
An NLP-Enhanced Approach to Test Comorbidities Risk Scoring
Based on Unstructured Health Data for Hospital Readmissions
Prediction
Tahir Hameed
1a
, Haris Jamal Khan
2
, Saad Khan
2
, Mutahira Khalid
2b
, Asim Abbas
3c
and Syed Ahmad Chan Bukhari
3d
1
Girard School of Business, Merrimack College, North Andover, MA, 01824, U.S.A.
2
School of Electrical Engineering and Computer Science, NUST, H-12, Islamabad, Pakistan
3
Division of Computer Science, St. John’s University, Queens, NY 11439, U.S.A.
mkhalid.msit20seecs@seecs.edu.pk, abbasa@stjohns.edu, bukharis@stjohns.edu
Keywords: Comorbidities, Hospital Readmissions, Clinical Decision Support Systems, Natural Language Processing,
Bert, Deep Learning.
Abstract: Hospital readmissions have emerged as a key healthcare quality indicator since the passing of the Affordable
Care Act in 2010. It is easier to predict the readmission risk of patients without complications, but
comorbidities, such as diabetes and cardiovascular diseases, make it difficult to accurately assess the
readmission risk. 30-days hospital readmissions (30DRA) risk models typically rely on demographic, socio-
economic, and medical variables from structured data, such as diagnosis, vitals, lab reports, and comorbidities,
etc. Comorbidity indices help in assessing overall disease burden by accounting for the disease codes in
electronic health records (EHRs). With the advent of natural language processing (NLP), there is a potential
to extract additional health related variables including the possibility of gleaning additional disease codes for
comorbidities in unstructured portion of the EHRs, such as clinical notes, medical history, and discharge
summaries. Whereas NLP has been applied heavily in healthcare information systems, to the best of our
knowledge, there is no research that identifies comorbidities from unstructured clinical texts. This paper
employs a Bidirectional Encoder Representation from Transfer (BERT) deep learning technique to predict
additional comorbid conditions in the unstructured portions of EHRs and evaluates the effectiveness in
comorbidity scoring. Comorbidity scores based on the NLP-predicted comorbidity codes (predicted) were
compared against the scores calculated from codes identified by the health providers (diagnosed), and also
against a combination of the two (diagnosed and predicted). We find NLP is effective in improving the
accuracy of comorbidity calculations, that in turn could improve predictive power of AI models for hospital
readmissions and mortality predictions. It is among the first papers employing NLP to predict ICD-10 codes
from unstructured EHRs for comorbidity index calculations.
1 INTRODUCTION
A good assessment of comorbidities aids healthcare
providers in performing better diagnosis and effective
treatments. Timely comorbidity assessment during
medical encounters also improves management of
risks and healthcare quality indicators, such as health
outcomes, mortality rates and hospital readmissions
rates (Goltz et.al, 2019; Hameed, 2020; Menendez et
a
https://orcid.org/0000-0002-6824-6803
b
https://orcid.org/0000-0001-8482-4004
c
https://orcid.org/0000-0001-6374-0397
d
https://orcid.org/0000-0002-6517-5261
al., 2014; Sharma et al, 2021). Comorbidity
assessment methods and their correct application,
therefore, have become critical in medical practice.
Two most popular measures to assess
comorbidities i.e. Charlson Comorbidity Index (CCI)
and Elixhauser Comorbidity Index (EHI) assess the
overall disease burden of primary diagnosed
conditions in a patient by accounting for any other
diseases among the most prevalent diseases and
Hameed, T., Khan, H., Khan, S., Khalid, M., Abbas, A. and Bukhari, S.
An NLP-Enhanced Approach to Test Comorbidities Risk Scoring Based on Unstructured Health Data for Hospital Readmissions Prediction.
DOI: 10.5220/0011944500003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 5: HEALTHINF, pages 649-659
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
649

conditions (Charlson et al, 1987, Elixhauser et.al,
1998). The occurence of multiple comorbid
conditions results in a higher CCI or EHI score
indicating higher overall diasese burden. Quite
intutively, the co-occurrence or severity of ceratin
diseases would imapct the health more negatively.
Some researchers have demonstrated that modified
summary CCI or EHI scores (a single number) based
on aggregated or weighted burden of comorbidities in
a patient are more valuable metrics of comorbidities
(Van Walraven et al, 2009, Thompson et al, 2015).
Higher the modified comorbidity score, higher the
risks to the patient’s health and vice versa. Even
though both frequency counting and weight adjusting
indices are effective when physicians make the
health decisions, it is not clear if they would be as
effective when it comes to AI-based clincal decision
support systems (CDSS). Moreover, it is not clear if
they would perform optimally in the wake of newer
data-driven healthcare delivery models such as
patient-centered healthcare models that require a lot
of individual patient details and customization, such
as value-based care and personalized medicine.
Electronic Health Records (EHRs) are the most
common data source containing the diagnosis of
primary diseases as well as comorbid conditions.
EHRs typically record ICD-9 or ICD-10 medical
codes as part of a classification structure titled
‘International Classification of Diseases’ (ICD). ICD-
9 codes are most prevalant as many healthcare IT
systems have trasnitioned or are in the process to
move to ICD-10 codes. ICD-11 codes have already
been annouced but they are not adopted yet. The most
common use of these codes is in medical billing
processes but with the advent of AI they have found
heavy use in CDSS.
Typically, physicians specify ICD-9 or ICD-10
codes of the primary disease in their clinical notes
along with other conditions which become part a
pateint’s medical history, often times part of EHRs.
Medical coders convert these verbal or textual notes
into ICD codes and verify them for claims and billing
purposes. So, in addition to sturctured data, EHRs
also contain lot of textual data, such as clinical notes,
discharge summaries and specialist reports. They
often contain additional information on a patient’s
comorbid conditions. It happens quite often a patient's
medical history and past health records are not
immediately visible to the physician during a medical
encounter. Also, the comorbidities inlcuded as text
but not as ICD codes maybe hard to identify upfront.
Lastly, older EHRs have ICD-9 codes that have not
been updated to the ICD-10 codes yet.
Since, the available Elixhauser comoridity
algorithms (EHI) are based on ICD codes only, they
are are being limited by losing valuabe uncoded
information. With the advent of AI and NLP
techniques in computer-assisted coding (CAC) area,
it is high time that the uncoded information in
unstructured portions of EHRs could also be
leveraged with an aim to improve the accuracy of
comorbidity algorithms. That, in turn, should
improve the predictive performance of CDSS
designed for health outcomes, mortality and hospital
readmissions rate, etc.
Our research aims to develop and test an NLP
based deep learning approach to glean (predict) ICD-
10 medical codes in the unstructured portions of
EHRs, generate Elixhauser Index comorbidity scores
(EHI) by including the newly predicted codes in
them, and test these richer comorbidity scores in 30-
days hospital readmissions (30DRA) prediction
models. To that end, we employed a Bidirectional
Encoder Representation from Transfer (BERT)
model for gleaning ICD-10 codes from the textual
part of the EHRs.
Once the codes were extracted, several tests were
performed on the EHI comorbidity soring and
30DRA classification. At first, chi-square test of
independence was used to assess the dependence of
30DRA on each one of the thirty diseases/conditions
in the EHI comorbidity scores with the newly
predicted codes. Next, we also performed principal
component analysis to assess the effect of each
comorbid condition in classifying the 30DRA and the
top components that explain most of the variance.
Multi-logistic regression-based feature scoring was
conducted to further clarify the influence (weight) of
each comorbid condition in 30DRA classification.
Finally, student T-tests were done to compare the
classification performance of existing codes, the
newly predicted codes and a combination of the two.
In general, we have found NLP approaches to be
effective in extracting comorbidity codes from
unsturctured portions of EHRs. They improve CDSS
predictive models that consider combobidties index
scores as feature/s. Detailed findings are included in
the results and analysis. This paper is a first in the
CDSS area employing NLP technique to predict
additional comorbid conditions. It is also among the
first in the healthcare area to predict ICD-10
comorbidity codes using NLP. Rest of the paper is
organized as follows. Section 2 covers current
litearure on relevant topics. Section 3 describes the
data and the methods. Section 4 presents the results
and analysis while section 5 concludes with pointers
to limitations and future research.
CCH 2023 - Special Session on Machine Learning and Deep Learning for Preventive Healthcare and Clinical Decision Support
650

2 LITERATURE REVIEW
2.1 Comorbidity Indices and Their
Application in Clinical Decision
Support Systems
The most commonly referred definition of
comorbidities identify them as, “Any distinct
additional entity that has existed or may occur during
the clinical course of a patient who has the index
disease under study” (Feinstein, 1970). So,
comorbidities are additional diseases or conditions a
patient is suffering from in addition to the disease or
condition they are being treated for. Another
approach is considering “the co-occurrence of
multiple chronic or acute diseases and medical
conditions within one person” disregarding direct
relationships between a primary disease and
comorbid diseases (Bayliss et al, 2008). Whether
primary or comorbid, the diseases and conditions are
identified by International Classification of Diseases
(ICD) Codes, commonly used for medical billing.
The current version of ICD-codes is ICD-10 with
ICD-11 codes already announced, but most health IT
systems are still using ICD-9 codes or they are
transitioning from ICD-9 to ICD-10 codes.
Comorbidity indexes typically use ICD codes in
EHRs to measure the frequency, co-occurrence and
severity of the diseases in a patient. The most
prevalent comorbidity scoring methods in clinical
practice include the Charlson Comorbidity Index
(CCI) and the Elixhauser Comorbidity Index (EHI).
CCI’s mortality risk was based on review of patient
medical records for the 17 diseases and conditions
that caused mortality (Charlson et al, 1987). EHI
takes ICD codes into accounts for mortality risk in 30
different diseases organized in multiple diagnosis
related groups (DRGs) recognized by the Centres for
Medicare and Medicare Services (CMS). Agency for
Healthcare Research and Quality (AHRQ) provides a
software algorithm on its website to calculate the
Elixhauser comorbidity score (AHRQ, n.d.)
Both these indices have been quite effective in
their prognostic value in clinical settings. Therefore,
they have been widely adopted in medical practice.
The performance of both CCI and EHI comorbidity
indices has been somewhat similar for ICD-9, ICD-9
CM and ICD-10 codes but Elixhauser has proven to
be better in terms of overall prognostic value (Quan
et.al, 2005). There have been some concerns raised
about the above-mentioned approaches on
discounting the nature of diseases and their
chronological order of progression, etc. (Valderas,
2009) but generally they have remained the
predominant tools for measuring comorbidity with
good prognostic outcomes.
There is a long list of other comorbidity indices
developed or calibrated according to the outcomes of
interest, relation to primary diseases, and regulations
governing regional health systems, etc. The
performance of the same comorbidity indices varies
when applied in different diseases. For instance, one
comorbidity index may perform well for a specific
type of cancer but not for hip-joint replacement. Some
other broader but prominent comorbidity indices
include Chronic Disease Score (CDS) by Von Korff
(1992), ICED Index of Co-existing Diseases by
Greenfield (1993), Health-related Quality of Life
Comorbidity Index (HRQL-CI) by Mukherjee et al
(2011), the National Institute on Aging (NIA) and
National Cancer Institute (NCI) comorbidity index
(Havlik et al, 1994), and the Adult Comorbidity
Evaluation-27 (ACE-27) by Piccirillo et al (2004).
These and several others are used by providers as a
single or combination comorbidity measures when
dealing with various medical areas or diseases.
Since the decision on diagnosis and treatments has
to come from the physician/s, the role of these
indices, whether calculated manually or
automatically, is mainly clinical decision support
during admission, on the patient bed-side, during
discharge and adherence monitoring, etc. Physicians
use comorbidity indices, especially CCI and EHI, to
measure severity of illness, mortality risk, prognosis,
treatment difficulty, need for intervention, required
resource intensity, and hospital readmissions risk, etc.
in these situations.
2.2 NLP in Healthcare AI and Clinical
Decision Support Systems
Current EHR-based health information systems are
able to streamline workflows, boost productivity and
improve doctor-patient interactions. They also play a
major role in emerging AI-based predictive analytics
and deep learning models for clinical decision
support. The diversity of information, however,
comes with the cost of varying structured and
unstructured storage formats. Patient demographics,
weight, height, blood pressure, binary lab results, and
administered medications are a few examples of the
structured data. Contrarily, narrative data found in
EHRs such as surgical records, clinical notes,
discharge summaries, and pathology and radiology
reports are not amenable to computational analysis
and it needs to be transformed or integrated with
structured data points to increase their usability. The
integration of diverse data types opens up avenues for
An NLP-Enhanced Approach to Test Comorbidities Risk Scoring Based on Unstructured Health Data for Hospital Readmissions Prediction
651

research but is not free from its own challenges.
These challenges include heterogeneous data formats,
non-flexible storage structures, and the lack of a big
data pipeline (Evans, 2016). Thus, the conversion of
unstructured health information into standards-
compliant, comparable, and consistent data are
essential for health informatics research.
Over the years, researchers have tapped into
Natural Language Processing (NLP) models for
comprehension of unstructured data in the medical
domain. NLP is a range of computational techniques
for the automatic analysis and representation of
human language (Cambria and White, 2014). They
allow computer systems to comprehend free text by
transforming it into a format that is machine-readable.
In the medical domain, NLP is in a period of robust
development, with 100 publications annually with an
incremental trend (Wang et al, 2020). Some notable
venues of NLP research have been discussed in the
following paragraphs.
Named Entity Recognition (NER) is a semantic
information extraction technique that locates and
categorizes relevant entity data from the text at scale
(Aya, 2020). Named entities include everything from
names, brands, addresses, locations, and virtually any
classifiable textual information. NER makes these
specific entities usable in NLP models. Unstructured
clinical notes often contain valuable information such
as tumor location, diagnosis explanations, and at
times ICD codes, that when present in a structured
format would allow for quicker analysis. Several
attempts have been made to convert these notes into
a structured format with the help of NER modeling.
In (Tome et al, 2017), a rule-based NER that consists
of two phases was used for dietary recommendations.
The first one involves the detection and determination
of the entities mentioned, and the second one involves
the selection and extraction of the entities. Bio-NER
(Soomro et al, 2017) is another NER model for
biomedical entities. Parts of speech tags and N-grams
were used to enhance the performance compared to
previously existing NERs. Panchendrarajan &
Amaresan’s (2018) NER technique combined deep
learning and Bidirectional LSTM-CRF model. This
model includes bidirectional LSTM with a
bidirectional Conditional Random Field that is able to
capture both the word level and sentence level
encodings along with the positional encodings of text.
Combined with the context from LSTM, the
encodings are then fed to the CRF for classification.
BI-CRF has improved performance in comaprison to
unidirectional CRF and backward CRF.
Transformer models are also being used for NER.
BERT (Bidirectional Encoder Representation from
Transfer), is one of the most popular transformer
models that produce a state-of-the-art result for NER
task (Devlin et.al., 2018). BioBERT is the biomedical
version of the BERT language model for the
biomedical text and is widely used by the biomedical
text-mining domain experts for NER, question
answering and summarization tasks (Lee et al., 2020).
The BERT model has been finetuned on Wikipedia,
PMC and PubMed articles. ClinicalBERT (Alsentzer
et al, 2019) is another extension of BioBERT and has
been further trained on the MIMIC III dataset. All
discharge summaries (880M words) were used to
finetune this BERT to create embeddings for tasks
associated with BERT.
Knowledge graphs (KG) are increasingly being
used to further enrich information contained in EHRs.
Vafajoo et al (2018) investigated the risk factors of
cancer and chronic disease by creating KGs from
biomedical literature. The suggested methodology
included KG, disease-specific word embedding using
NLP approaches, and literature-based discovery
(LBD). The developed KG revealed that the clinical
characteristics were the main emphasis of the breast
cancer literature rather than the conventional
chemical recommendations. KG built from EHRs has
been offered as a diagnostic tool also Chaudhri,
2022). Using string matching on the two datasets
provided by Beth Israel Deaconess Medical Center.
the mentions of diseases and their symptoms were
manually retrieved from both organised and
unstructured data. Google Health Knowledge Graph
(GHKG) was used to compare the disease-symptom
edge that was generated for the constructed KG,
based on evaluation metrics including recall and
precision. Esteban et al (2017) proposed the Clinical
Knowledge Graph (CKG), an open-source platform
with more than 16 million nodes and 220 million
relationships, to represent the experimental data, the
literature, and public databases. Using CKG with
statistical and machine learning approaches
significantly accelerated the analysis and
interpretation of conventional proteomics procedures.
NLP is also being used to deduce or predict
patient health outcomes from clinical texts so that
appropriate interventions could be made in time.
Conventional prognostic scores usually require
predefined clinical variables to predict health
outcomes (Sung et al, 2021). They have used free text
on the history of the present illness and computed
tomography reports to build NLP-based machine
learning models to predict the poor functional
outcome at 90 days post-stroke. Similarly, Arnaud et
al, (2021) employed convolutional neural networks
CCH 2023 - Special Session on Machine Learning and Deep Learning for Preventive Healthcare and Clinical Decision Support
652

(CNN) to provide an early prediction of the medical
specialities at hospital admission.
Lastly, NLP techniques can also be seen in models
aiming to improve the quality of life of patients and
assist the overall healthcare system. For instance,
deep learning techniques have been regularly applied
to clinical notes to predict their ICD labels (Bao et al,
2021). Moreover, Le et al (2020) aimed to evaluate
the hypothesis that possible future medical concepts
can be predicted from a patient’s EHR. By using
time-based prefixes and suffixes, where each prefix
or suffix was a set of medical concepts from a medical
ontology, comparisons between prefixes of patients in
the collection were done with the state of the current
patient using various interpatient distance measures.
Their study shows that indications of future events
using this methodology are feasible.
In the above sections, not only have we discussed
the criticality of comorbidities in clinical decicions
but we have also comprehensivley discussed several
applications of traditional and emerging NLP
approaches in the healthcare information systems
area. We did notice a few cases of medical codes
extraction and labelling from narrative portions of
EHRs. However, we are not aware of, to the best of
our knowledge, research that has attempted to extract
ICD-10 codes for comorbidities from unsturctured
data. In the next section, we attempt at developing and
testing such an approach.
3 DATA AND METHODS
3.1 NLP-Enhanced Approach to
Calculate Comorbidity Scores
based on Extracted ICD-10 Codes
Our proposed model consists of four major steps. The
first step involves extracting and pre-processing patient
dataset for hospital readmissions with ample structured
variables of intertest also containing rich unstructured
texts such as clinical notes or discharge summaries.
Second step involves extracting additional ICD-10
codes from unstructured text using an NLP technique.
To that end, we planned to employ an HLAN
(Hierarchical Label-wise Attention Network) deep
learning technique. However, in this particular
instance, we have used a transfomer-based BERT
model to predict additional comorbid conditions. The
third step focuses on re-organizing data in a way that
enables calculations, and comparative evaluations. The
highlight of this part is to assign, clean and organize the
extracted comorbid codes to patient records in a clean
and consistent manner. The final step requires that
comorbidity scores for each disease included in the
selected comorbidity index are calculated and made
available as features of the target variable in our model
i.e. hospital readmissions. That involves
operationalization of either tested summary
comborbidity indices such as Elixhauser (EHI), VW
(vanWalraven, 2009) and Thompson score
(Thompson,2015), or devising own algorithms based
on diseases and outcomes of interest. Since our
research focuses on all-cause hospital readmissions
and general adminsitrative data, therefore we limit
oursleves to diseases/comorbidities specified in EHI
comorbdity index for now. But we do plan to devise
own weighted adjustments in the future.
For testing and evaluation step, that is not a regular
part CDSS pipeline , we plan to employ statistical
testing. At first, chi-squared test of independence will
identify the dependence of the 30-days hospital
readmissions on each of the thirty Elixhauser index
cormobidities or otherwise. Logistic regression based
feature scoring will then calculate the weighted
influence of each comorbidity on the 30-days hospital
readmissions classification. Principal component
analysis will show the top 5 components affecting
hosiptal readmissions prediction and alignment of
identified comorbisities with those key components.
They help us in comparing infleunetial comorbidities
in different datasets as well as comparative evaluation
of the variance explained by multiple models (based
on datasets explained in the next sub-section). It is
dreiterate that the purpose of PCA is not to suggest new
prinicipal components but deomstrate the alignment of
various diseases/comorbidities included in Elixhauser
model with the logistic regression feature scores
(weights). Lastly, a t-test of summary Elixhauser
comorbidity scores calculated from these datasets
based on the diagnosed codes, NLP-extracted codes,
and a combination of the two types of codes, enable a
comparative assessment of each.
3.2 Data Preparation
We used MIMIC-III clinical database containing over
58976 all-cause admissions records of aorund 40,000
patients staying at Beth Israel Deaconess Hospital
between 2001 and 2012 (Johnson et al, 2016, 2019).
The database is anonymized and the calendars of all
the events have been off-set. The dataset is open to
public with terms and conditions of use. It contains
sturctued EHR data on patient’s admissions, labs,
treatments, medicines, transfer, and discharges as
well as unstructured textual discharge summaries
dictateted or narrated by physicians.
An NLP-Enhanced Approach to Test Comorbidities Risk Scoring Based on Unstructured Health Data for Hospital Readmissions Prediction
653

Records with 30-days readmissions were identified
by scanning multiple admission IDs for the same
patient ID and the days between the discharge and next
admission. Patients readmitted within 30 days of
discharge were labelled ‘1’ while others were labelled
‘0’. Since it is mainly an emergency room database,
filters were applied to exclude NEWBORN or
ELECTIVE admissions, leaving only 37812 records.
After counting the frequency of admitted patients
against specific diseases, the records were filtered only
for those diseases where the patient count was 100 or
more, leaving us with only 29528 records.
Clinical notes for each admission were then
extracted from NOTEEVENTS. Even though they
contain lab reports, discharge summaries, and nursing
reports, etc., we only considered discharge summaries
for each admission since they contain all the
information for this analysis. If there were multipe
discharge summaries against an admission ID, only
the most recent one was kept. That brought down the
dataset to 21368 entries. After balancing the numbers
of readmitted and non-readmitted patients for the
final sample dataset, it was reduced to 3213 patients
with discharge summaries.
3.2.1 ICD-10 Code Extraction (Prediction)
from Unstructured Texts
Predicting appicable ICD-10 codes correctly from the
discharge summaries played a key role in calculating
Elixhauser comorbidity score. Since the process of
training a BERT would take a lot of time and
computational resources, a pre-trained BERT model
was acquired from the Hugging face community to
predict ICD-10 codes from the discharge summaries
against each admission (Devlin et,a;, 2018; Hugging
face, n.d.). The BERT model extracted multiple codes
successfully from the unstructured text of each
discharge summary.
We noted a couple of limitations working with
BERT here. The first one was the limited size of
unstructured text a BERT model could take as an
input. To overcome that, larger discharge summaries
were broken down into smaller blocks and serially
processed through BERT. Second, if certain portions
or types of textual summaries are richer than others in
terms of terminologies related to the diseases, they
will generate more codes, hence lot of overlapping
codes to be dealt with. In ICD-10 codes, it is common
to have extended codes adding depth to the diseases
classification. Therefore, keeping the codes used for
predictions uniform is another challenge.
Extended codes were removed from the extracted
codes and results were saved as ‘PREDICTED_
COMORBIDITIES’ dataset. Similarly, ICD-10 data
was obatined for the specific diagnosis made by the .
The ICD-10 codes were the saved as ‘DIAGNOSED’
dataset. A third data set was constucted by combing
the two above datasets. This combined dataset
was titled ‘DIAGNOSED + PREDICTED
COMORBIDITIES’.
3.2.2 Final Data Parsing for Comorbidities
Risk Calculation and Hospital
Readmissions Prediction Model
Next, each of the 30 diseases listed in the Elixhauser
comoribidity index was separtaetd in a column
against 3213 patient-admission rows. If an ICD-10
code was found for a disease, it was marked 1
otherwise 0. In this way, not only multiple ICD-10
codes for the diseases in the same disease related
groups (DRGs) were eliminated in a pateint, but the
correct count of comoborbidities a patient had was
also recorded in a table format that could be readily
input to a classfier.
Since the above data pasing was done first for the
codes of diseases directly entered into the EHRs, it
only prepared one dataset DIAGNOSED. The above
parsing steps were repeated on BERT extracted codes
and saved as PREDICTED_CORMORBIDITIES’
dataset with its own 30 columns for the diseases and
count of comorbidities based on the extracted codes.
Lastly, the codes from both the above datasets were
logically “ORed” to keep one instance from either
DIANOSED or PREDICTED_CORMORBIDITIES’
and the newly formed dataset was saved as
DIAGNOSED+PREDICTED_ COMORBIDITIES’.
Elixhauser comoridity score was also calculated
for each patient-admission in three separate columns
just for comparitive evaluation of the three scenarios
covered in the above datasets.
Our holistic 30-days hospital readmissions
prediction model takes into account variables
(features) from multiple areas. However, for the
purpose of measuring direct effect of each comorbidity
listed in Elixhausers’s index on hospital readmissions,
we used simple logistic regression model with 30DRA
as target variable and all the thirty comorbid diseases
as features. For compartiave evaluation purposes
between physician diagnosed, NLP extracted and a
combination of both, the model was tested on the three
above datasets separately. In this way, it was a reduced
model of the original just to test the effectiveness of
NLP-extracted comorbidities from unsturctued text.
The aim is to incoporate a summary cormobdity score
as one a single feature into the holistic 30-days-
hospital-readmissions prediction model.
CCH 2023 - Special Session on Machine Learning and Deep Learning for Preventive Healthcare and Clinical Decision Support
654

4 RESULTS AND ANALYSIS
As mentioned earlier, several statistical tests were
conducted on the diseases/comorbidities datasets
including chi-square test for independence, logistic
regression feature scoring and principal component
analysis (PCA). Many statistical tests were performed
mainly because we wanted to leverage the pros of
multiple techniques to analyse the performance from
various angles. The above tests were conducted on
three different datasets mentioned above i.e.
DIAGNOSED, PREDICTED COMORBIDITIES and
DIAGNOSED + PREDICTED COMORBIDITIES
datasets in relation with 30-days hospital readmissions
predictions. Table 1 presents the complete results of
these tests attached at the end of the paper due to its
size and format. The results reported in Table 2 have
been sorted (rank-ordered) as per logistic regression
feature scores of the comorbidities based on NLP-
predicted codes.
4.1 Comorbidities Diagnosed by the
Physicians
A chi-square test of independence was performed to
test the relation between each Elixhauser comorbid
condition and 30-days hospital re-admission. With N
= 3213, df = 30, the critical value for the chi-square
distribution comes out at 43.773. For the
DIAGNOSED scenario, the names of the diseases
(along with their ICD-9 does) are diagnosed by the
physicians. All values of chi-square statistics are
below the critical value indicating significant
relationship between all comorbidity measures in the
EHI and hospital readmissions at p<0.5 significance
level. The logistic features scores on its right-side
highlight Hypertension, Congestive Heart Failure,
Hyperthyroidism, Renal Failures and Peripheral
Vascular Disorders as the top five contributing
comorbidities to hospital readmissions. However,
there are at least 10 features that have NaN values
which implies there are several conditions that are not
commonly diagnosed (or ignored) by the physicians.
There is a possibility that this could be due to the
Emergency Room data of MIMIC-III that certain
types of diseases or conditions were not common
among this group. Furthermore, even though top 5
principal components identified by PCA analysis
explain 74% of the variance (See Table 1), they are
not very well-aligned with the key features
highlighted by the logistic regression.
Table 1: Cumulative variance explained by principal
components.
Cumulative variance
1 PC 2 PCs 3 PCs 4 PCs 5 PCs
DIAGNOSED 0.29 0.44 0.58 0.68 0.74
PREDCITED_
COMORBIDITIES
0.25 0.47 0.6 0.71 0.78
DIAGNOSED +
PREDCITED
COMORBIDITIES
0.27 0.47 0.61 0.71 0.77
4.2 Comorbidities Extracted by NLP
The Chi-square test performed on the PREDICTED
COMORBIDITIES dataset also returns chi-square
statistics for all the co-morbidities well below the
critical value. Therefore, the relation between EHI
comorbidities predicted by the NLP is significant at
p<0.05. However, there are conspicuous differences
in the diseases/comorbidities highlighted as most
influential ones in this dataset in comparison to the
DIAGNOSED dataset. Except for the AIDS/HIV
Lymphoma which is life-threatening, rest of them
appear to be chronic but slowly developing conditions
gradually affecting patient’s quality of life. It is not
surprising that Psychoses, Hypothyroidism,
Rheumatoid Arthritis, Peptic ulcer diseases, and drug
abuse have been noted in the unstructured parts of the
EHRs but not in the main diagnosis. The doctors
would typically note the most prevalent and billed
comorbidities in their diagnoses while discussing the
other notable historical conditions in the notes.
It shows the effectiveness of NLP techniques in
identifying conditions that are otherwise side-lined.
But what is even more notable are the 78% variance
explained by principal components as well as much
better alignment between the logistic regression
features and the principal components. This looks like
a stronger model than the first one. But at the same, it
is surprising that well-known comorbidities leading
to early mortality, such as congestive heart failure and
hypertension have not been identified as key factors
behind early hospital readmissions. There are few
overlaps between the significant principal
components of the two datasets.
4.3 Combined Comorbidities
The Chi-square test performed on this combined
dataset yields the same significant results for p<0.5
that all considered comorbidities are related to
hospital readmissions. However, these are even better
An NLP-Enhanced Approach to Test Comorbidities Risk Scoring Based on Unstructured Health Data for Hospital Readmissions Prediction
655
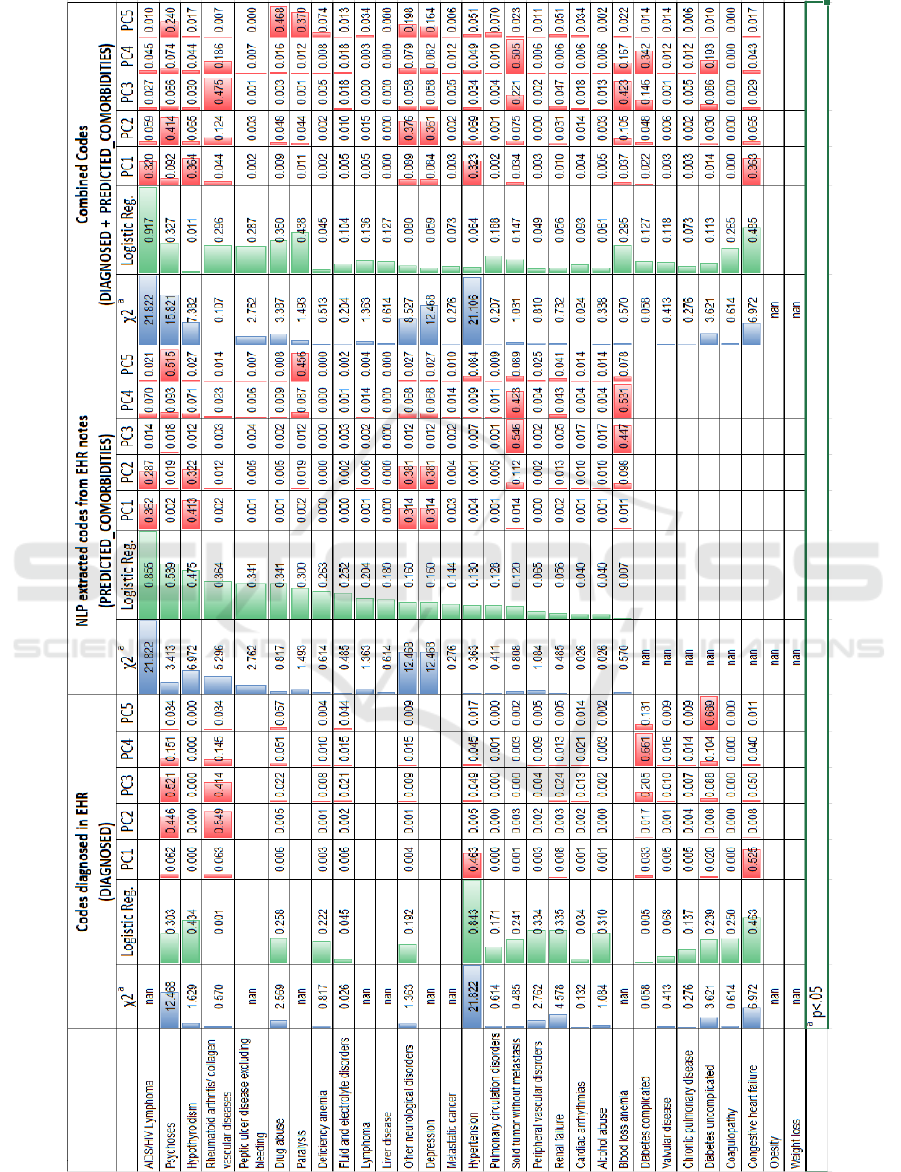
Table 2: Results of Chi-square, Logistic Regression Feature Scoring and Principal Component Analysis – sorted on Logistic
Regression Features Scores of Predicted Codes.
CCH 2023 - Special Session on Machine Learning and Deep Learning for Preventive Healthcare and Clinical Decision Support
656
since quite intuitively, this dataset has fewer NaN
values i.e. only two compared to the eight and ten of
the other two models. In other words, there are more
significant relations between comorbidities and
hospital readmissions in this dataset. The logistic
regression feature scores definitely overcome the
limitations shown by each of the earlier models. For
instance, the top five influential features include
Congestive Heart Failure and AIDS/HIV lymphoma
on one hand and on the other hand they also check
slowly progressing diseases like psychoses, paralysis
and drug abuse. The combined variance explained by
top 5 principal components is not improved further
staying at 77% but it is evident visually that many
more features are contributing to an align with these
principal components.
4.4 Comparative Classification
Performance of the Three EHI
Models
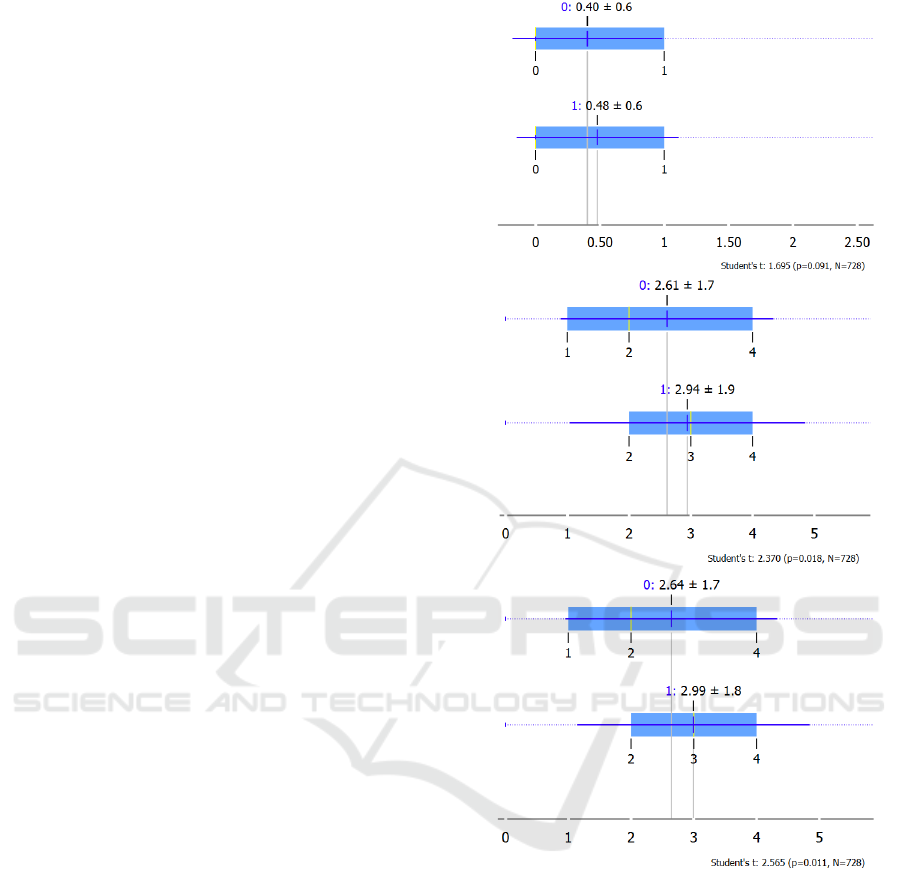
The student T-tests for independent samples were
performed on the three variants of the Elixhauser
comorbidity scoring models discussed in the previous
sub-sections. Each data set (N=3123) was broken
into a 75% training (n= 2395) and 25% test dataset
(n=728). A logistic regression classifier was trained
with the training dataset and then tested for
classification performance (0,1) with the test dataset.
The results are shown in Figs 1 a-c.
The t-statistic of classification done by scoring
model using diagnosed codes appears better but with
p>.05 it becomes insignificant. Scoring models based
on predicted codes (stand-alone or combination) are
significant at p<0.5 hence they tend to perform better
for hospital readmissions classification.
Overall, it is abundantly clear that NLP-enhanced
comorbidity scoring has merit and potential to
improve performance of the CDSS employing
comorbidities and comorbidity summary scores.
5 CONCLUSIONS
Improved comorbidity scoring is valuable as one of
the features in AI-based decision models. This
research aimed to test if comorbidity scoring can be
improved using NLP-enhanced approaches,
especially by gleaning additional ICD-10 codes from
unstructured portions of the EHRs. Such
improvements have huge implications for better
healthcare delivery, cost savings, and patient
outcomes.
Figure 1: a, b and c: Classification performance of the three
datasets based on calculated Elixhauser comorbidity scores
(a) Diagnosed, (b) Predicted_Comorbidities, and (c)
Combination of Diagnosed + Predicted_Comorbidities.
We employed a BERT model to glean additional
ICD-10 codes from unstructured portions of the
MIMIC-III patient EHRs. The comorbidities
represented by these codes and included in Elixhauser
index were then tested for hospital readmission
classification, separately as well as in combination
with existing codes diagnosed by the physians. We
noted improvements both in comorbidity
measurements as well as 30-days hospital
readmissions predictions. It is anticipated that better
NLP techniques, such as HLAN and KGs will offer
An NLP-Enhanced Approach to Test Comorbidities Risk Scoring Based on Unstructured Health Data for Hospital Readmissions Prediction
657

even more improvements. We also demostrated any
improvements in Computer-assisted coding (CAC)
especially for ICD-10 and ICD-11 codes will also
support other venues in the CDSS area. There is room
for building user trust in CDSS.
Like every research study, we faced limitations.
At first, the MIMIC-III dataset is primarily an
emergency admissions database. The physicians’
healthcare goals in emergency settings are different
from those in a general admissions so they may view
comorbidites and some variables differently from the
physicians handling regular admissions. The high
variations in logistic regression features scores
between DIAGNOSED and
PREDICTED_COMORBIDITIES could partly be
stemming from such differences. Another limitation
could be using a binary classifier for 30-days hospital
readmissions. The performance and effects would
have been more practical if a 30-days risk scoring
model was used using comorbidity summary score/s
as feature/s.
Methods and pipelines for testing and analysis can
also be improved. The effects of most diseases are
caclulated separately. Multimorbidity indices are
considered more relevant for assessing risks in some
medical areas. Co-occurene and covariances would
have be to accounted in comborbidy risk calculations.
We do intend to revisit and improve this research
using an HLAN (Hierarchical Label-wise Attention
Network) technique and a holistic 30-days risk
scoring model (including medical, demographic and
socio-economic features) as the final output of our
hospital readmission risks score cosidering alternate
care facilities.
REFERENCES
AHRQ, n.d., Agency for Healthcare Research and Quality
– Healthcare Cost and Utilization Project (AHRQ-
HCUP), Available at: https://www.hcup-us.ahrq.gov/
toolssoftware/comorbidityicd10/comorbidity_icd10.js
p, Last accesses: Nov 12, 2022.
Alsentzer, E., Murphy, J. R., Boag, W., Weng, W. H., Jin,
D., Naumann, T., & McDermott, M. (2019). Publicly
available clinical BERT embeddings. arXiv preprint
arXiv:1904.03323.
Arnaud, É., Elbattah, M., Gignon, M., & Dequen, G. (2021,
August). NLP-Based Prediction of Medical Specialties
at Hospital Admission Using Triage Notes. In 2021
IEEE 9th International Conference on Healthcare
Informatics (ICHI) (pp. 548-553). IEEE.
Aya (2020) Named entity recognition, Named Entity
Recognition, NER in NLP, NER Annotation. Available
at: https://www.ayadata.ai/service/named-entity-
recognition (Accessed: November 12, 2022).
Bao, W., Lin, H., Zhang, Y., Medical code prediction via
capsule networks and ICD knowledge. BMC Med
Inform Decis Mak 21 (Suppl 2), 55 (2021).
https://doi.org/10.1186/s12911-021-01426-9
Bayliss, E. A., Edwards, A. E., Steiner, J. F., & Main, D. S.
(2008). Processes of care desired by elderly patients
with multimorbidities. Family practice, 25(4), 287-293.
Bottle, A., & Aylin, P. (2011). Comorbidity scores for
administrative data benefited from adaptation to local
coding and diagnostic practices. Journal of clinical
epidemiology, 64(12), 1426-1433.
Cambria, E., & White, B. (2014). Jumping NLP curves: A
review of natural language processing research. IEEE
Computational intelligence magazine, 9(2), 48-57. doi:
10.1109/MCI.2014.2307227.
Chaudhri, V., Baru, C., Chittar, N., Dong, X., Genesereth,
M., Hendler, J., & Wang, K. (2022). Knowledge
Graphs: Introduction, History and, Perspectives. AI
Magazine, 43(1), 17-29.
Charlson, M. E., Pompei, P., Ales, K. L., & MacKenzie, C.
R. (1987). A new method of classifying prognostic
comorbidity in longitudinal studies: development and
validation. Journal of chronic diseases, 40(5), 373-383.
De Giorgi, A., Di Simone, E., Cappadona, R., Boari, B.,
Savriè, C., López-Soto, P. J., & Fabbian, F. (2020).
Validation and Comparison of a Modified Elixhauser
Index for Predicting In-Hospital Mortality in Italian
Internal Medicine Wards. Risk Management and
Healthcare Policy, 13, 443.
Devlin, J., Chang, M. W., Lee, K., & Toutanova, K. (2018).
Bert: Pre-training of deep bidirectional transformers for
language understanding. arXiv preprint
arXiv:1810.04805.
Elixhauser, A., Steiner, C., Harris, D. R., & Coffey, R. M.
(1998). Comorbidity measures for use with
administrative data. Medical care, 8-27.
Esteban-Gil, A., Fernández-Breis, J. T., & Boeker, M.
(2017). Analysis and visualization of disease courses in
a semantically-enabled cancer registry. Journal of
biomedical semantics, 8(1), 1-16.
Evans, R. Scott. “Electronic health records: then, now, and
in the future.” Yearbook of medical informatics 25.S 01
(2016): S48-S61.
Fabbian, F., De Giorgi, A., Maietti, E., Gallerani, M., Pala,
M., Cappadona, R., & Fedeli, U. (2017). A modified
Elixhauser score for predicting in-hospital mortality in
internal medicine admissions. European Journal of
Internal Medicine, 40, 37-42.
Feinstein, A. R. (1970). The pre-therapeutic classification
of co-morbidity in chronic disease. Journal of chronic
diseases, 23(7), 455-468.
Gasparini, A. (2018). Comorbidity: An R package for
computing comorbidity scores. Journal of Open Source
Software, 3(23), 648.
Ghazalbash, S., Zargoush, M., Mowbray, F., &
Papaioannou, A. (2021). Examining the predictability
and prognostication of multimorbidity among older
Delayed-Discharge Patients: A Machine learning
analytics. International Journal of Medical Informatics,
156, 104597.
CCH 2023 - Special Session on Machine Learning and Deep Learning for Preventive Healthcare and Clinical Decision Support
658

Greenfield, S., Apolone, G., McNeil, B. J., & Cleary, P. D.
(1993). The importance of co-existent disease in the
occurrence of postoperative complications and one-
year recovery in patients undergoing total hip
replacement: comorbidity and outcomes after hip
replacement. Medical care, 31(2), 141-154.
Goltz, D. E., Ryan, S. P., Howell, C. B., Attarian, D.,
Bolognesi, M. P., & Seyler, T. M. (2019). A weighted
index of Elixhauser comorbidities for predicting 90-day
readmission after total joint arthroplasty. The Journal
of arthroplasty, 34(5), 857-864.
Hameed, T., & Bukhari, S. A. C. (2020). Predicting 30-days
All-cause Hospital Readmissions Considering
Discharge-to-alternate-care-facilities. In HEALTHINF
(pp. 864-873).
Havlik, R. J., Yancik, R., Long, S., Ries, L., & Edwards, B.
(1994). The National Institute on Aging and the
National Cancer Institute SEER collaborative study on
comorbidity and early diagnosis of cancer in the
elderly. Cancer, 74(S7), 2101-2106.
Huggingface, n.d., Huggingface BERT model, available at:
https://huggingface.co/docs/transformers/model_doc/b
ert, [Last accessed: Nov 12, 2022]
Johnson, A. E., Pollard, T. J., Shen, L., Lehman, L. W. H.,
Feng, M., Ghassemi, M., ... & Mark, R. G. (2016).
MIMIC-III, a freely accessible critical care database.
Scientific data, 3(1), 1-9.
Johnson, A. E., Pollard, & Mark, R. G. (2019). MIMIC-III
Clinical Database Demo (version 1.4). PhysioNet.
http://doi.org/10.13026/C2HM2Q.
Le, N., Wiley, M., Loza, A., Hristidis, V., & El-Kareh, R.
(2020). Prediction of Medical Concepts in Electronic
Health Records: Similar Patient Analysis. JMIR
Medical Informatics, 8(7), e16008. https://doi.org/
10.2196/16008
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., &
Kang, J. (2020). BioBERT: a pre-trained biomedical
language representation model for biomedical text
mining. Bioinformatics, 36(4), 1234-1240.
Menendez, M. E., Neuhaus, V., van Dijk, C. N., & Ring, D.
(2014). The Elixhauser comorbidity method
outperforms the Charlson index in predicting inpatient
death after orthopaedic surgery. Clinical Orthopaedics
and Related Research®, 472(9), 2878-2886.
Moore, B. J., White, S., Washington, R., Coenen, N., &
Elixhauser, A. (2017). Identifying increased risk of
readmission and in-hospital mortality using hospital
administrative data. Medical care, 55(7), 698-705.
Mukherjee, B., Ou, H. T., Wang, F., & Erickson, S. R.
(2011). A new comorbidity index: the health-related
quality of life comorbidity index. Journal of clinical
epidemiology, 64(3), 309-319.
Piccirillo, J. F., Tierney, R. M., Costas, I., Grove, L., &
Spitznagel Jr, E. L. (2004). Prognostic importance of
comorbidity in a hospital-based cancer registry. Jama,
291(20), 2441-2447.
Quan, H., Sundararajan, V., Halfon, P., Fong, A., Burnand,
B., Luthi, J. C., & Ghali, W. A. (2005). Coding
algorithms for defining comorbidities in ICD-9-CM
and ICD-10 administrative data. Medical care, 1130-
1139.
Quan, H., Li, B., Couris, C. M., Fushimi, K., Graham, P.,
Hider, P., & Sundararajan, V. (2011). Updating and
validating the Charlson comorbidity index and score for
risk adjustment in hospital discharge abstracts using
data from 6 countries. American journal of
epidemiology, 173(6), 676-682.
Panchendrarajan, R., & Amaresan, A. (2018). Bidirectional
LSTM-CRF for named entity recognition. In
Proceedings of the 32nd Pacific Asia Conference on
Language, Information and Computation.
Sharabiani, M. T., Aylin, P., & Bottle, A. (2012).
Systematic review of comorbidity indices for
administrative data. Medical care, 1109-1118.
Sharma, N., Schwendimann, R., Endrich, O., Ausserhofer,
D., & Simon, M. (2021). Comparing Charlson and
Elixhauser comorbidity indices with different
weightings to predict in-hospital mortality: an analysis
of national inpatient data. BMC health services
research, 21(1), 1-10.
Soomro, P. D., Kumar, S., Shaikh, A. A., & Raj, H. (2017).
Bio-NER: biomedical named entity recognition using
rule-based and statistical learners. International
Journal of Advanced Computer Science and
Applications, 8(12).
Sung, S. F., Chen, C. H., Pan, R. C., Hu, Y. H., & Jeng, J.
S. (2021). Natural Language Processing Enhances
Prediction of Functional Outcome After Acute
Ischemic Stroke. Journal of the American Heart
Association, 10(24), e023486. https://doi.org/10.1161/
JAHA.121.023486
Tome Eftimov, Barbara Koroušić Seljak, and Peter
Korošec. “A rule-based named-entity recognition
method for knowledge extraction of evidence-based
dietary recommendations”. In: PloS one 12.6 (2017),
e0179488
Vafajoo, A., Salarian, R., & Rabiee, N. (2018).
Biofunctionalized microbead arrays for early diagnosis
of breast cancer. Biomedical Physics & Engineering
Express, 4(6), 065028.
Valderas, J. M., Starfield, B., Sibbald, B., Salisbury, C., &
Roland, M. (2009). Defining comorbidity: implications
for understanding health and health services. The
Annals of Family Medicine, 7(4), 357-363.
van Walraven, C., Austin, P. C., Jennings, A., Quan, H., &
Forster, A. J. (2009). A modification of the Elixhauser
comorbidity measures into a point system for hospital
death using administrative data. Medical care, 626-633.
Von Korff, M., Wagner, E. H., & Saunders, K. (1992). A
chronic disease score from automated pharmacy data.
Journal of clinical epidemiology, 45(2), 197-203.
Wang, J., Deng, H., Liu, B., Hu, A., Liang, J., Fan, L.,
Zheng, X., Wang, T., & Lei, J. (2020). Systematic
Evaluation of Research Progress on Natural Language
Processing in Medicine Over the Past 20 Years:
Bibliometric Study on PubMed. Journal of medical
Internet research, 22(1), e16816.
https://doi.org/10.2196/16816
Wolff, J. L., Starfield, B., & Anderson, G. (2002).
Prevalence, expenditures, and complications of
multiple chronic conditions in the elderly. Archives of
internal medicine, 162(20), 2269-2276.
An NLP-Enhanced Approach to Test Comorbidities Risk Scoring Based on Unstructured Health Data for Hospital Readmissions Prediction
659
