were then subjected to molecular docking
simulations. The results of the screening and
molecular docking simulations can be seen in Table
1. Among the isoflavonoid compounds, Rotenone
showed the highest docking score (-8.62 kcal/mol),
while Piscerythramine exhibited the lowest docking
score (-1.49 kcal/mol).
Figure 1: a) Pink color: hydrogen bonding zone, green color: hydrophobic zone, blue color: polarity zone. (b) Redocking
results of the E2N603 ligand. The red color represents the native ligand E2N603, while the green color represents the
redocking results of the native ligand E2N603. The redocking results obtained a rmsd value of 0.9 Å.
(a)
(b)
In Silico Virtual Screening Studies Using Molecular Docking of Isoflavonoid Compounds as Potential Antimalarials on the Plasmodium
Falciparum Dihydroorotate Dehydrogenase (PfDHODH) Enzyme
133

Figure 2: (a) Overlay of native ligand E2N603 interactions before docking and after redocking. (b) E2N603 ligand interactions
at the PfDHODH enzyme receptors.
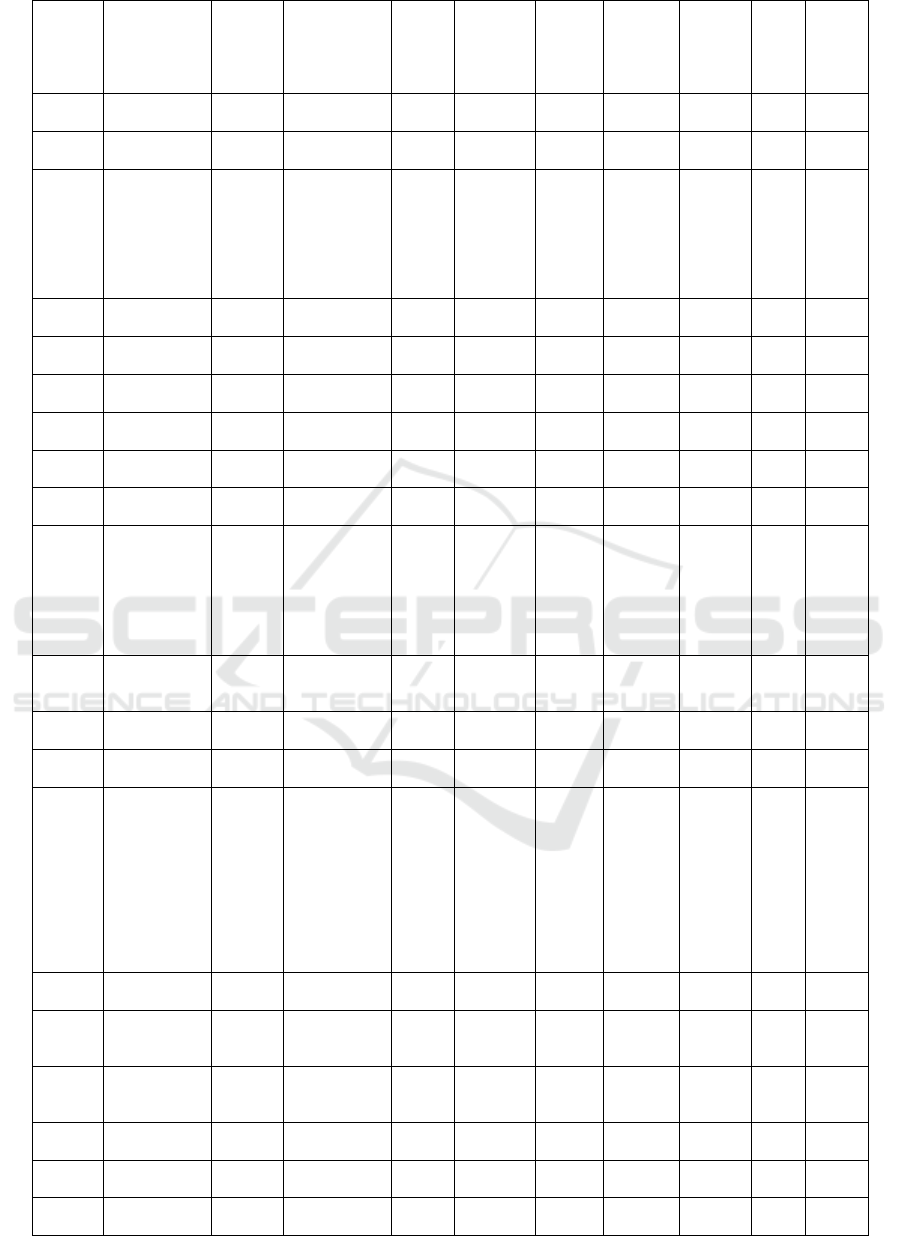
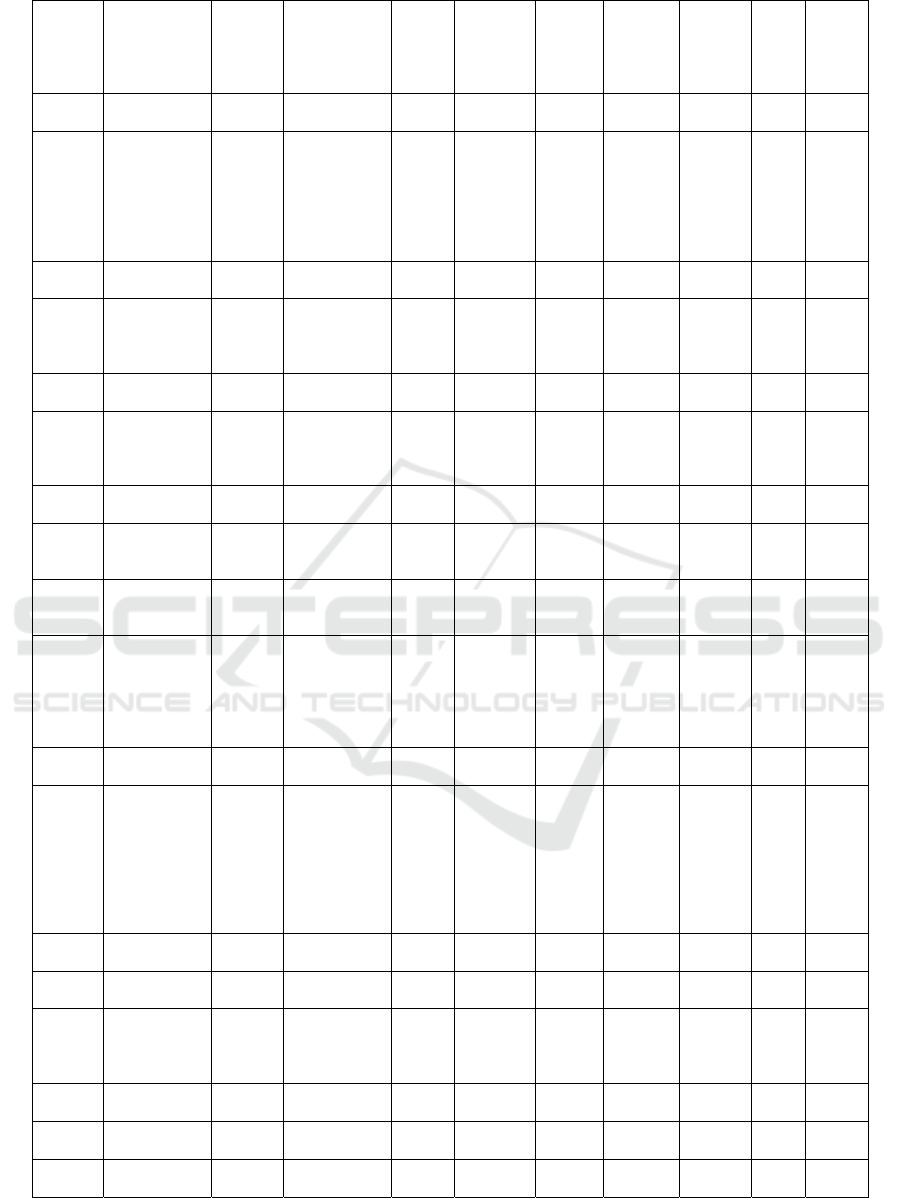
Table 1: The result of docking isoflavonoid compounds at the PfDHODH enzyme receptor
Com-
pounds
ID
Names
Mole-
cular
Mass
(>500
Dalton)
Formula
N
Rota-
te
Bond
N Hbond
Acceptors
(<10)
N
Hbond
Donors
(<5)
Molar
refract-
tivity
(<130)
TPSA
(<140A)
W-
logP
Dock-
ing
Score
(Kcal
/mol)
PSCdb
00419
Rotenone 394.42 C23H22O6 3 6 0 106.15 63.22 3.7 -8.62
PSCdb
02048
Rotenonone 406.38 C23H18O7 3 7 0 112.31 88.11 3.95 -8.43
PSCdb
01996
Deguelin 394.42 C23H22O6 2 6 0 106.98 63.22 3.9 -8.28
PSCdb
02015
12a-
Hydroxyrote
none
410.42 C23H22O7 3 7 1 107.2 83.45 2.7 -8.12
PSCdb
02059
Toxicarol;
alpha-
Toxicarol
410.42 C23H22O7 2 7 1 109.01 83.45 3.6 -8.12
PSCdb
02057
Tephrosin 410.42 C23H22O7 2 7 1 108.03 83.45 2.89 -8.09
PSCdb
01831
Cristacarpin 354.4 C21H22O5 3 5 2 97.93 68.15 3.19 -7.91
PSCdb
02032
Mucronulato
l
302.32 C17H18O5 3 5 2 82.11 68.15 2.83 -7.80
PSCdb
02025
Lotisoflavan 302.32 C17H18O5 3 5 2 82.11 68.15 2.83 -7.80
PSCdb
02034
Ononin;
Formononeti
n 7-O-
glucoside
430.4 C22H22O9 5 9 4 108.56 138.82 0.65 -7.78
PSCdb
02550
7-Hydroxy-
2',4',5'-
trimethoxyis
oflavone
328.32 C18H16O6 4 6 1 89.42 78.13 3.19 -7.75
PSCdb
02019
Irisolidone 314.29 C17H14O6 3 6 2 84.95 89.13 2.89 -7.73
PSCdb
02031
Millettone 378.37 C22H18O6 0 6 0 100.06 63.22 3.61 -7.65
PSCdb
02052
Sophoraisofl
avanone A
370.4 C21H22O6 4 6 3 101.78 96.22 3.68 -7.63
(a) (b)
ICOMESH 2023 - INTERNATIONAL CONFERENCE ON MEDICAL SCIENCE AND HEALTH
134
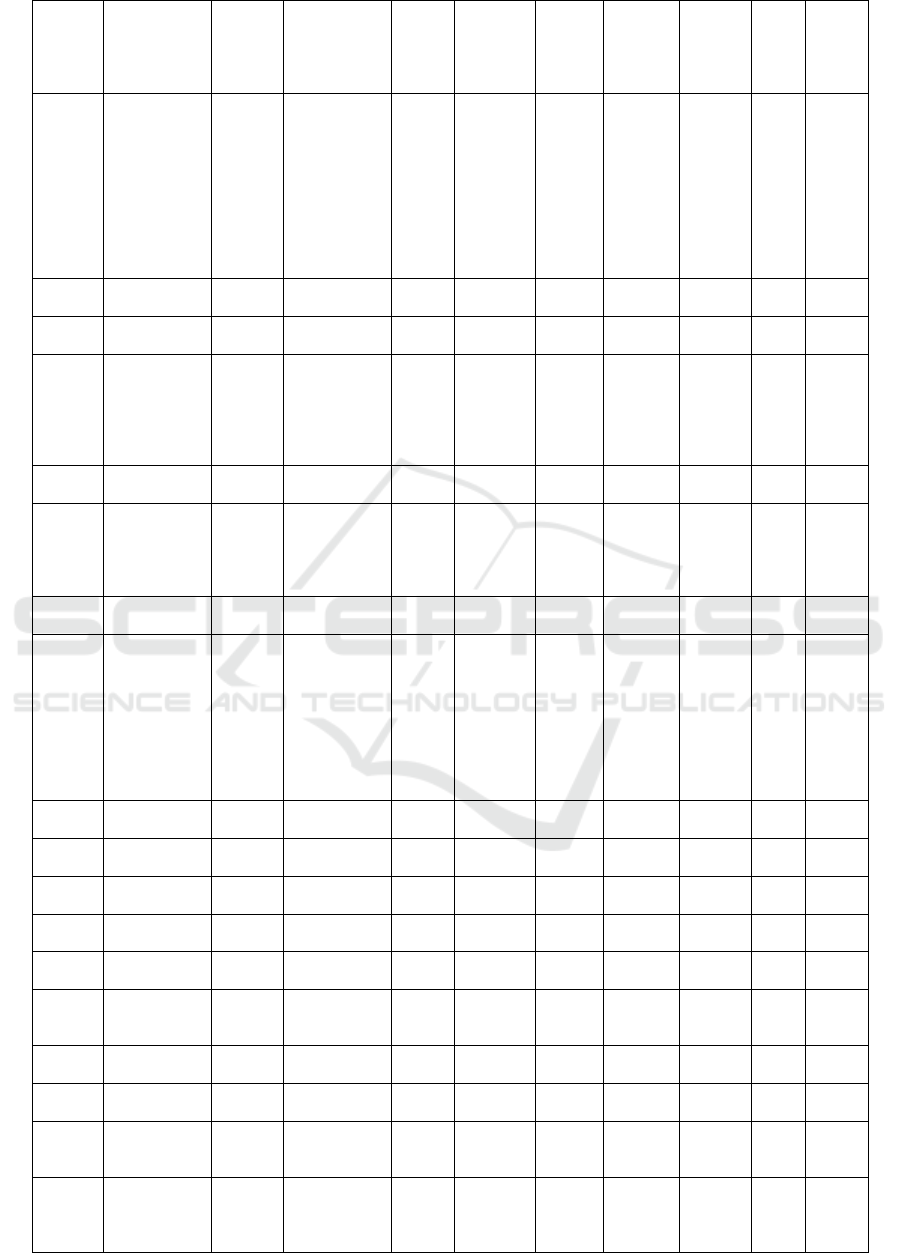
Com-
pounds
ID
Names
Mole-
cular
Mass
(>500
Dalton)
Formula
N
Rota-
te
Bond
N Hbond
Acceptors
(<10)
N
Hbond
Donors
(<5)
Molar
refract-
tivity
(<130)
TPSA
(<140A)
W-
logP
Dock-
ing
Score
(Kcal
/mol)
PSCdb
02036
Pachyrrhizon
e
366.32 C20H14O7 1 7 0 92.21 76.36 3.29 -7.54
PSCdb
01826
Betavulgarin 312.27 C17H12O6 2 6 1 82.5 78.13 2.9 -7.54
PSCdb
02022
Licoisoflavo
ne A;
2',4',5,7-
Tetrahydrox
y-3'-(3,3-
dimethylallyl
) isoflavone
354.35 C20H18O6 3 6 4 99.73 111.13 3.79 -7.50
PSCdb
02053
(-)-
Sparticarpin
300.31 C17H16O5 2 5 1 79.66 57.15 2.69 -7.45
PSCdb
02001
(-)-
Glyceollin II
338.35 C20H18O5 0 5 2 91.84 68.15 2.75 -7.42
PSCdb
00770
Moracin A 286.28 C16H14O5 3 5 2 78.68 72.06 3.53 -7.41
PSCdb
02049
(-)-Sativan 286.32 C17H18O4 3 4 1 80.09 47.92 3.13 -7.39
PSCdb
02002
Hildecarpin 330.29 C17H14O7 1 7 2 80.27 86.61 1.41 -7.38
PSCdb
02033
(-)-Nissolin 286.28 C16H14O5 1 5 2 75.19 68.15 2.39 -7.37
PSCdb
02530
Glyceocarpin
; 2-
Dimethylally
l-(6aS,11aS)-
3,6a,9-
trihydroxypt
erocarpan
340.37 C20H20O5 2 5 3 93.46 79.15 2.89 -7.32
PSCdb
02038
(-)-
Phaseollin;
Phaseolin
322.35 C20H18O4 0 4 1 90.8 47.92 3.75 -7.30
PSCdb
02562
(-)-Sophorol 300.26 C16H12O6 1 6 2 75.61 85.22 2.19 -7.30
PSCdb
02026
Luteone 354.35 C20H18O6 3 6 4 99.73 111.13 3.79 -7.28
PSCdb
01998
Ferreirin;
2,3-Dihydro-
5,7-
dihydroxy-3-
(2-hydroxy-
4-
methoxyphe
nyl)-4H-1-
benzopyran-
4-one
302.28 C16H14O6 2 6 3 78.06 96.22 2.17 -7.24
PSCdb
02050
Sayanedine 298.29 C17H14O5 3 5 1 82.93 68.9 3.18 -7.17
PSCdb
00045
(-)-
Vestitone;
Vestitone
286.28 C16H14O5 2 5 2 76.04 75.99 2.47 -7.14
PSCdb
02013
4-
Hydroxyhom
opterocarpin
300.31 C17H16O5 2 5 1 79.66 57.15 2.69 -7.14
PSCdb
02040
Pisatin; (+)-
Pisatin
314.29 C17H14O6 1 6 1 78.25 66.38 1.7 -7.13
PSCdb
02532
Glyceollin
III
338.35 C20H18O5 1 5 2 91.01 68.15 2.55 -7.13
PSCdb
01824
Afrormosin 298.29 C17H14O5 3 5 1 82.93 68.9 3.18 -7.11
In Silico Virtual Screening Studies Using Molecular Docking of Isoflavonoid Compounds as Potential Antimalarials on the Plasmodium
Falciparum Dihydroorotate Dehydrogenase (PfDHODH) Enzyme
135
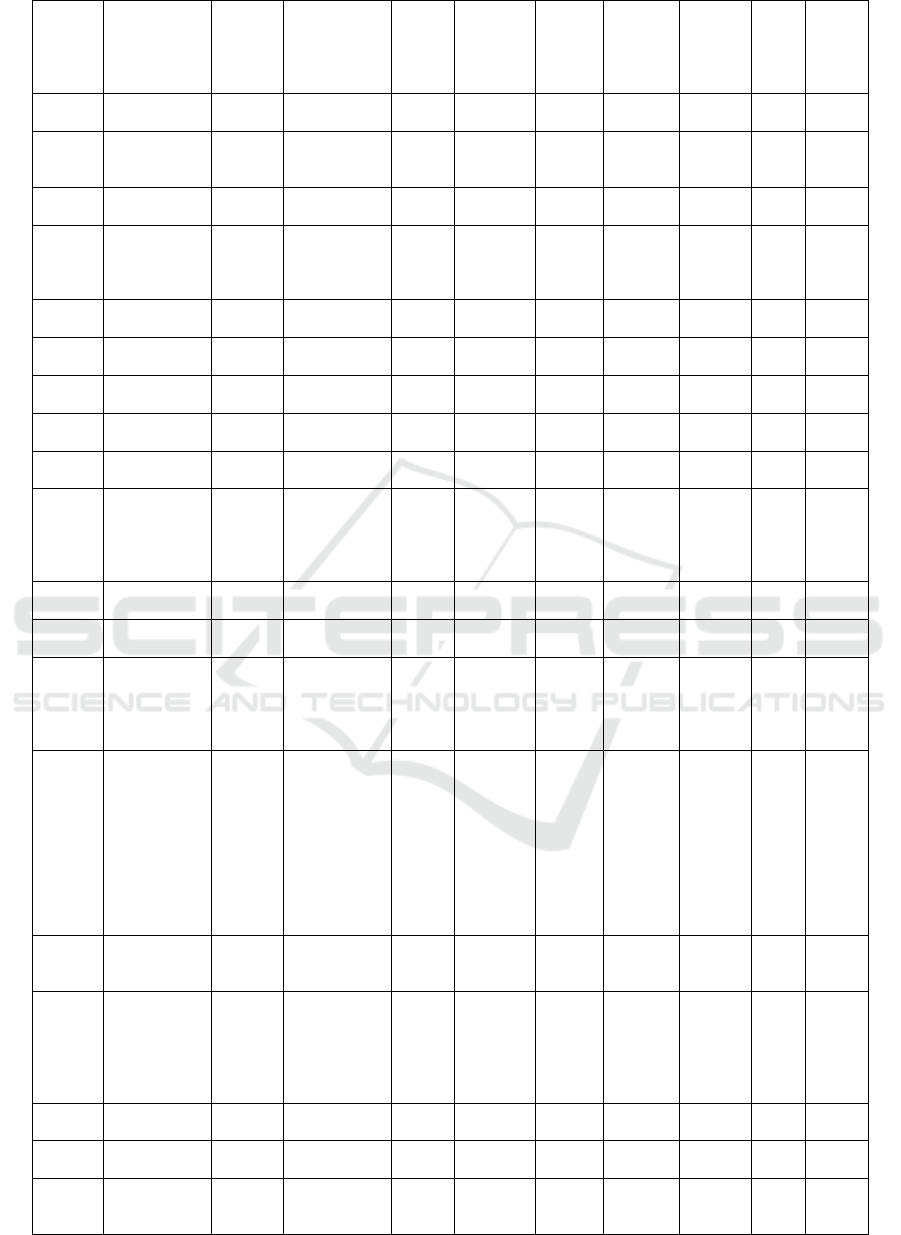
Com-
pounds
ID
Names
Mole-
cular
Mass
(>500
Dalton)
Formula
N
Rota-
te
Bond
N Hbond
Acceptors
(<10)
N
Hbond
Donors
(<5)
Molar
refract-
tivity
(<130)
TPSA
(<140A)
W-
logP
Dock-
ing
Score
(Kcal
/mol)
PSCdb
00685
Albafuran A;
4-((2E)-3,7-
Dimethyl-
2,6-
octadienyl)-
5-(6-
hydroxy-2-
benzofuranyl
)-1,3-
b
enzenediol
378.46 C24H26O4 6 4 3 115 73.83 6.45 -7.11
PSCdb
02559
(-)-Vestitol 272.3 C16H16O4 2 4 2 75.62 58.92 2.83 -7.10
PSCdb
02054
Sumatrol 410.42 C23H22O7 3 7 1 108.18 83.45 3.41 -7.09
PSCdb
02557
(-)-
Medicocarpi
n;
Medicarpin
3-O-
glucoside
432.42 C22H24O9 4 9 4 105.29 127.07 0.16 -7.09
PSCdb
02043
Pratensein 300.26 C16H12O6 2 6 3 80.48 100.13 2.59 -7.09
PSCdb
02448
2'-
Hydroxybioc
hanin A;
Dehydroferre
irin
300.26 C16H12O6 2 6 3 80.48 100.13 2.59 -7.05
PSCdb
02017
Irilone 298.25 C16H10O6 1 6 2 78.03 89.13 2.6 -7.05
PSCdb
02549
7,2'-
Dihydroxy-
4'-methoxy-
isoflavanol;
DMI;
(3R,4R)-4'-
Methoxyisofl
avan-2',4,7-
triol
288.3 C16H16O5 2 5 3 76.78 79.15 1.99 -7.05
PSCdb
02046
Psoralidin 336.34 C20H16O5 2 5 2 97.53 83.81 4.61 -7.04
PSCdb
01827
Bowdichione 298.25 C16H10O6 2 6 1 77.53 93.81 1.56 -7.04
PSCdb
02062
Vestitol 272.3 C16H16O4 2 4 2 75.62 58.92 2.83 -7.04
PSCdb
02561
(+)-Sophorol 300.26 C16H12O6 1 6 2 75.61 85.22 2.19 -7.03
PSCdb
00895
Sainfuran 286.28 C16H14O5 3 5 2 78.68 72.06 3.53 -7.02
PSCdb
00116
(-)-
Glyceollin I;
Glyceollin
338.35 C20H18O5 0 5 2 91.84 68.15 2.75 -7.00
PSCdb
02063
Wedelolacto
ne
314.25 C16H10O7 1 7 3 82.32 113.27 2.82 -7.00
PSCdb
02056
Tectorigenin 300.26 C16H12O6 2 6 3 80.48 100.13 2.59 -6.99
PSCdb
02027
(-)-
Maackiain;
Inermin
284.26 C16H12O5 0 5 1 72.74 57.15 2.41 -6.99
PSCdb
02560
2',7-
Dihydroxy-
4',5'-
methylenedi
298.25 C16H10O6 1 6 2 78.03 89.13 2.6 -6.98
ICOMESH 2023 - INTERNATIONAL CONFERENCE ON MEDICAL SCIENCE AND HEALTH
136
Com-
pounds
ID
Names
Mole-
cular
Mass
(>500
Dalton)
Formula
N
Rota-
te
Bond
N Hbond
Acceptors
(<10)
N
Hbond
Donors
(<5)
Molar
refract-
tivity
(<130)
TPSA
(<140A)
W-
logP
Dock-
ing
Score
(Kcal
/mol)
oxyisoflavon
e; DMI
PSCdb
02039
(-)-
Phaseolliniso
flavan
324.37 C20H20O4 1 4 2 93.25 58.92 3.89 -6.98
PSCdb
00911
Vignafuran 270.28 C16H14O4 3 4 1 76.66 51.83 3.82 -6.97
PSCdb
00182
Dihydrobioc
hanin A; 2,3-
Dihydrobioc
hanin A
286.28 C16H14O5 2 5 2 76.04 75.99 2.47 -6.96
PSCdb
00050
Biochanin A 284.26 C16H12O5 2 5 2 78.46 79.9 2.88 -6.95
PSCdb
02061
(-)-Variabilin 300.31 C17H16O5 2 5 1 78.68 57.15 1.98 -6.93
PSCdb
02051
Sojagol 336.34 C20H16O5 0 5 1 95.59 72.81 4.5 -6.92
PSCdb
00104
Calycosin 284.26 C16H12O5 2 5 2 78.46 79.9 2.88 -6.92
PSCdb
01829
Cajanol 316.31 C17H16O6 3 6 2 82.53 85.22 2.47 -6.92
PSCdb
01997
5-
Deoxykievit
one; (+-)-5-
Deoxykievit
one
340.37 C20H20O5 3 5 3 95.29 86.99 3.67 -6.91
PSCdb
02045
Pseudobaptig
enin
282.25 C16H10O5 1 5 1 76.01 68.9 2.89 -6.87
PSCdb
01828
Cajanin 300.26 C16H12O6 2 6 3 80.48 100.13 2.59 -6.86
PSCdb
02551
2,7-
Dihydroxy-
4'-
methoxyisofl
avanone
286.28 C16H14O5 2 5 2 75.18 75.99 2.08 -6.86
PSCdb
02531
4-
Glyceollidin;
4-
Dimethylally
lglycinol; 4-
Dimethylally
l-(6aS,11aS)-
3,6a,9-
trihydroxypt
erocarpan
340.37 C20H20O5 2 5 3 93.46 79.15 2.89 -6.86
PSCdb
02028
Medicarpin;
(-)-
Medicarpin
270.28 C16H14O4 1 4 1 73.17 47.92 2.69 -6.85
PSCdb
00193
2'-
Hydroxyfor
mononetin;
2'-
Hydroformo
nonetin
284.26 C16H12O5 2 5 2 78.46 79.9 2.88 -6.83
PSCdb
02064
Wighteone 338.35 C20H18O5 3 5 3 97.71 90.9 4.09 -6.82
PSCdb
00108
Kievitone 356.37 C20H20O6 3 6 4 97.31 107.22 3.38 -6.79
PSCdb
01823
(-)-
Acanthocarp
an
328.27 C17H12O7 0 7 1 77.82 75.61 1.42 -6.77
In Silico Virtual Screening Studies Using Molecular Docking of Isoflavonoid Compounds as Potential Antimalarials on the Plasmodium
Falciparum Dihydroorotate Dehydrogenase (PfDHODH) Enzyme
137
Com-
pounds
ID
Names
Mole-
cular
Mass
(>500
Dalton)
Formula
N
Rota-
te
Bond
N Hbond
Acceptors
(<10)
N
Hbond
Donors
(<5)
Molar
refract-
tivity
(<130)
TPSA
(<140A)
W-
logP
Dock-
ing
Score
(Kcal
/mol)
PSCdb
02522
Glycitein;
7,4'-
Dihydroxy-
6-
methoxyisofl
avone
284.26 C16H12O5 2 5 2 78.46 79.9 2.88 -6.75
PSCdb
02044
Prunetin 284.26 C16H12O5 2 5 2 78.46 79.9 2.88 -6.74
PSCdb
02444
Isoformonon
etin; 4'-
Hydroxy-7-
methoxyisofl
avone
268.26 C16H12O4 2 4 1 76.43 59.67 3.17 -6.71
PSCdb
00055
Formononeti
n
268.26 C16H12O4 2 4 1 76.44 59.67 3.17 -6.70
PSCdb
00401
7-O-
Methylluteon
e
368.38 C21H20O6 4 6 3 104.2 100.13 4.09 -6.69
PSCdb
01994
(+-)-
Dalbergioidi
n
288.25 C15H12O6 1 6 4 73.59 107.22 1.87 -6.69
PSCdb
01832
Cyclokievito
ne
354.35 C20H18O6 1 6 3 95.69 96.22 3.24 -6.68
PSCdb
02058
Texasin; 6,7-
Dihydroxy-
3-(4-
methoxyphe
nyl)-4-
b
enzopyrone
284.26 C16H12O5 2 5 2 78.46 79.9 2.88 -6.66
PSCdb
00173
2'-
Hydroxydaid
zein; 3-(2,4-
Dihydroxyph
enyl)-7-
hydroxychro
men-4-one
270.24 C15H10O5 1 5 3 73.99 90.9 2.58 -6.59
PSCdb
02035
Orobol 286.24 C15H10O6 1 6 4 76.01 111.13 2.28 -6.58
PSCdb
02447
2'-
Hydroxygeni
stein;
2',4',5,7-
Tetrahydrox
yisoflavone
286.24 C15H10O6 1 6 4 76.01 111.13 2.28 -6.57
PSCdb
02564
(+)-6a-
Hydroxymaa
ckiain
300.26 C16H12O6 0 6 2 73.78 77.38 1.4 -6.57
PSCdb
00206
2'-
Hydroxydihy
drodaidzein;
2'-Hydroxy-
2,3-
dihydrodaidz
ein
272.25 C15H12O5 1 5 3 71.57 86.99 2.16 -6.56
PSCdb
02000
Glabridin 324.37 C20H20O4 1 4 2 93.25 58.92 3.89 -6.53
PSCdb
00262
Phaseollidin;
(-)-
Phaseollidin
324.37 C20H20O4 2 4 2 92.42 58.92 3.89 -6.51
PSCdb
02488
2-Hydroxy-
2,3-
288.25 C15H12O6 1 6 4 72.73 107.22 1.48 -6.50
ICOMESH 2023 - INTERNATIONAL CONFERENCE ON MEDICAL SCIENCE AND HEALTH
138
Com-
pounds
ID
Names
Mole-
cular
Mass
(>500
Dalton)
Formula
N
Rota-
te
Bond
N Hbond
Acceptors
(<10)
N
Hbond
Donors
(<5)
Molar
refract-
tivity
(<130)
TPSA
(<140A)
W-
logP
Dock-
ing
Score
(Kcal
/mol)
dihydrogenis
tein
PSCdb
00228
3,9-
Dihydroxypt
erocarpan;
(6aR,11aR)-
3,9-
Dihydroxypt
erocarpan
256.25 C15H12O4 0 4 2 68.7 58.92 2.38 -6.47
PSCdb
02563
(+)-
Maackiain
284.26 C16H12O5 0 5 1 72.74 57.15 2.41 -6.46
PSCdb
02566
2,6,7,4'-
Tetrahydrox
yisoflavanon
e
288.25 C15H12O6 1 6 4 72.73 107.22 1.48 -6.44
PSCdb
02514
Equol 242.27 C15H14O3 1 3 2 69.13 49.69 2.82 -6.40
PSCdb
00387
Genistein;
5,7,4'-
Trihydroxyis
oflavone
270.24 C15H10O5 1 5 3 73.99 90.9 2.58 -6.40
PSCdb
02003
Hispaglabrid
in A
392.49 C25H28O4 3 4 2 116.97 58.92 5.4 -6.38
PSCdb
02536
2,7,4'-
Trihydroxyis
oflavanone
272.25 C15H12O5 1 5 3 70.71 86.99 1.78 -6.38
PSCdb
02519
3',4',7-
Trihydroxyis
oflavone
270.24 C15H10O5 1 5 3 73.99 90.9 2.58 -6.30
PSCdb
00119
(+)-
Medicarpin;
3-Hydroxy-
9-
methoxypter
ocarpan
270.28 C16H14O4 1 4 1 73.17 47.92 2.69 -6.29
PSCdb
01833
Daidzein 254.24 C15H10O4 1 4 2 71.97 70.67 2.87 -6.28
PSCdb
00075
3,6,9-
Trihydroxypt
erocarpan;
(6aS,11aS)-
3,6a,9-
Trihydroxypt
erocarpan; (-
)-Glycinol
272.25 C15H12O5 0 5 3 69.74 79.15 1.38 -6.25
PSCdb
00772
Mulberrofura
n A
392.49 C25H28O4 7 4 2 119.47 62.83 6.75 -6.08
PSCdb
01830
Coumestrol 268.22 C15H8O5 0 5 2 73.81 83.81 3.1 -5.99
PSCdb
01825
Anhydroglyc
inol; 3,9-
Dihydroxypt
erocarpen
254.24 C15H10O4 0 4 2 70.23 62.83 3.25 -5.93
PSCdb
02042
Pomiferin 420.45 C25H24O6 3 6 3 121.83 100.13 5.16 -5.73
PSCdb
02023
Lonchocarpe
nin
448.51 C27H28O6 5 6 1 130.77 78.13 5.77 -5.12
PSCdb
02041
Piscerythram
ine
451.51 C26H29NO6 6 6 4 132.33 126.15 5.19 -1.49
In Silico Virtual Screening Studies Using Molecular Docking of Isoflavonoid Compounds as Potential Antimalarials on the Plasmodium
Falciparum Dihydroorotate Dehydrogenase (PfDHODH) Enzyme
139
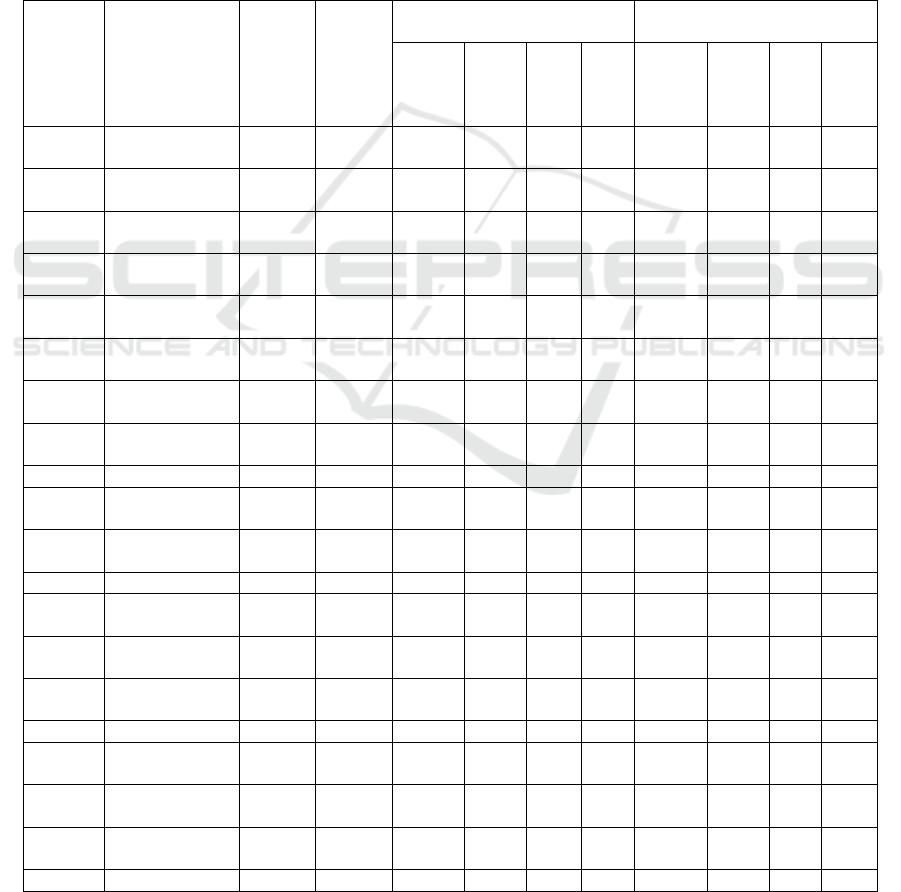
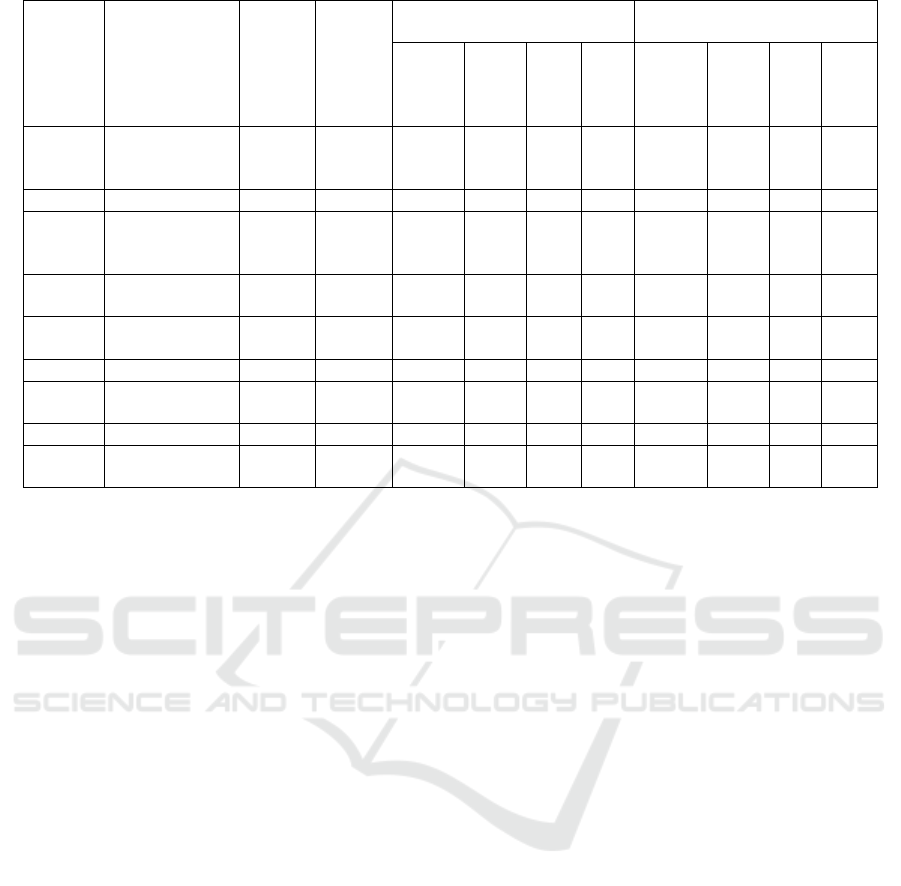
The results of molecular docking simulations of
isoflavonoid compounds were then screened based on
docking score values above -7.87 kcal/mol, which is
the score of the E2N603 ligand inhibitor. This
screening resulted in 7 isoflavonoid compounds with
the lowest docking scores. The interactions of these
isoflavonoid compounds from the docking simulation
with the PfDHODH enzyme complex included
hydrogen bond donors involving residues CYS184
and LEU531, as well as Pi-H interactions with the
amino acid VAL532, while no ionic interactions were
found in the complex. The complete interactions of
the PfDHODH complex with the isoflavonoid
compounds can be seen in Table 2. The results of the
molecular docking simulations for the isoflavonoid
compounds did not show the same amino acid
interactions as the native ligand E2N603. The
screening results of the molecular docking
simulations identified seven isoflavonoid compounds
with lower docking scores than the native ligand. The
differences in hydrogen bond, ionic, and Pi-H
interactions formed in the PfDHODH complex with
the test compounds can be considered for further in
vitro experimental testing to determine whether these
compounds are capable of inhibiting the activity of
the PfDHODH enzyme.
Table 2: Molecular interactions of isoflavonoid compounds docked on the PfDHODH enzyme receptor
Com-
pounds
ID
Names
Dock-
ing
Score
(Kcal/
mol)
Resep-
tor
Interactions distance (A)
Energy interactions
(kcal/mol)
Hbon
d
accep-
tor
Hbo
nd
dom
or
Ioni
c
Pi-
H
Hbon
d
accept
or
Hbo
nd
dom
or
Ion
ic
Pi-
H
Native
li
g
an
d
E2N603 -7.87 ARG
265
3.14 - - - -4.4 - - -
HIS
185
2.79 - - - -4.7 - - -
TYR
258
3.11 - - - -3.4 - - -
HIS
185
- - 2.79 - - - -6.1 -
HIS
185
- - 3.39 - - - -2.3 -
PHE
188
- - - 4.39 - - - -0.5
PHE
188
- - - 3.68 - - - -0.6
VAL
532
- - - 3.65 - - - -0.7
PSCdb
00419
Rotenone -8.62 CYS
184
- 3.48 - - - -0.7 - -
LEU
531
- 3.48 - - - -0.7 - -
PSCdb
02048
Rotenonone -8.43 CYS
184
- 3.61 - - - -0.5 - -
VAL
532
- - - 3.97 - - - -0.6
VAL
532
- - - 4.21 - - - -0.5
PSCdb
01996
Deguelin -8.28 CYS
184
- 3.42 - - - -0.8 - -
LEU
531
- 3.25 - - - -0.7 - -
VAL
532
- - - 4.18 - - - -0.5
ICOMESH 2023 - INTERNATIONAL CONFERENCE ON MEDICAL SCIENCE AND HEALTH
140
Com-
pounds
ID
Names
Dock-
ing
Score
(Kcal/
mol)
Resep-
tor
Interactions distance (A)
Energy interactions
(kcal/mol)
Hbon
d
accep-
tor
Hbo
nd
dom
or
Ioni
c
Pi-
H
Hbon
d
accept
or
Hbo
nd
dom
or
Ion
ic
Pi-
H
PSCdb
02015
12a-
Hydroxyrote-
none
-8.12 CYS
184
- 3.29 - - - -0.8 - -
PSCdb
02059
Toxicarol;
alpha-
Toxicarol
-8.12 CYS
184
- 3.43 - - - -0.8 - -
LEU
531
- 3.29 - - - -0.6 - -
VAL
532
- - 4.18 -0.6
PSCdb
02057
Tephrosin -8.09 CYS
184
- 3.19 - - - -0.8 - -
PSCdb
01831
Cristacarpin -7.91 LEU
531
- 2.69 - - - -1.1 - -
4 CONCLUSION
The screening results of molecular docking of
isoflavonoid compounds against the PfDHODH
enzyme yielded seven compounds: Retonone,
Retononone, Degueline, 12a-hydroxyrotanenone,
Toxicarol, Tephrosin, and Cristacarpin. These
isoflavonoid compounds have lower docking scores
than the native ligand 2EN603. Further in vitro
experimental research can be conducted on these
compounds to test their inhibitory activity against
the PfDHODH enzyme.
REFERENCES
Abhimanyu, Srivastava, P. and Jain, C.K. (2022) ‘In-
Silico Investigation of Plant-Derived Natural
Allosteric Compounds Towards Enhanced Drug-
Protein Interaction of MOA Protein Complex in
Depression Based on Molecular Docking and
Molecular Dynamic Simulation Approaches’,
Current Trends in Biotechnology and Pharmacy,
16(4), pp. 529–539. Available at:
https://doi.org/10.5530/ctbp.2022.4.86.
Belén Cassera, M., Zhang, Y., Hazleton, K.Z. and
Schramm, V.L. (2011) ‘Purine and Pyrimidine
Pathways as Targets in Plasmodium falciparum’, Top
Med Chem, 11(16), pp. 2103–2115.
Frame, I.J., Deniskin, R., Arora, A. and Akabas, M.H.
(2015) ‘Purine import into malaria parasites as a target
for antimalarial drug development’, Annals of the
New York Academy of Sciences, 1342(1), pp. 19–28.
Available at: https://doi.org/10.1111/nyas.12568.
Hebbar, S., Nandan, S.K., Shetty, C.R., Nayak, P., Dhas,
N. and Naha, A. (2022) ‘In-silico screening and
molecular docking studies of active constituents of
Withania somnifera to investigate its kinase inhibitory
activities’, Journal of Pharmaceutical Negative
Results, 13(4), pp. 1342–1349. Available at:
https://doi.org/10.47750/pnr.2022.13.04.187.
Hevener, K.E., Zhao, W., Ball, D.M., Babaoglu, K., Qi,
J., Stephen, W. and Lee, R.E. (2010) ‘Validation of
Molecular Docking Programs for Virtual Screening
against Dihydropteroate Synthase’, J Chem Inf
Model, 49(2), pp. 444–460.
Hoelz, L.V., Calil, F.A., Nonato, M.C., Pinheiro, L.C. and
Boechat, N. (2018) ‘Plasmodium falciparum
dihydroorotate dehydrogenase: A drug target against
malaria’, Future Medicinal Chemistry. Future
Medicine Ltd., pp. 1853–1874. Available at:
https://doi.org/10.4155/fmc-2017-0250.
Leartsakulpanich, U., Imwong, M., Pukrittayakamee, S.
and Yuthavong, Y. (2002) ‘Molecular
characterization of dihydrofolate reductase in relation
to antifolate resistance in Plasmodium vivax’,
Molecular & Biochemical Parasitology 119, 119, pp.
63–73.
Lipinski, C.A. (2000) ‘Drug-like properties and the
causes of poor solubility and poor permeability’,
Journal of Pharmacological and Toxicological
Methods, 44, pp. 235–249.
Lipinski, C.A. (2004) ‘Lead- and drug-like compounds:
The rule-of-five revolution’, Drug Discovery Today:
Technologies, pp. 337–341. Available at:
https://doi.org/10.1016/j.ddtec.2004.11.007.
In Silico Virtual Screening Studies Using Molecular Docking of Isoflavonoid Compounds as Potential Antimalarials on the Plasmodium
Falciparum Dihydroorotate Dehydrogenase (PfDHODH) Enzyme
141

Löffler, M., Fairbanks, L.D., Zameitat, E., Marinaki,
A.M. and Simmonds, H.A. (2005) ‘Pyrimidine
pathways in health and disease’, Trends in Molecular
Medicine, 11(9), pp. 430–437. Available at:
https://doi.org/10.1016/j.molmed.2005.07.003.
Phillips, M.A. and Rathod, P.K. (2010) ‘Plasmodium
dihydroorotate dehydrogenase: a promising target for
novel anti-malarial chemotherapy’, Infect Disord
Drug Targets, 10(3), pp. 226–239. Available at:
http://www.mmv.org/.
Rachman, A. and Mutalib, A. (2008) ‘Molecular Docking
Fevicordin on Human Estrogen α Receptor Using
MOE Software’, in Proceeding of The International
Seminar on Chemistry 2008, pp. 325–331. Available
at: www.intel.com.
Sarwar, M.W. (2013) ‘Insilico Characterization and
Homology Modeling of Arabitol Dehydrogenase
(ArDH) from Candida albican’, Bioinformation,
9(19), pp. 952–957. Available at:
https://www.researchgate.net/publication/259224502
Shivanika, C., S, D.K., Ragunathan, V., Sumitha, A. and
P, B.D. (2020) ‘Molecular docking, validation,
dynamics simulations , and pharmacokinetic
prediction of natural compounds against the SARS-
CoV-2 main- protease’, Journal of Biomolecular
Structure and Dynamics, pp. 1–27. Available at:
https://doi.org/10.1080/07391102.2020.1815584.
Valdés-Jiménez, A., Peña-Varas, C., Borrego-Muñoz, P.,
Arrue, L., Alegría-Arcos, M., Nour-Eldin, H., Dreyer,
I., Nuñez-Vivanco, G. and Ramírez, D. (2021) ‘Psc-
db: A structured and searchable 3d-database for plant
secondary compounds’, Molecules, 26, p. 1124.
Available at:
https://doi.org/10.3390/molecules26041124.
Vyas, V.K. and Ghate, M. (2011) ‘Recent Developments
in the Medicinal Chemistry and Therapeutic Poten-
tial of Dihydroorotate Dehydrogenase (DHODH)
Inhibitors’, Reviews in Medicinal Chemistry, 11, pp.
1039–1055.
Vyas, V.K., Qureshi, G., Ghate, M., Patel, H. and Dalai,
S. (2016) Identification of novel PfDHODH inhibitors
as antimalarial agents via pharmacophore-based
virtual screening followed by molecular docking and
in vivo antimalarial activity, SAR and QSAR in
Environmental Research. Taylor and Francis Ltd.
Available at:
https://doi.org/10.1080/1062936X.2016.1189959.
WHO (2021) World malaria report 2021. 1st edn.
Geneva: World Health Organization.
Wink, M. (2003) ‘Evolution of secondary metabolites
from an ecological and molecular phylogenetic
perspective’, Phytochemistry, 64(1), pp. 3–19.
Available at: https://doi.org/10.1016/S0031-
9422(03)00300-5.
World Health Organization (2023) Malaria Fact Sheet,
Malaria. Available at: https://www.who.int/en/news-
room/fact-sheets/detail/malaria.
ICOMESH 2023 - INTERNATIONAL CONFERENCE ON MEDICAL SCIENCE AND HEALTH
142
