
The Importance of Bioinformatics Tools in Medical Field: From
Prediction to Prescription
Syarif Hidayat, Khariri, Setyo Adiningsih, Rina Isnawati, Nona Rahmaida Puetri, Putri Reno Intan,
Uly Alfi Nikmah, Sunarno and Novaria Sari Dewi Panjaitan
Center for Biomedical Research, Research Organization for Health, National Research and Innovation Agency (BRIN),
Cibinong Science Center, Bogor Km. 46, West Java, Indonesia
Keywords:
Bioinformatic, Bioinformatic Tools, Prediction, Antibiotic Resistance, AMR.
Abstract:
The bioinformatics study was a modernly designed study that enabled scientists worldwide, especially those
working in genetic in living organisms, to do deeper analysis in researches. In recent reported studies regarding
the bioinformatics tools used in the analysis, the issues regarding how to utilize well-developed bioinformatics
tools in the studies requiring wet laboratories works were much discussed. Even though, those tools were being
utilized in the preliminary step as a prediction, the accurate prediction predicted by the bioinformatics analysis
was often guiding the research itself to a more directing research pathway. The growing issues and efforts of
‘personalized medicine’ was a good example of how important bioinformatics tools used in it for physicians
in order to decide the exactly needed prescription subjectively. In this article, a brief descriptive review of the
uses of several bioinformatics tools in the antibiotic resistance in bacterial infection cases. By an advanced
search in PubMed, 78.563 articles regarding bioinformatics and antibacterial resistance were found without
any limitations set on the publication date. However, due to the aim of this article was to briefly overview the
importance of bioinformatics study in helping physicians making the right prescription, especially regarding
the antibiotic use and antibiotic resistance based on recent reports, the limitations was set for only from 2018
to February 2023. Based on the filter set in MesH and title/abstract terms and publication date, a total of
45.435 articles in this search were found. However, from those 45.435 articles, there were approximately
181 systematic review articles found. These searches showed the needs in scientific communications between
researcher using the bioinformatics tools and physicians for deciding the most appropriate way to well utilize
the available tools to make better prescription worldwide, as one of many efforts in optimizing the antibacterial
agent uses and against the antibacterial resistances.
1 ANTIBIOTIC RESISTANCES
AND ITS SERIOUSNESS
Antibiotic resistance occurs when bacteria change
over time and become unresponsive to drugs, mak-
ing the undergoing occurred infections more difficult
to treat, spreading disease, and causing severe dis-
ease which highly possibly increase the risk of ill-
ness and death of the patients. Antibiotic resistance
demonstrated as the worldwide problem by World
Health Organization (WHO) was a health problem
which requires real action to combat. In their Fact
Sheet, WHO had explained why this matter has be-
come a global health burden and mentioned that the
antibiotic-resistant pathogenic bacteria had acquired
novel resistance mechanisms which should be unrav-
eled in the studies studied globally.1 In addition, due
to One Health policy, WHO also considered the im-
portance of a coordinated action which could be func-
tion as an approach in combating the antibiotic resis-
tance.
Certain programs were designed by WHO and
widely performed in many countries worldwide, such
as Global Action Plan on Antimicrobial Resistance
(GAP) since 2015, Tripartite joint Secretariat on An-
timicrobial Resistance declared in United Nations
committee meetings in 2016, World Antimicrobial
Awareness Week (WAAW) held annually since 2015,
the Global Antimicrobial Resistance and Use Surveil-
lance System (GLASS) launched also in 2015 in pur-
pose to fulfill the knowledge gaps and to inform pos-
sibly and thoughtful strategies, Global Research and
Development priority setting on Antimicrobial Resis-
tance started in 2017, and lastly the Global Antibiotic
166
Hidayat, S., Khariri, ., Adiningsih, S., Isnawati, R., Puetri, N., Intan, P., Nikmah, U., Sunarno, . and Panjaitan, N.
The Importance of Bioinformatics Tools in Medical Field: From Prediction to Prescription.
DOI: 10.5220/0012445900003848
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 3rd International Conference on Advanced Information Scientific Development (ICAISD 2023), pages 166-171
ISBN: 978-989-758-678-1
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

Research and Development Partnership (GARDP)
(Organization, 2021).
In several narrative and descriptive review arti-
cles reported recently, several priorities due to the
resistance towards antibiotic was well written and
discussed. Most of the antibiotic resistant-bacterial
isolates were those commonly causing the blood-
stream infections, Acinetobacter baumanii (BaiB and
HuangW, 2022). A. baumannii is an emerging bacte-
rial pathogen that provokes certain types of infectious
diseases in hospitals occurred and reported around
the world. By utilizing the available bioinformat-
ics tools, such as GenVision a component of DNAS-
TAR’s Lasergene Core Suite, the genome of A. bau-
manii isolates which consists of chromosomes and
plasmids were studied.3 A. baumanii isolates often
carry plasmids vary in size and are widespread. Many
of them are associated with the acquisition of an-
tibiotic resistance genes, which could be the reasons
for their ability to well survive in the patients under
treatment with commonly used antibiotics (Salgado-
camargo et al., 2020).
The resistance cases found in patients diagnosed
with tuberculosis (TB) is one of resistance issue that
should not be neglected. Mycobacterium tuberculo-
sis, the causative infectious agent of TB, is one of
the world’s leading killers, but there are a significant
number of antibiotics directed against tuberculosis.
This is primarily due to drug resistance mechanisms
present in bacteria leading to multidrug-resistant tu-
berculosis (MDR-TB) (BiswasSS and BorahVV, ). A
variety of computational tools are available that are
used for virtual screening of large numbers of certain
compounds, either synthetic or the plant extracts, in
order to combat the occurring resistance in this bac-
terium. However, till nowadays, there is still no re-
port reported highly successful approach for handling
this matter while the number of resistance case was
reported to continuously increase (YuwonoA et al., ).
Take the antibiotic resistance of Klebsiella pneu-
moniae, a common cause of health-care associated in-
fections known as nosocomial infections, as another
example. K. pneumoniae is one of biofilm-well form-
ing bacteria (LiY and ZhangL, 2022). The ability
of this bacterium in forming the biofilms could be
the reason of largely resistance found in isolated K.
pneumoniae isolates. The ability to form biofilms has
been shown to be highly resistant to antibiotics (LiY
and ZhangL, 2022; Panjaitan, 2019). Especially in K.
pneumoniae, the genetic information difference and
the different expression levels of certain genes in this
bacterium was reported to play an important role in
regulating its ability to form the biofilm, which surely
affects its resistance towards certain antibiotic. Et-
cABC, the novel clustered genes in an operon, was
previously reported and proposed to be the genes
regulating the biofilms formation in K. pneumoniae
STU-1 (Panjaitan, 2019). However, etcABC is not the
only genes regulating virulence factors of K. pneu-
moniae. The components of phosphoenol-pyruvate
phosphotransferase system (PTS) in K. pneumoniae
was reported also to affect the formation of bacterial
biofilms (HorngYT et al., 2018).
Many approaches had been developed and pro-
cessed by WHO worldwide. However, till nowadays,
the approach that could be utilized as a way out still
stays being a question. What approach could it be?
Although predictions are reasonably accurate, medi-
cal interventions to improve patient outcomes are of-
ten too late to be effective. To advance medical care
of our patients, this study aims to identify micro-
bial characteristics associated with poor clinical out-
comes.
2 BIONFORMATICS TOOLS
USED FOR PREDICTING AND
ADDRESSING THE
ANTIBIOTIC RESISTANCES
In recent years, the amount of molecular informa-
tion attributed to clinical data has increased signif-
icantly, thanks to the implementation of approaches
and resources that enable an ’omics’ view of dis-
eases. Bioinformatics is therefore seen as essential
for managing vast amounts of data for better diagno-
sis and treatment of rare and complex diseases. In-
deed, bioinformatics approaches are capable of find-
ing relationships between genomics, transcriptomics,
proteomics, metabolomics, interactomics, and other
’omics’ data that can elucidate complex interactions
between different levels and timescales. The tech-
niques of data mining enable the simulation of com-
plex systems and the construction of dynamic net-
works with the aim of developing predictive, preven-
tive and personalized medicine.
In order to further explore the molecular and ge-
nomic characteristics of bacterial isolates with resis-
tance towards antibacterial agents, whole-genome se-
quencing and analysis was widely utilized nowadays.
The use of next generation sequencing (NGS), a se-
quencing platform, started to be very useful for AMR
analysis (SabatAJ and AkkerboomV, 3 01). Next-
generation DNA sequencing machines are generat-
ing sequence data at unprecedented speeds, but tra-
ditional single-processor sequence alignment algo-
rithms struggle to keep up.
The Importance of Bioinformatics Tools in Medical Field: From Prediction to Prescription
167
Two sequencing platforms, Ion Torrent and Illu-
mina, were compared previously by using MiSeq and
Bioinformatics for Analysis of AMR Genes data anal-
ysis pipeline after considering all differences between
two platforms.10 However, the results of that partic-
ular study suggested that whether the platform (Ion
Torrent or Illumina) or sequencing chemistry used has
little impact on the outcome of the AMR data (SoniT
et al., 2022). For any certain region of genetic in-
formation or protein sequences which could not be
found by annotation or BLAST searches; an exper-
imental approach is needed to identify such replica-
tion regions. The advance method and strategy of
next-generation sequencing has enabled researchers
to analyze whole genomes or multiple genes simul-
taneously for mutation detection or gene expression
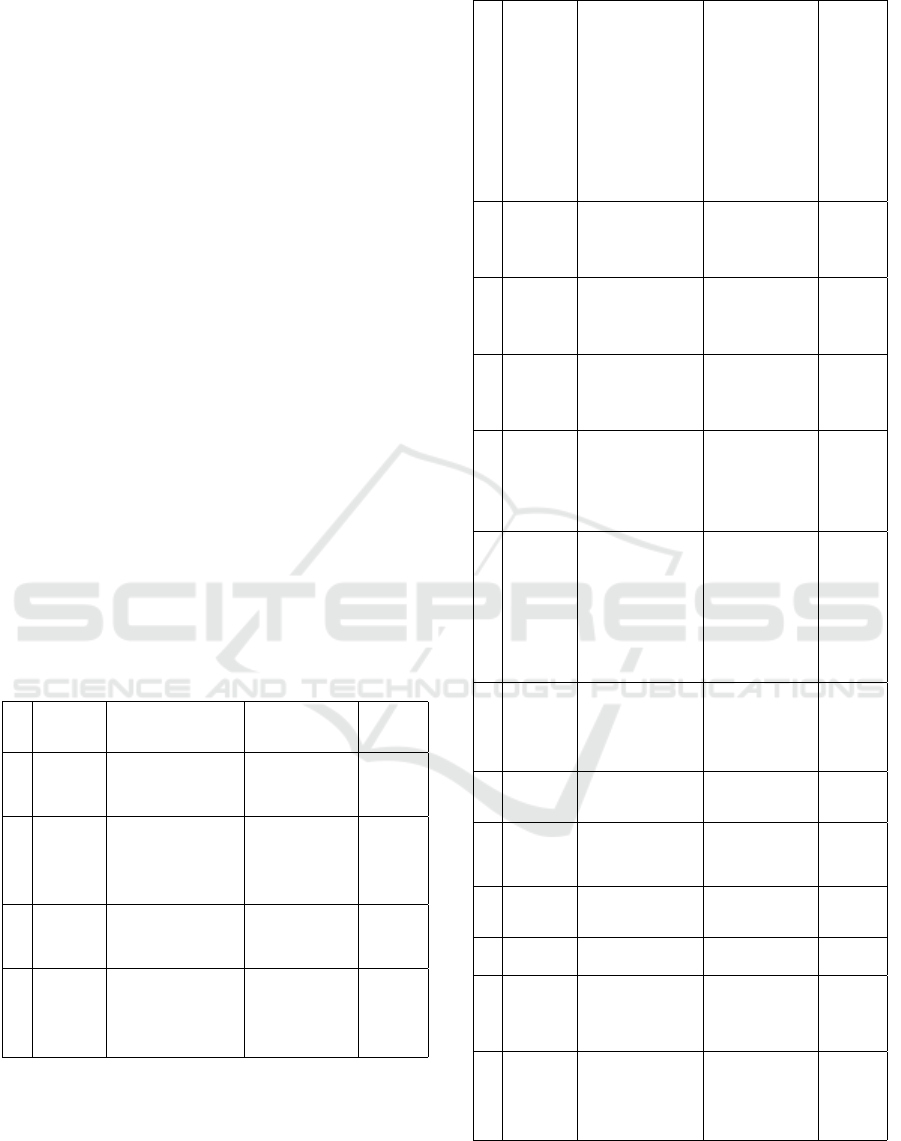
studies (Mohini et al., 2023). A table regarding the
well-developed and useful bioinformatics tools for
the genomic based data analysis was provided in Ta-
ble 1. Till nowadays, there are several bioinformat-
ics tools which had been developed and being uti-
lized in studies worldwide, such as AzureBlast, Blas-
tReduce, CloudBLAST, CloudAligner, CloudBurst,
Crossbow, DistMap, Eoulsan, FX, GATK, Hadoop-
BAM, HadooZiper, Seal, SeqWare, and TIARA (Ta-
ble 1). These tools were created for different purposes
in metagenomics study. However, those tools are con-
sidered and reported useful.
Table 1: Bioinformatic tools for genomic based data analy-
sis (Mohini et al., 2023).
No.Available
bioinfor-
matics
tools
Definitions Description and
functions in previ-
ous study
Ref.
1. AzureBlast A case study of develop-
ing science applications
on the cloud
Parallel BLAST
running on the
Microsoft Azure
cloud computing
platform
(LuW,
2010)
2. BlastReduce New parallel read map-
ping algorithms opti-
mized for matching se-
quence data from these
machines to reference
genomes
BLAST-based on
Hadoop
(SchatzMC,
2009a;
KhawlaT
and
Azed-
dineZ,
2018)
3. CloudBLASTCombination of MapRe-
duce and Virtualiza-
tion on Distributed
Resources for Bioinfor-
matics Applications
Cloud-based
BLAST imple-
mentation
(Calabre-
seB,
2021)
4. CloudAligner fast and full-featured
MapReduce based tool
for sequence mapping
method
Genomic sequence
mapping (se-
quence mapping
technology based
on MapReduce
that is full- fea-
tured and rapid)
(NguyenT
and
RudenD,
2011)
5. CloudBurst A highly sensitive
read mapping with
MapReduce, a new
parallel read map-
ping algorithms
optimized for map-
ping next-generation
sequence data to
the human genome
and other reference
genomes for use in a
variety of biological
analyses, including
SNP discovery,
genotyping, and
personal genomics
MapReduce-based
genomic sequence
mapping using
very sensitive
short reads
(SchatzMC,
2009b)
6. Crossbow Analysis of genomic
sequences utilizing
cloud computing,
including read
mapping and SNP
calling
Genotyping, SNP
detection, Read
mapping, Service
composition
(Crossbow,
)
7. DistMap A Toolkit for Dis-
tributed Short Read
Mapping on a
Hadoop Cluster
An integrated
workflow for short
read mapping
against a user-
specified reference
genome.
(PandeyRV,
2013)
8. Eoulsan A Cloud Computing-
Based Framework
Facilitating High
Throughput Se-
quencing Analyses
RNA sequencing
analysis based on
a scalable, flexible
framework built
on the Hadoop
platform
(JourdrenL
et al., 4
10)
9. FX An RNA-Seq analy-
sis tool that runs in
parallel on a cloud
computing infras-
tructure to estimate
gene expression lev-
els and call genomic
variants.
Purposed for
RNA-seq data
analysis
(HongD
and soo,
2012)
10. GATK The industry stan-
dard for identifying
SNPs and indels in
germline DNA and
RNAseq data. Its
scope now includes
somatic short vari-
ant calls and is being
extended to address
copy number (CNV)
and structural varia-
tion (SV)
Next-generation
resequencing data
management for
sequence files,
a gene analysis
toolset
(BathkeJ,
)
11. HadoopBAM The integration be-
tween analytic soft-
ware and BAMfiles
Management of
sequence files,
Directly ma-
nipulating next
generation se-
quencing data in
the cloud
(NiemenmaaM
et al.,
2012)
12. HadooZiper A cloud envi-
ronment for bio-
informatics data
analysis
(BretaudeauA
and
CollinO,
2012)
13. Seal A distributed short
read mapping and
duplicate removal
tool
Genomic sequence
mapping (using
Hadoop’s
(PiredduL
and
Zanet-
tiG,
2011)
No. Available
bioinfor-
matics
tools
Definitions Description and
functions in
previous study
Ref.
duplication re-
moval and short
read pair mapping)
14. SeqWare The query engine
enabling information
from databases,
storing and searching
sequence data in the
cloud
Management of
sequence files
(ConnorBDO
and Nel-
sonSF,
2010)
15. TIARA A database for ac-
curate analysis of
multiple personal
genomes based on
cross-technology
A user-friendly
genome browser,
which retrieves
read-depths (RDs)
and log2 ratios
from NGS and
CGH arrays
(HongD
and
JuYS,
2011)
ICAISD 2023 - International Conference on Advanced Information Scientific Development
168
3 THE POSSIBILITY OF
BIOINFORMATICS TOOLS
BEING USED BY PHYSICIANS
The remaining question is that could the available
bioinformatics tools and bioinformatics based anal-
ysis be really manifested and utilized by working
physicians in helping them making the better deci-
sions in prescribing the needed medications, partic-
ularly antibacterial agents? There are a lot of con-
sideration and thoughts made by a physician while
handling a patient, especially one diagnosed with in-
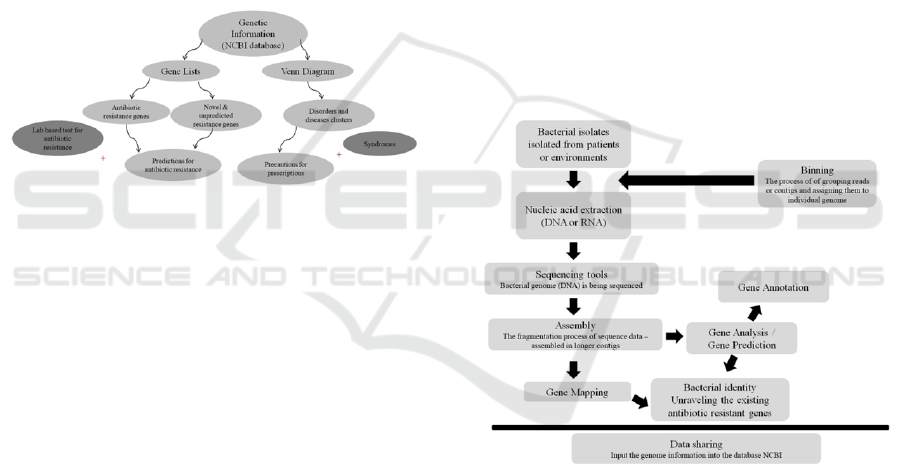
fectious diseases. A scheme describing a pathway as
an example of how useful bioinformatics tools could
be utilized in deciding a prescription, particularly re-
garding the antibiotic resistance in bacterial infectious
diseases, was provided in Figure 1.
Figure 1: Scheme describing the simple useful example of
how bioinformatic analysis was utilized in making a deci-
sion for antibiotic prescription (ConnorBDO and NelsonSF,
2010).
A previous behavior study had reported their re-
sults regarding the acceptance of bioinformatics ap-
plications by biomedical researchers, the decision-
making processes underlying the selection of tools
for primer design and microarray analysis, and the
long-term impact of training on these behaviors.28
The results of that particular study suggested a num-
ber of possible roles for medical libraries in support-
ing bioinformatics implementations, including infras-
tructure support, consulting, and training. Addition-
ally, libraries can provide services and initiate col-
laborative research on complex tasks.28 Further in-
vestigation of how bioinformatics can be integrated
into biomedical research and to develop training mod-
ules for improved bioinformatics uptake was also pro-
posed (ShachakA and FineS, 2007).
Bioinformatics, ”the science of managing and an-
alyzing biological data using advanced computational
techniques”, is indeed a useful tool since it could
reveal the unknown genetic materials in infectious
agents. However, a useful tool could not be consid-
ered useful unless most of people know how to use it
regardless of the field. Therefore, the question raised
here is how to improve the use of bioinformatics tools
by physicians whom probably are not involved in re-
searches?
4 PIPELINE OF HOW
BIOINFORMATICS TOOLS
ARE BEING USED IN MEDICAL
EFFORTS AND ACTIVITIES
The bioinformatics analysis pipelines were usually
consisting continuously dependent processes which
consist of nucleic acid material (DNA/RNA) isolation
from the clinically or environmental isolated bacterial
isolates, assemble sequence data (directly from en-
vironmental samples) to build contiguous sequences
(contigs and scaffolds), prediction of genes (and puta-
tive proteins) based on the assembled data, prediction
of putative proteins predicting domains, functions and
pathways. These pipelines were widely utilized in
metagenomics analysis (RoumpekaDD et al., 2017).
Figure 2: Scheme of a typical bioinformatics tools used in
genomic study. Genomic material (taken directly from en-
vironmental samples) is sequenced and processed using as-
sembly, gene prediction, and gene annotation tools. Finally,
the results are shared among scientific groups around the
world. (RoumpekaDD et al., 2017).
The annotated sequences and identified genomic
information such as genes and regulatory elements
are the next steps of metagenomic analysis pipeline.
The short reads generated by NGS (sequencing pro-
cess) are usually hard to be assembled. Even suc-
cessfully being assembled, the resulted contigs are of-
ten too short and fragmented to be analyzed. High-
throughput platforms may require more samples to be
sequenced in parallel to
The Importance of Bioinformatics Tools in Medical Field: From Prediction to Prescription
169

provide an economic advantage over smaller se-
quencing platforms. One of final goals of these conti-
nous pipelines is to generate and develop the diagnos-
tic kits for certain infectious agents which could be
detected through a simple detection method due to the
genomic information available.30 Therefore, the fu-
ture studies in bionfomatics are continuously needed
to be improved, for its actual use in medical field.
The final step of these pipelines is sharing the
metagenomic sequences or data. Data sharing under-
pins reproducible science, but expectations and best
practices are often vague. The funder, researcher,
and publisher communities continue to wrestle with
what should be requested or encouraged. We focus on
stakeholders in the scientific community to shed light
on the reasons for data sharing, technical challenges,
and social and cultural challenges. In biomedical re-
search, participants are important to these stakehold-
ers. Ethical sharing must consider both the value of
the research effort and her cost of participant privacy.
The current best practices for different types of ge-
nomic data and how coordinating incentives fosters
ethical data sharing that accelerates science has been
discussed elsewhere(ByrdJB et al., 2020).
5 DATABASE SEARCH
STRATEGY
The articles related to bioinformatics tools used
in studies regarding antibacterial resistances were
searched in PubMed through an advanced search by
(((antibacterial drug resistance [MeSH Terms]) OR
(antibacterial drug resistance [Title/Abstract])) AND
(bioinformatics [MeSH Terms]) OR (bioinformatics
[Title/Abstract]). From this search, 78.563 articles
were found. In addition, the date publication was set
for 2018 to 2023 which was followed by a filter for
systematic review. Respectively, 45.435 articles and
181 articles were found for each search (Filters: Sys-
tematic Review, in the last 5 years. Sort by: Most
Recent). Manually, the latest issues of relevant publi-
cations and a reference list of included text and related
articles were screened.
6 CONCLUSIONS
Effective use of bioinformatics in biomedical research
has important implications for discovering the under-
lying mechanisms and potential treatments for nu-
merous diseases. Bioinformatics knowledge and de-
velopment of bioinformatics based tools has made a
possible way to access the unknown genes regulating
the bacterial phenotypes, such as antibacterial agents
resistances, genetic information, novel proteins and
many other features of a sample at a genome-wide
level, which presents considerable opportunities for
secondary analysis and its utilization in biomedical
researches and medical field. In this article, several
useful bioinformatics tools were listed and briefly dis-
cussed. The difficulties of manifestation of bioinfor-
matics tools use in medical or health field were also
openly discussed. The importance of bioinformatics
tools in biomedical research had been realized in im-
proving the target of research, especially in antibac-
terial resistance issue, even to personalized medicine.
Therefore, the improved –user friendly bioinformat-
ics tools development is highly recommended for be-
ing further studied.
ACKNOWLEDGEMENTS
All authors would like to acknowledge to National
Research and Innovation Agency (BRIN) for the
given support for the researchers and to convey grat-
itude to all researchers in the Center of Biomedical
Research for the hard working and efforts in biomed-
ical researches.
REFERENCES
BaiB, E. and HuangW (2022). Clinical and genomic anal-
ysis of virulence-related genes in bloodstream infec-
tions caused by acinetobacter baumannii. Virulence,
13(1):1920–1927.
BathkeJ, L. Ovarflow : a resource optimized gatk 4 based
open source variant calling workflow. bioRxiv. Pub-
lished online 2021.
BiswasSS, B. and BorahVV, R. In silico approach for
phytocompound-based drug designing to fight efflux
pump-mediated multidrug-resistant mycobacterium
tuberculosis. Published online, 2021:1757–1779.
BretaudeauA, S. and CollinO (2012). Hadoopizer: A cloud
environment for bio-informatics data Analysis HAL
Id, hal-00766066.
ByrdJB, G., PrasadDV, J., and GreeneCS (2020). Respon-
sible, practical genomic data sharing that accelerates
research. Nat Rev Genet, 21(10):615–629.
CalabreseB (2021). Cloud-Based Bioinformatics Tools Pro-
vided for non-commercial research and educational
use.
ConnorBDO, M. and NelsonSF (2010). Seqware query
engine : storing and searching sequence data in the
cloud. BMC Bioinformatics, 11(Suppl 12).
Crossbow, U. John Hopkins Univer-
ICAISD 2023 - International Conference on Advanced Information Scientific Development
170

sity. Published 2013. https://bowtie-
bio.sourceforge.net/crossbow/index.shtml
HongD, P. s. and JuYS, K. (2011). Tiara : a database for ac-
curate analysis of multiple personal genomes based on
cross-technology. Nucleic Acid Res, 39:D883-D888.
HongD, R. and soo, P. (2012). Fx: an rna-seq analysis tool
on the cloud. Genome Anal, 28(5):721–723.
HorngYT, W., ChungWT, C., and ChenYY, S. (2018). Phos-
phoenolpyruvate phosphotransferase system compo-
nents positively regulate klebsiella biofilm formation.
J Microbiol Immunol Infect, 51(2):174–183.
JourdrenL, B., agn
`
es, D., and CromSLe (2012-04-10).
Eoulsan: A cloud computing-based framework facil-
itating high throughput eoulsan. A cloud computing-
based framework facilitating high throughput se-
quencing analyses. Bioinformatics.
KhawlaT, F. and AzeddineZ, S. (2018). Sciencedirect sci-
encedirect the first international conference on intelli-
gent computing in data sciences a blast implementa-
tion in hadoop mapreduce using low cost commodity
hardware. Procedia Comput Sci, 127:69–75.
LiY, K. and ZhangL, W. (2022). Klebsiella pneumonia
and its antibiotic resistance: A bibliometric analysis.
Biomed Res Int, 2022.
LuW, B. (2010). Azureblast : A case study of developing
science applications on the cloud azureblast : A case
study of developing science applications on the cloud.
Life and Medical Sciences.
Mohini, D., Dixit, A., Kaur, R., Jayan, A., Dutta, S.,
and TharakkalS (2023). A review on bioinformatics
tools for transcriptomics ngs data analysis. Lett Appl
NanoBioscience, 12(4).
NguyenT, S. and RudenD. Cloudaligner: A fast and full-
featured mapreduce based tool for sequence mapping.
Published online 2011.
NiemenmaaM, K., SchumacherA, K., and KorpelainenE,
H. (2012). Hadoop-bam: Directly manipulating next
generation sequencing data in the cloud hadoop-bam,
directly manipulating next generation sequencing data
in the cloud. Seq Anal, 28(6):876–877.
Organization, W. H. (2021). Antimicrobial resistance. Fact
Sheet. Published. Accessed February14, 2023.
PandeyRV, S. (2013). Distmap: A toolkit for distributed
short read mapping on a hadoop cluster.
Panjaitan, N. S. D. (2019). Horngy tze, chengs wen,
chungw ting, smithcm. EtcABC, a Putative EII Com-
plex , Regulates Type 3 Fimbriae via CRP-cAMP Sig-
naling in Klebsiella pneumoniae, 10(July):1–15.
PiredduL, L. and ZanettiG, P. (2011). Seal: A distributed
short read mapping and duplicate removal tool. Seq
nalysis, 27(15):2159–2160.
RoumpekaDD, W., EscalettesF, F., and WatsonM (2017).
A review of bioinformatics tools for bio-prospecting
from metagenomic sequence data. Front Genet,
8(MAR):1–10.
SabatAJ, Z. and AkkerboomV (2017-03-01). Targeted
next-generation sequencing of the 16s-23s rrna region
for culture-independent bacterial identification - in-
creased discrimination of closely related species.
Salgado-camargo A.D., Semiramis Castro-Jaimes, Jes
´
us
Silva-Sanchez, J. P
´
erez-oseguera
´
A., Volkow P.,
Miguel A. Cevallos.(2020). Structure and Evolution
of Acinetobacter baumannii Plasmids,11(June):1–21.
doi: 10.3389/fmicb.2020.01283. eCollection 2020.
SchatzMC (2009a). Blastreduce: High performance
short read mapping with mapreduce. Seq Anal,
25(11):1363–1369.
SchatzMC (2009b). Cloudburst: highly sensitive read map-
ping with mapreduce. Seq Anal, 25(11):1363–1369.
ShachakA, S. and FineS (2007). Barriers and enablers to
the acceptance of bioinformatics tools: A qualitative
study. J Med Libr Assoc, 95(4):454–458.
SoniT, P., BlakeD, J., and JoshiM (2022). Journal of global
antimicrobial resistance comparative analysis of two
next-generation sequencing platforms for analysis of
antimicrobial resistance genes. J Glob Antimicrob Re-
sist, 31:167–174.
YuwonoA, I., IdCP, S., and LestariBW. Factors affecting
outcome of longer regimen multidrug-resistant tuber-
culosis treatment in west java indonesia: A retrospec-
tive cohort study. Published online, 2021:1–13.
The Importance of Bioinformatics Tools in Medical Field: From Prediction to Prescription
171
