
Natural Language Explanatory Arguments for Correct and Incorrect
Diagnoses of Clinical Cases
Santiago Marro
a
, Benjamin Molinet
b
, Elena Cabrio
c
and Serena Villata
d
Universit
´
e C
ˆ
ote d’Azur, Inria, CNRS, I3S, France
Keywords:
Natural Language Processing, Information Extraction, Argument-based Natural Language Explanations,
Healthcare.
Abstract:
The automatic generation of explanations to improve the transparency of machine predictions is a major chal-
lenge in Artificial Intelligence. Such explanations may also be effectively applied to other decision making
processes where it is crucial to improve critical thinking in human beings. An example of that consists in the
clinical cases proposed to medical residents together with a set of possible diseases to be diagnosed, where
only one correct answer exists. The main goal is not to identify the correct answer, but to be able to explain
why one is the correct answer and the others are not. In this paper, we propose a novel approach to generate
argument-based natural language explanations for the correct and incorrect answers of standardized medical
exams. By combining information extraction methods from heterogeneous medical knowledge bases, we pro-
pose an automatic approach where the symptoms relevant to the correct diagnosis are automatically extracted
from the case, to build a natural language explanation. To do so, we annotated a new resource of 314 clinical
cases, where 1843 different symptoms are identified. Results in retrieving and matching the relevant symp-
toms for the clinical cases to support the correct diagnosis and contrast incorrect ones outperform standard
baselines.
1 INTRODUCTION
Explanatory Artificial Intelligence (XAI) is a main
topic in AI research nowadays, given, on the one side,
the predominance of black box methods, and on the
other side, the application of these methods to sen-
sitive scenarios like medicine. Among the huge set
of contributions in this area (Tjoa and Guan, 2019;
Saeed and Omlin, 2021), some approaches highlight
the need to build explanations which are clearly in-
terpretable and possibly convincing, leading to the in-
vestigation of the generation of argument-based ex-
planations (Cyras et al., 2021). These explanations
are intended to be not only rational, but “manifestly”
rational (Johnson, 2000), such that arguers can see
for themselves the rationale behind inferential steps
taken. This task becomes even more challenging if we
target the generation of natural language argument-
based explanations (Cyras et al., 2021; Vassiliades
et al., 2021).
a
https://orcid.org/0000-0001-6220-0559
b
https://orcid.org/0000-0002-8208-2139
c
https://orcid.org/0000-0002-8208-2139
d
https://orcid.org/0000-0003-3495-493X
In this paper, we tackle this challenging task, fo-
cusing on a specific application scenario, i.e., the gen-
eration of explanatory natural language arguments in
medicine. More precisely, our goal is to automati-
cally generate natural language argument-based ex-
planations to be used for educational purposes to train
medical residents. These students are trained through
tests where first there is the description of a clinical
case (i.e., symptoms experienced by the patient, re-
sults of clinical exams and analysis, and some further
information concerning the patient herself like age,
gender, or population group), and they need to an-
swer the following question: ”Which of the following
is the most likely diagnosis?”. The test is composed
of a number of possible answers to this question, i.e.,
potential diagnoses, among which, one of them is the
correct diagnosis, and the others are incorrect. The
solution consists in selecting the correct answer. In
addition, medical residents are asked to justify their
answer through an explanation. In order to autom-
atize this training phase, we address the task of au-
tomatically generating explanations of the kind: ”The
patient is affected by [diagnosis
x
] because the follow-
ing relevant symptoms have been identified: [correct
438
Marro, S., Molinet, B., Cabrio, E. and Villata, S.
Natural Language Explanatory Arguments for Correct and Incorrect Diagnoses of Clinical Cases.
DOI: 10.5220/0011927000003393
In Proceedings of the 15th International Conference on Agents and Artificial Intelligence (ICAART 2023) - Volume 1, pages 438-449
ISBN: 978-989-758-623-1; ISSN: 2184-433X
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

diagnosis symptoms]. The [diagnosis
y
] is incorrect
because the patient is not showing the symptoms [in-
correct diagnosis symptoms]”.
To address this task, a full pipeline needs to be
designed in order to (i) detect the symptoms in the
clinical case description, (ii) match them with the
symptoms, in a medical knowledge base, to iden-
tify to which diseases they are associated with, and
what is their frequency, and finally, (iii) generate
pattern-based natural language explanations employ-
ing the elements identified in the two previous steps.
To do so, we first annotate a new resource of 314
unique clinical cases in English, with the symptoms
which are relevant to derive the correct and incor-
rect diagnoses. These symptoms are extracted from
the Human Phenotype Ontology (HPO) knowledge
base (K
¨
ohler et al., 2021), where each disease is as-
sociated with the list of symptoms that can be mani-
fested in this disease.
Relying on contextual embedding search, our con-
tribution is threefold: (i) we detect in the clinical case
description, the symptoms from a newly annotated
resource; (ii) we automatically match the symptoms
with those available on HPO, with the aim to asso-
ciate them to the correct and incorrect diagnoses, and
(iii) natural language explanatory arguments are auto-
matically generated. We address an extensive evalua-
tion of this new full pipeline to generate natural lan-
guage explanations for clinical cases, obtaining very
promising results. The work we present in this paper
is motivated by the lack of existing medical textual
resources annotated with symptoms associated with
diagnoses and the need for effective methods to ad-
dress natural language explanations in medicine. To
the best of our knowledge, this is the first approach to
generate such a kind of natural language explanations
in the medical domain for educational purposes, i.e.,
to train medical residents to generate effective natural
language explanations about the correct and incorrect
diagnosis of a clinical case.
2 RELATED WORK
Since the introduction of BERT (Devlin et al., 2019),
transformer-based models have recently had a ma-
jor impact on most NLP tasks. Multiple models
evolved from it with different design choices, like
RoBERTa (Liu et al., 2019), ELECTRA (Clark et al.,
2020) and ALBERT (Lan et al., 2019). These models
are trained on a large amount of data from multiple
sources and domains, which means that they are not
necessarily prepared for the biomedical domain.
In recent years, a great number of resources
and NLP tools have been developed specifically for
the biomedical domain. For entity extraction, the
most popular datasets are BC4CHEMD (Krallinger
et al., 2015), B5CDR-Chem (Li et al., 2016), NCBI-
Disease (Do
˘
gan et al., 2014), BC2GM (Smith et al.,
2008), JNLPBA (Kim et al., 2004), where the an-
notations range from drug-disease interactions to the
identification of diseases, genes, and molecular enti-
ties such as protein, DNA, RNA. Symptom detection,
i.e., the task we address in this paper, can be seen as a
sub-task of the broader task of medical entity extrac-
tion.
Off-the-shelf NLP tool-kits such as Spacy (Honni-
bal and Montani, 2017), MedSpacy (Eyre et al., 2021)
and CLAMP (Soysal et al., 2018) provide multiple
modules for text processing. In particular, MedSpacy
is built on top of Spacy specifically for clinical natural
language processing, while CLAMP offers a method
for named entity recognition (NER) as well as a visual
interface for annotating and labeling clinical text.
Most of the recent approaches treat NER as a se-
quence labeling task where specialized transformer-
based models hold the best results. For exam-
ple, (Naseem et al., 2021) showed that pre-training the
ALBERT model on a huge biomedical corpus ensured
that the model captured better biomedical context-
dependent NER. Results outperform non-specialized
models obtaining SOTA results in a lot of datasets.
Similar results can be seen in (raj Kanakarajan et al.,
2021), where the authors pre-train a biomedical lan-
guage model using biomedical text and vocabulary
with the technique proposed by ELECTRA. Other
specialized models based on BERT have been pro-
posed by (Beltagy et al., 2019), (Lee et al., 2020)
and (Gu et al., 2020) and BioMed-RoBERTa (Guru-
rangan et al., 2020) based on RoBERTa.
(Michalopoulos et al., 2020) propose UmlsBERT,
a contextual embedding model that integrates domain
knowledge from the Unified Medical Language Sys-
tem (UMLS) (Bodenreider, 2004), taking into consid-
eration structured expert domain knowledge. They
show that UmlsBERT can associate different clini-
cal terms with similar meanings in the UMLS knowl-
edge base and create meaningful input embeddings by
leveraging information from the semantic type of each
word. In our work, we compare the representation of
the symptoms found in the clinical case with different
contextual embeddings with the goal to find a repre-
sentation which matches the one provided in the Hu-
man Phenotype Ontology (HPO).
Ngai et al. (Ngai and Rudzicz, 2022) also tackle
the problem of finding relevant clinical information,
where among the entities they also identify symp-
toms. In contrast to our work, they only focus on 6
Natural Language Explanatory Arguments for Correct and Incorrect Diagnoses of Clinical Cases
439

specific diagnoses. Furthermore, their goal is to pre-
dict the correct diagnosis and explain these predic-
tions using feature attribution methods, whilst ours is
to generate high-quality explanations in natural lan-
guage for educational purposes, i.e., to improve med-
ical residents’ skills in explaining their answer to the
test.
Besides detecting symptoms from clinical cases,
in our work, we also aim to accurately map them
to medical ontologies, such as the Human Pheno-
type Ontology (HPO), to identify the relationship be-
tween the symptoms (originally described in layper-
son terms) and diseases. Recent work by (Manzini
et al., 2022) proposes a tool for automatically translat-
ing between layperson terminology and HPO, using
a vector space and a neural network to create vector
representations of medical terms and compare them
to layperson versions. However, this approach has a
limitation in that it translates layperson terms with-
out considering their context, potentially missing rel-
evant information that may change the semantics of
the term. In our work, we propose a method that takes
into account the context in which the layperson term
is introduced, leading therefore to an accurate map-
ping to an HPO term.
Natural language explanation generation has re-
ceived a lot of attention in recent years, grounding
on the progress of generative models to train specific
models for explanations. (Camburu et al., 2018) gen-
erate explanations by justifying a relation (entailment,
contradiction or neutral) for a premise-hypothesis
pair by training a Bi-LSTM on their e-SNLI dataset,
i.e., the Stanford Natural Language Inference (Bow-
man et al., 2015) dataset augmented with an explana-
tion layer which explains the SLNI relations. (Kumar
and Talukdar, 2020) propose to generate short expla-
nations with GPT-2 (Radford et al., 2019), learned to-
gether with the input by a classifier to improve the
final label prediction, using e-SNLI (Camburu et al.,
2018). These solutions are not applicable to our use
case given that explaining a medical diagnosis is a
more challenging task than restraining the explana-
tions to the three basic relations considered by (Cam-
buru et al., 2018) and (Kumar and Talukdar, 2020).
(Narang et al., 2020) propose an approach based on
the T5 model (Raffel et al., 2019) to generate an ex-
planation after prediction. Again, this solution is not
applicable to the specific medical scenario we tar-
get, where explanations require to be structured fol-
lowing precise argumentative structures (Josephson
and Josephson, 1994; Campos, 2011; Dragulinescu,
2016) and to ground on medical knowledge, like the
one we inject through the HPO.
Other approaches use explanations via tem-
plates (Reiter and Dale, 1997), e.g., (Abujabal et al.,
2017) uses templates and inject the reasoning steps
and query of their Q&A system. To the best of our
knowledge, no related work generates natural lan-
guage post-hoc explanations for the medical domain.
3 DATASET
To train and evaluate the proposed approach to build
natural language explanatory arguments, we rely on
the MEDQA dataset (Jin et al., 2021), which contains
a set of clinical case descriptions together with a set
of possible questions and answers on the correct di-
agnosis. The questions and their associated answers
were collected from the National Medical Board Ex-
amination in the USA (USMLE), Mainland China
(MCMLE), and Taiwan (TWMLE). In this work, we
only focus on the clinical cases and the questions in
English (i.e., USMLE). In total, the MEDQA-USMLE
dataset consists of 12,723 unique questions on differ-
ent topics, ranging from questions like “Which of the
following symptoms belongs to schizophrenia?” to
questions about the most probable diagnosis, treat-
ment or outcomes for a certain clinical case which is
described (Jin et al., 2021). To reach our goal, we ex-
tract the clinical cases belonging to the latter group,
which are intended to test medical residents to make
the correct diagnosis. We end up with 314 unique
clinical cases associated with the list of possible di-
agnoses.
Annotation of the MEDQA-USMLE Clinical
Cases. To annotate the clinical cases from the
MEDQA-USMLE dataset, we rely on the labels from
the Unified Medical Language System (UMLS) (Bo-
denreider, 2004) Semantic Types, making it consis-
tent with standard textual annotations in the medi-
cal domain (Campillos-Llanos et al., 2021; Albright
et al., 2013; Mohan and Li, 2019). In particular, we
annotate the following elements in the clinical case
descriptions: Sign or Symptom, Finding, No Symp-
tom Occurrence, Population Group, Age Group, Lo-
cation and Temporal Concept. In this paper, we use
only the symptoms, but we addressed a complete an-
notation to employ these data for future work. Quan-
tifiers defining a symptom have not been annotated
(e.g., we can find “moderate pain”, where we only
annotate “pain”). The labels Sign or Symptom and No
Symptom Occurrence are associated only to the text
snippet defining the symptom in a sentence. Findings
consist of such information discovered by direct ob-
servation or measurement of an organism’s attribute
or condition. For instance, components in ”Her tem-
NLPinAI 2023 - Special Session on Natural Language Processing in Artificial Intelligence
440

perature is 39.3°C (102.8°F), pulse is 104/min, respi-
rations are 24/min, and blood pressure is 135/88 mm
Hg”. Location refers to the location of a symptom in
the human body, and Temporal Concept is used to tag
time-related information, including duration and time
intervals. Population Group and Age Group highlight
information on the age and gender of the patient.
To address the annotation process of the MEDQA-
USMLE dataset, we first carried out a semi-automatic
annotation relying on the UMLS database. We pro-
cessed each clinical case through the UMLS database
and obtained all the entities detected along their Con-
cept Unique Identifiers (CUI) and their semantic type.
The semantic type is then used to disambiguate the
entities and generate the pre-annotated files. After the
definition of the detailed annotation guidelines (sum-
marized above) in collaboration with clinical doctors,
three annotators with a background in computational
linguistics carried out the annotation of the 314 clin-
ical cases. To ensure the reliability of the annotation
task, the inter-annotator agreement (IAA) has been
calculated on an unseen shared subset of 10 clinical
cases annotated by four annotators, obtaining a Fleiss’
kappa (Fleiss, 1971) of 0.70 for all of the annotated
labels, 0.61 for Sign or Symptom, 0.94 for Location,
0.71 for Population Group, 0.66 for Finding, 0.96 for
Age Group and 0.96 for No Symptoms Occurrence.
We can see a substantial agreement for Sign or Symp-
tom, Finding and Population Group, and an almost
perfect agreement for Location, Age Group and No
Symptoms Occurrence.
Table 1 reports on the statistics of the final dataset,
named MEDQA-USMLE-Symp.
1
The accuracy of
the annotations provided by the three annotators has
been validated from a medical perspective with a clin-
ical doctor. Of the seven entity labels, only three con-
tain medical vocabulary (Sign or Symptom, Finding,
and No Symptom Occurrence) and they have been
evaluated by this expert. More specifically, we ran-
domly sampled 10% of the data (i.e., 30 cases) and
we asked the clinician to verify whether the entity was
correctly labeled and whether there were any missing
or extra words. The results of the validation showed
that 98% of the data was labeled correctly. Less than
2% of the instances were evaluated as incorrectly la-
beled (e.g., a Finding that was labeled as a Sign or
Symptom or vice versa).
Knowledge Base of Diseases and Relevant Symp-
toms. To collect the medical knowledge needed to
define whether a detected symptom is relevant with
1
https://github.com/Wimmics/
MEDQA-USMLE-Symp
Table 1: Statistics of the MEDQA-USMLE-Symp dataset.
Label # Entities
Sign or Symptom 1579
Finding 1169
Temporal Concept 567
Location 498
Population Group 364
Age Group 304
No Symptom Occurrence 264
respect to a given disease, we employ the HPO knowl-
edge base to retrieve (i) the relevant information of
each diagnosis which is proposed as an option to an-
swer the question ”Which of the following is the most
likely diagnosis?”, and (ii) the symptoms (named
terms in HPO) associated to each diagnosis. This
knowledge base also includes information on the fre-
quency
2
of the occurrence of symptoms, defined in
collaboration with ORPHA
3
as follows: Excluded
(0%); Very rare (1-4%); Occasional (5-29%); Fre-
quent (30-79%); Very frequent (99-80%). Obligate
(100%); HPO integrates different sources of symp-
toms, including ORPHA and OMIM
4
. This knowl-
edge base is quite rich and contains also links and
hierarchical links between symptoms (Symptom S2
subclass of Symptom S1), genes or definitions.
4 PROPOSED FRAMEWORK
An overview of the framework we propose to address
automatic symptom relevancy assessment and match-
ing to build our natural language explanations is visu-
alized in Figure 1. Starting from the clinical cases in
which the correct and incorrect diagnosis are already
identified, the goal is to assess the relevant symptoms
present in the case such that these symptoms can be
used to explain why a certain diagnosis is the correct
one and why the incorrect ones have to be discarded.
In order to accurately diagnose a patient’s condi-
tion, it is important to identify the symptoms that are
most relevant to the possible diagnoses. This means
looking at all of the symptoms that have been detected
and determining which ones are most likely to be re-
lated to the underlying cause of the patient’s condi-
tion. This can be done by considering the individual
symptoms and their potential connections to the pos-
sible diagnoses. It is also important to consider any
additional information that may be available, such as
2
https://hpo.jax.org/app/browse/term/HP:0040279
3
https://www.orpha.net/consor/cgi-bin/index.php?lng=
FR
4
https://www.omim.org/
Natural Language Explanatory Arguments for Correct and Incorrect Diagnoses of Clinical Cases
441

the patient’s medical history and other relevant fac-
tors, in order to be able to fully explain the diagnosis.
Our work focuses on identifying relevant symptoms
in order to accurately diagnose a patient’s condition.
The relevancy assessment model associates, when
possible, the pertinent symptoms mentioned in the
clinical case description with a symptom of a diagno-
sis found in the HPO knowledge base. The proposed
framework consists of two different steps, where: (i)
we retrieve from HPO the required diagnosis informa-
tion (i.e., the symptoms and how common they are),
then the symptoms in the case are detected and ex-
tracted using an attention-based neural architecture
which relies on the clinical case text only; (ii) the
relevancy of each symptom is assessed by matching
the detected symptoms with the ones retrieved from
HPO. The matched symptoms are then used to gener-
ate natural language argument-based explanations for
correct and incorrect diagnoses. In the following, we
explain in detail each sub-task in the pipeline:
Symptoms Detection, consisting in detecting the dif-
ferent symptoms described in the clinical case (medi-
cal terms or symptoms described by the patient’s own
words). In order to detect these entities, we propose
a neural approach based on pre-trained Transformer
Language Models.
Symptoms Alignment, to align a symptom detected
in the clinical case with an identical term in HPO.
We first compute an embedding vector for each found
symptom and then calculate the cosine distance with
each term in HPO. We then assign the closest concept
to that symptom. We evaluated both static and con-
textual embedding methods.
Explanation Generation We propose template-
based explanations based solely on the symptoms that
are relevant to explain the diagnosis. To do this we
propose several templates that tackle different kinds
of explanations, going from explaining why a patient
was given a certain diagnosis to explaining why the
alternatives cannot be considered viable options. We
support our explanations with statistical information
obtained from HPO such as the frequency of each
symptom incidence, and we propose to look for pos-
sible symptoms that were not detected by the system
but are frequent for a certain disease.
5 EXPERIMENTS
In this section, we report on the experimental setup,
the obtained results and the error analysis for the
symptom detection and symptom alignment methods.
Setup. For the symptom detection task, we exper-
imented with different transformer-based Language
Models (LMs) such as BERT (Devlin et al., 2019),
SciBERT (Beltagy et al., 2019), BioBERT (Lee et al.,
2020), PubMedBERT (Gu et al., 2020) and Umls-
BERT (Michalopoulos et al., 2020) initialized with
their respective pre-trained weights. All the mod-
els we employ are specialized in the biomedical do-
main, with the exception of BERT which will serve
us as a baseline. We cast the symptom detection prob-
lem as a sequence tagging task. Following the BIO-
tagging scheme, each token is labeled as either be-
ing at the Beginning, Inside or Outside of a compo-
nent. This translates into a sequence tagging problem
with three labels, i.e., B-Sign-or-Symptom, I-Sign-or-
Symptom and Outside. To fine-tune the LMs, we use
the PyTorch implementation of huggingface (Wolf
et al., 2020) (v4.18). For BERT, we use the un-
cased base model with 12 transformer blocks, a hid-
den size of 768, 12 attention heads, and a learning
rate of 2.5e-5 with Adam optimizer for 3 epochs. The
same configuration was used to fine-tune SciBERT
BioBERT, PubMedBERT and UmlsBERT. For SciB-
ERT, we use both the cased and uncased versions, and
for BioBERT we use version 1.2. Batch size was 8
with a maximum sequence length of 128 subword to-
kens per input example.
Regarding the matching module, we experimented
with two different methods to align our detected
symptoms with terms in HPO by (i) directly compar-
ing the computed embeddings of the detected symp-
toms with the embeddings of the terms in HPO, and
(ii) by taking into account the context in which the
symptoms are detected and applying the same context
to every term in HPO.
To align our detected symptoms (in the clinical
case) with the equivalent HPO terms, we calculate the
cosine distance of each embedding of the HPO terms
with respect to the embedding of the detected symp-
tom. In the experimental setting of (i) and (ii), we use
the static pre-trained embeddings GloVe 6B as well
as BERT, SciBERT, BioBERT and UmlsBERT in the
same configurations as in the symptom detection task.
For (ii), it is necessary to calculate the context embed-
dings “on the fly” because each context is unique and
depends on the clinical case where it was detected. It
is not reasonable to recalculate all HPO term embed-
dings on the fly for each new context since the on-
tology contains 10,319 unique terms, so we propose
to generate all the HPO terms embedding at once and
save them. Therefore, this module takes as input the
symptoms detected by the previous module and finds
the context
5
of these symptoms in the clinical case.
5
The context consists of the sentence(s) containing the
NLPinAI 2023 - Special Session on Natural Language Processing in Artificial Intelligence
442
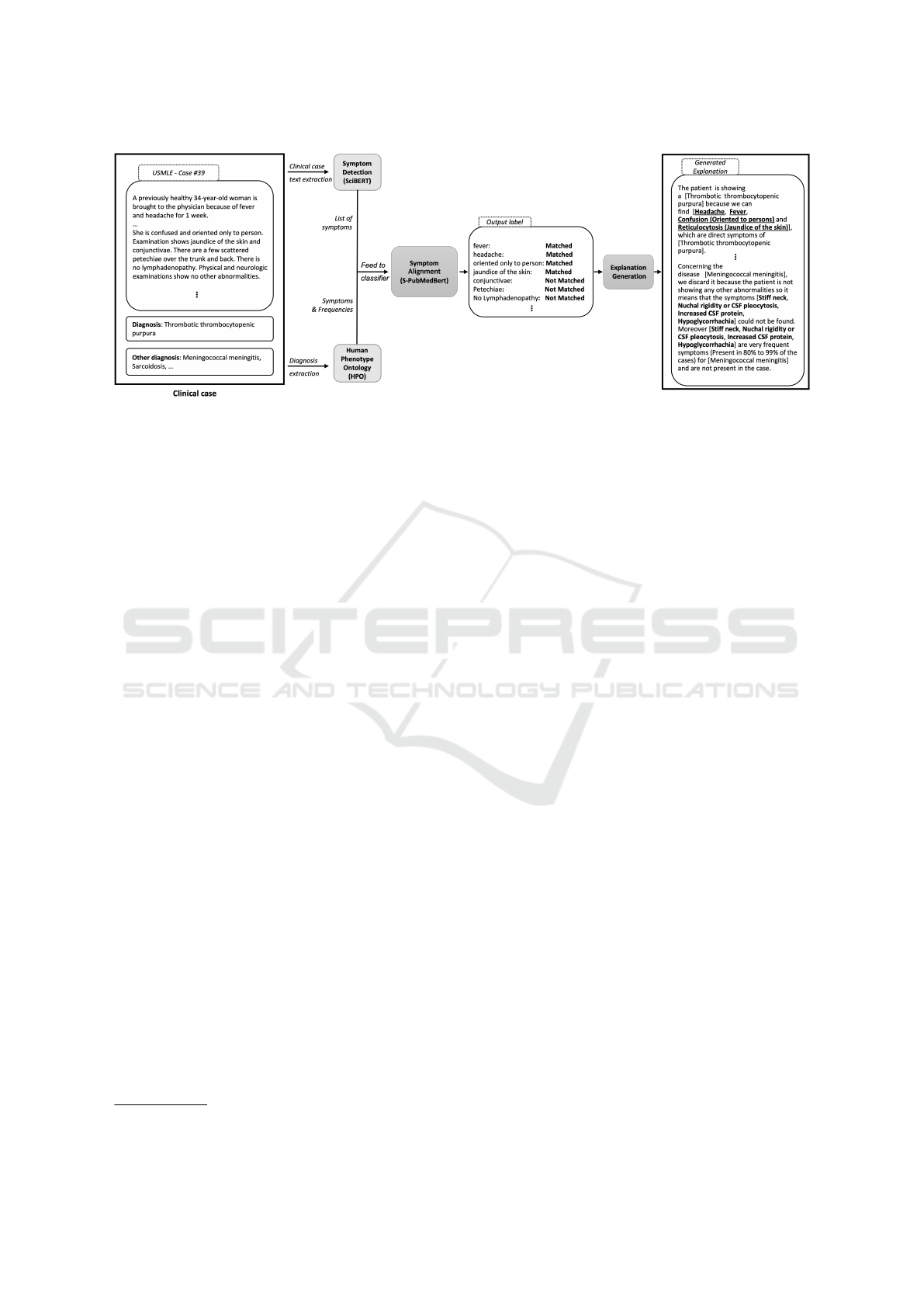
Figure 1: Overview of our full pipeline for symptom prediction and alignment, and NL explanation generation module.
The context C is embedded using sentence embed-
ding methods and saved separately from the symp-
tom S, and the two embeddings are added together
(C + S) to form the reference R. This same context
embedding C is added in the same way to each HPO
term embedding T
1
, T
2
, . . . , T
i
to form the candidates
C
1
, C
2
, . . . , C
i
.
We compute and retrieve the five best cosine dis-
tances between C and R to address a fair comparison
with other systems.
We defined a test set of 23 cases where (i) we re-
trieved from HPO the symptoms related to the dis-
eases for each case, and (ii) we manually aligned the
annotated symptoms in the case to the concepts from
HPO. This resulted in 162 symptoms aligned to a spe-
cific term in HPO that serve us as a testing set for our
matching module.
As mentioned in Section 2, the system proposed
by (Manzini et al., 2022) offers a similar approach to
translating layperson terms to medical terms in HPO.
However, their work does not take into account the
context in which a symptom is found. To the best
of our knowledge, this system constitutes the state-
of-the-art when translating layperson terms to HPO
terms so we decided to compare our proposal with
theirs. However, due to the unavailability of their
model, we rely on their online demo, which outputs
only the top 5 ranking of the HPO terms that are clos-
est to the input symptom. To perform a compari-
son with our pipeline, we first compute the accuracy
of the aligned symptoms using our symptoms align-
ment module and then replaced it with (Manzini et al.,
2022) proposed system (DASH). Results are shown in
Table 4.
Since a symptom can be composed of several
words (e.g., “shortness of breath”), we split the
symptom and the entire clinical case.
symptom into words that we encode by either us-
ing each word as an input on Glove (Pennington
et al., 2014), or extracting directly from the con-
textualized models the representation of the symp-
tom by summarizing the hidden states of the last
four layers in the model. We then sum the vec-
tors of each word to get an n-gram representation
of the symptom. We also explore sentence embed-
dings, by making use of Sentence-BERT (Reimers
and Gurevych, 2019), a new model that derives
semantically meaningful sentence embeddings (i.e.,
semantically similar sentences are close in vector
space) that can be compared using cosine similar-
ity. Sentence-BERT can be used with different pre-
trained models, in this work we focus on the models
BERT (Devlin et al., 2019) , SciBERT (Beltagy et al.,
2019), UMLSBERT (Michalopoulos et al., 2020) and
S-PubMedBert by (Deka et al., 2022). The first repre-
sents a competitive baseline in our experiments since
it is the SOTA model for comparing sentences cross-
domain, while the three latter models are pre-trained
on scientific or medical data or both.
To tackle both tasks we make use of our annotated
dataset (Section 3). The annotations are converted
into two datasets, one for each part of the pipeline.
The first dataset is used for the symptom detection
task, and it is in the CoNLL format for token-wise
labels. The second dataset, for the symptom align-
ment task, is converted into a csv format, where each
symptom in the clinical case description and avail-
able related knowledge (i.e., the list of symptoms and
their frequencies for each possible diagnosis associ-
ated with the case) extracted from HPO are paired.
Results. Results for the symptom detection task are
shown in Table 2 in macro multi-class precision, re-
call, and F1 score. We can observe that all models
Natural Language Explanatory Arguments for Correct and Incorrect Diagnoses of Clinical Cases
443

Table 2: Results for entity recognition in macro multi-class
precision, recall, and F1-score.
Model P R F1
BERT 0.85 0.84 0.84
BioBERT v1.2 0.84 0.85 0.84
UmlsBERT 0.85 0.85 0.85
PubMedBERTbase 0.83 0.84 0.83
SciBERT cased 0.85 0.85 0.85
SciBERT uncased 0.85 0.86 0.86
perform similarly, with the best results from the spe-
cialized SciBERT (Beltagy et al., 2019) model. The
biggest difference in performance is given by com-
paring SciBERT uncased with PubMedBERT, with
the SciBERT model performing better. Interestingly,
BERT performs closely to the specialized models,
and, in some cases, it outperforms them. This may be
due to the fact that the clinical cases from our dataset
are written for medical exams at the med school. They
contain some technical specialized words, but overall
the symptoms are described in layperson terms.
It is worth noticing that the majority of our labels
do not pertain to medical terminology (e.g. Age and
Population Group, Location and Temporal Concept).
Sign or Symptom and Finding are the only labels that
require specialized vocabulary.
Overall, SciBERT uncased is the best-performing
model (in bold) with a macro F1-score of 0.86, out-
performing the other approaches for each of the cate-
gories. In Table 3 we report the performances for each
entity with the best-performing model. The Sign or
Symptom detection task obtains a 0.82 F1 score. In the
work of (Ngai and Rudzicz, 2022), the authors also
detect symptoms obtaining an F1 score of 0.61. How-
ever, these results can not be directly compared since
the datasets on which both models were fine-tuned are
different: we train on clinical cases, while they use
dialogues between doctors and patients. Moreover,
given that the dataset they use is not released, we can
not evaluate our approach to their data.
The results of the symptoms alignment module
experiments are summarised in Table 4. As baseline
models, we propose to use the same methods but with-
out the context of the symptom, similarly to (Manzini
et al., 2022) DASH. In Table 4 we show only the best-
performing baseline PubMedBERT no context obtain-
ing similar results to DASH (0.41 and 0.37, respec-
tively). Adding contextual representation to the em-
beddings results in a significant improvement (up to
0.53 in accuracy) supporting the hypothesis that con-
text plays an important role when translating layper-
son terms to formal medical terms.
Table 3: Results for entity recognition using our best per-
forming model (SciBERT uncased) in P, R, and F1-score.
Entity P R F1
Other 0.93 0.91 0.92
Age Group 1.00 0.97 0.98
Finding 0.85 0.88 0.86
Location 0.74 0.80 0.77
No Symptom Occurrence 0.79 0.72 0.75
Population Group 0.88 0.95 0.91
Sign or Symptom 0.83 0.82 0.82
Temporal Concept 0.78 0.87 0.82
Weighted avg 0.89 0.89 0.89
Macro avg 0.85 0.86 0.86
Table 4: Results for DASH and our symptom alignment
method using different embeddings with and without con-
text (accuracy score).
Model Accuracy
DASH 0.37
PubMedBERT no context 0.41
BERT + context 0.38
SciBERT + context 0.39
UMLSBERT + context 0.44
S-PubMedBERT + context 0.53
Error Analysis. HPO has limitations with respect
to the number of symptoms associated with each di-
agnosis. For some diagnoses, we have multiple symp-
toms, while for others we can have only one or none.
We notice that in those cases where the diagnosis is a
mental disease, the model tends to make more mis-
takes. Inspecting HPO for this kind of diagnoses,
we find that either the diagnosis does not appear in
the HPO ontology or the symptoms tend to be more
general, including a lot of common symptoms like
changes in appetite or low energy, that alone may not
be relevant but all together indicate a precise diagno-
sis. Moreover, some relevant symptoms may not be
described explicitly but encoded in the clinical cases
as Findings. These findings often refer to a relevant
symptom that is not explicitly mentioned in the case,
like in the example introduced in Section 3 about find-
ings, where we have ”respirations are 24/min” that,
combined with the fact that the patient is a 34-year-
old woman, means that she has dyspnea. Automat-
ically deriving this implicit knowledge remains an
open challenging issue. Given that we rely on HPO
only, some diseases or diagnoses are not present in the
knowledge base, preventing us to generate the associ-
ated explanations. Combining HPO with more spe-
cialized medical knowledge bases is a future direc-
tion for this work, both to complete the information
we have, and also to integrate new diagnoses.
NLPinAI 2023 - Special Session on Natural Language Processing in Artificial Intelligence
444

6 NATURAL LANGUAGE
EXPLANATION GENERATION
In the previous section, we described the first steps
of our pipeline for automatically identifying the rel-
evant symptoms which occur in the clinical case de-
scription and then matching them with the symptoms
associated with the diseases in the medical knowl-
edge base HPO. We move now to the last step of the
pipeline, i.e., the generation of natural language ex-
planatory arguments, according to the identified rel-
evant symptoms for the correct and incorrect diag-
noses. Given the specificity of the clinical data we
are dealing with, we decide to address this task by
generating explanations through the definition of ex-
planatory patterns (Josephson and Josephson, 1994;
Campos, 2011; Dragulinescu, 2016). We have there-
fore defined different patterns which take into account
the different requirements of our use case scenario,
where we aim at (i) explaining the correct answer by
the detected symptoms and their frequency, (ii) ex-
plaining why the incorrect options cannot hold, and
(iii) highlighting the relevant symptoms not explicitly
mentioned in the clinical case. Let us consider the fol-
lowing clinical case, where in bold we highlight the
symptoms and we underline the relevant symptoms
supporting the correct answer.
Clinical Case. A previously healthy 34-year-old
woman is brought to the physician because of fever and
headache for 1 week. She has not been exposed to any
disease. She takes no medications. Her temperature
is 39.3°C (102.8°F), pulse is 104/min, respirations are
24/min, and blood pressure is 135/88 mm Hg. She is
confused and oriented only to person. Examination
shows jaundice of the skin and conjunctivae. There
are a few scattered petechiae over the trunk and back.
There is no lymphadenopathy. Physical and neurologic
examinations show no other abnormalities. Test of the
stool for occult blood is positive. Laboratory studies
show: Hematocrit 32% with fragmented and nucleated
erythrocytes Leukocyte count 12,500/mm3 Platelet count
20,000/mm3 Prothrombin time 10 sec Partial thromboplas-
tin time 30 sec Fibrin split products negative Serum Urea
nitrogen 35 mg/dL Creatinine 3.0 mg/dL Bilirubin Total
3.0 mg/dL Direct 0.5 mg/dL Lactate dehydrogenase 1000
U/L Blood and urine cultures are negative. A CT scan of
the head shows no abnormalities. Which of the following
is the most likely diagnosis?
The correct diagnosis is Thrombotic thrombocy-
topenic purpura, whilst the other (incorrect) options
are Disseminated intravascular coagulation, Immune
thrombocytopenic purpura, Meningococcal meningi-
tis, Sarcoidosis and Systemic lupus erythematosus.
Why Pattern. We focus here on the correct diagno-
sis explanation pattern, which allows explaining why
this is the correct diagnosis. We define the follow-
ing template to generate our natural language expla-
nations:
Template 1. (Why for correct diagnosis) The pa-
tient is showing a [CORRECT DIAGNOSIS] as these fol-
lowing symptoms [PERFECT MATCHED SYMPTOMS,
MATCHED SYMPTOMS] are direct symptoms of [COR-
RECT DIAGNOSIS].
Moreover, [OBLIGATORY SYMPTOMS] are obliga-
tory symptoms (always present, i.e., in 100% of the cases)
and [VERY FREQUENT SYMPTOMS] are very frequent
symptoms (holding on 80% to 99% of the cases) for [COR-
RECT DIAGNOSIS] and are present in the case descrip-
tion.
6
In Template 1, the [CORRECT DIAGNOSIS]
represents the correct answer to the question ”Which
of the following is the most likely diagnosis?” and
therefore the correct diagnosis of the described
disease. The [SYMPTOMS] in bold represent the
symptoms automatically detected through the first
module of our pipeline, and they are also underlined
when they are considered as relevant by our matching
module, i.e., they are listed among the symp-
toms for the disease in the HPO knowledge base.
Both [PERFECT MATCHED SYMPTOMS] and
[MATCHED SYMPTOMS] in Template 1 are con-
sidered relevant but they differ in the confidence level
the system assigns to the matched symptoms. This
allows us to integrate a notion of granularity in our
explanations and to rely on the symptoms detected in
the clinical case that strongly match with a symptom
in HPO. If the system does not detect any relevant
symptom, no explanation is generated for the correct
answer. Furthermore, we employ the information
about the symptom frequencies (retrieved through
HPO) in the [OBLIGATORY SYMPTOMS
] and
[VERY FREQUENT SYMPTOMS] to generate
stronger evidence to support our natural language
argumentative explanations. Sometimes the frequen-
cies are not available in the HPO, in which case we
do not display them in our final explanation.
We present now some examples of explanatory ar-
guments automatically generated by our system.
Example 1. The patient is showing a [Thrombotic
thrombocytopenic purpura] as these following symptoms
[Headache, Fever, Confusion (Oriented to persons) and
Reticulocytosis (Jaundice of the skin)] are direct symp-
toms of [Thrombotic thrombocytopenic purpura].
Moreover [Reticulocytosis (Jaundice of the skin)] are
very frequent symptoms (holding on 80% to 99% of the
cases) for [Thrombotic thrombocytopenic purpura] and are
present in the case description.
6
Sources from HPO: https://hpo.jax.org/app/browse/
term/HP:0040279
Natural Language Explanatory Arguments for Correct and Incorrect Diagnoses of Clinical Cases
445

When filling the [SYMPTOMS]
span in Template 1, we inject only the
symptoms matched in the HPO for the
[PERFECT MATCHED SYMPTOMS], and
we combine the HPO symptoms with the
symptoms detected in the case description for
the [MATCHED SYMPTOMS] in this form:
[matched symptom in HPO (detected symp-
tom in the clinical case)] (e.g., in Exam-
ple 1: Confusion (Oriented to persons) and
Reticulocytosis (Jaundice of the skin))
Why not Template. Explaining why a diagnosis is
the correct one is important, but it is also necessary to
be able to say why the other options are not correct
as possible diagnoses for the clinical case under in-
vestigation (Miller, 2019). We, therefore, propose to
provide explanations based on the relevant symptoms
for the incorrect options by contrasting them with the
clinical case at hand.
Template 2. (Why not for incorrect diagnosis) Con-
cerning the [INCORRECT DIAGNOSIS] diagnosis, it has
to be discarded because the patient in the case descrip-
tion is not showing [INCORRECT DIAGNOSIS SYMP-
TOMS FROM HPO (MINUS DETECTED SYMPTOMS
IN CASE)] symptoms.
Despite [SHARED CORRECT SYMPTOMS] symp-
toms shared with the [CORRECT DIAGNOSIS] correct
diagnosis, the [INCORRECT DIAGNOSIS] diagnosis is
based on [INCORRECT DIAGNOSIS SYMPTOMS].
Moreover, [OBLIGATORY SYMPTOMS] are obliga-
tory symptoms (always present, i.e., in 100% of the cases)
and [VERY FREQUENT SYMPTOMS] are very frequent
symptoms (holding on 80% to 99% of the cases) for [IN-
CORRECT DIAGNOSIS], and they are not present in the
case description.
Template 2 can be applied to each incorrect pos-
sible answer of the case, individually. The incor-
rect answer corresponds to the [INCORRECT DIAG-
NOSIS] and [INCORRECT DIAGNOSIS SYMP-
TOMS] are all relevant symptoms associated with
this disease in the HPO knowledge base, without the
symptoms in common with the correct answer. Again,
in the template, we use the frequencies provided by
HPO to provide further evidence to make our explana-
tory arguments more effective. The template includes
therefore with [OBLIGATORY SYMPTOMS] and
[VERY FREQUENT SYMPTOMS] the mandatory
and very frequent symptoms of the incorrect diagno-
sis, which are missing in the clinical case description.
The following explanations are automatically gener-
ated for (one of) the incorrect diagnoses of the clinical
case we introduced at the beginning of this section.
Example 2. Concerning the [Meningococcal meningitis]
diagnostic, it has to be discarded because the patient in the
case description is not showing [Stiff neck, Nuchal rigidity
or CSF pleocytosis, Increased CSF protein, Hypoglycor-
rhachia] symptoms.
Despite [Petechiae, Fever, Headache] symptoms
shared with the [Thrombotic thrombocytopenic purpura]
correct diagnosis, the [Meningococcal meningitis] diagno-
sis is based on [Stiff neck, Nuchal rigidity or CSF pleocy-
tosis, Increased CSF protein and Hypoglycorrhachia].
Moreover, [Stiff neck, Nuchal rigidity, CSF pleocy-
tosis, Increased CSF protein or Hypoglycorrhachia] are
very frequent symptoms (holding on 80% to 99% of the
cases) for [Meningococcal meningitis] and are not present
in the case description.
Example 2 shows the NL explanation of why the
possible answer [Meningococcal meningitis] is not
the correct diagnosis given the symptoms discussed
in the clinical case description. In case the disease is
not found in HPO, we do not generate the associated
explanation.
Additional Explanatory Arguments. In order to
enrich our explanations with additional explanatory
arguments to improve critical thinking in the medi-
cal residents, we also generate another template. In-
deed, in some clinical cases, it is possible that the
symptoms are not sufficient to explain the diagnosis
or sometimes the symptom has to be combined with
vital signs or other characteristics of the patient to be
correctly interpreted. Some of these signs represent
potentially important symptoms for the diagnosis, as
in the previous example, where the sentence respira-
tions are 24/min could be associated with the symp-
tom of Dyspnea in HPO. Template 3 aims at drawing
the medical residents’ attention to (statistically) im-
portant symptoms that are missing or not explicitly
mentioned in the clinical case description:
Template 3. Furthermore, [CORRECT DIAGNOSIS
VERY FREQUENT SYMPTOMS (MINUS MATCHED
SYMPTOMS)] are also frequent symptoms for [CORRECT
DIAGNOSIS] and could be found in the findings of the clin-
ical case.
Example 3 is generated by our system and brings at-
tention to Dyspnea. This additional explanatory ar-
gument complements the explanation we generate for
the correct and incorrect diagnoses in the case pre-
sented at the beginning of this section.
Example 3. Furthermore, [Dyspnea, Thrombocytope-
nia, Generalized muscle weakness, Reticulocytosis, and
Microangiopathic hemolytic anemia] are also frequent
symptoms for [Thrombotic thrombocytopenic purpura] and
could be found in the findings of the clinical case.
Limitations. Our work aims to generate template-
based natural language explanations to explain from a
symptomatic point of view why a diagnosis is correct
and why the remaining ones are incorrect. Template-
based explanations are limited in several ways. First,
NLPinAI 2023 - Special Session on Natural Language Processing in Artificial Intelligence
446

they are design-dependent, which means that if the
templates are not well-designed, they are not helping
the user in getting a better understanding of the rea-
son behind a correct/incorrect diagnosis. This can re-
duce the user’s overall satisfaction with the program
and make it less effective at achieving its intended
goals, i.e. supporting medical residents’ training. In
our case, we tried to build our template in collabora-
tion with doctors to have a result compliant with their
expectations and requirements. Templates are also in-
flexible and are fixed in advance, they may not be able
to adapt to changing circumstances or to new informa-
tion. This can make them less effective in dynamic or
rapidly-changing environments. Again, this is not a
serious issue in our case because we are using only
the symptoms as the source of data for the moment,
which are not evolving.
7 CONCLUSION
In this paper, we present a full pipeline to generate
natural language explanatory arguments for correct
and incorrect diagnoses in clinical cases. More pre-
cisely, based on a novel annotated linguistic resource,
our pipeline first automatically identifies in a clinical
case description the relevant symptoms and matches
them to the HPO medical knowledge base terms to
associate symptoms to the correct and incorrect diag-
noses proposed as potential answers to the test, and
second, automatically generates a natural language
explanatory argument which highlights why a certain
answer is the correct diagnoses and why the others are
not. Extensive experiments on a dataset of 314 clini-
cal cases in English on various diseases show good re-
sults (0.86 F1-Score on symptom detection and 0.53
Accuracy on relevant symptom alignment for Top 5
matches), outperforming competitive baselines and
SOTA approaches.
Several future work lines arise from this work.
First, we plan to address a user evaluation with medi-
cal residents. Even though clinical doctors have been
involved in the definition of the annotation guidelines
we defined, a user evaluation with medical residents
is required to get their feedback on our explanatory
arguments. Second, we plan to make these explana-
tions interactive to address a rule-based dialogue with
the student to focus on precise aspects of the clinical
case and go into more precise or generic explanations
if required by the student.
ACKNOWLEDGEMENTS
This work has been supported by the French govern-
ment, through the 3IA C
ˆ
ote d’Azur Investments in
the Future project managed by the National Research
Agency (ANR) with the reference number ANR- 19-
P3IA-0002. This work was supported by the CHIST-
ERA grant of the Call XAI 2019 of the ANR with the
grant number Project-ANR-21-CHR4-0002.
REFERENCES
Abujabal, A., Roy, R. S., Yahya, M., and Weikum, G.
(2017). Quint: Interpretable question answering over
knowledge bases. In Proceedings of the 2017 Con-
ference on Empirical Methods in Natural Language
Processing: System Demonstrations, pages 61–66.
Albright, D., Lanfranchi, A., Fredriksen, A., Styler IV,
W. F., Warner, C., Hwang, J. D., Choi, J. D., Dligach,
D., Nielsen, R. D., Martin, J., et al. (2013). Towards
comprehensive syntactic and semantic annotations of
the clinical narrative. Journal of the American Medi-
cal Informatics Association, 20(5):922–930.
Beltagy, I., Lo, K., and Cohan, A. (2019). Scibert: A
pretrained language model for scientific text. arXiv
preprint arXiv:1903.10676.
Bodenreider, O. (2004). The unified medical language sys-
tem (umls): integrating biomedical terminology. Nu-
cleic acids research, 32(suppl 1):D267–D270.
Bowman, S. R., Angeli, G., Potts, C., and Manning, C. D.
(2015). A large annotated corpus for learning natural
language inference. arXiv preprint arXiv:1508.05326.
Camburu, O.-M., Rockt
¨
aschel, T., Lukasiewicz, T., and
Blunsom, P. (2018). e-snli: Natural language infer-
ence with natural language explanations. In NeurIPS.
Campillos-Llanos, L., Valverde-Mateos, A., Capllonch-
Carri
´
on, A., and Moreno-Sandoval, A. (2021). A clin-
ical trials corpus annotated with umls entities to en-
hance the access to evidence-based medicine. BMC
medical informatics and decision making, 21(1):1–19.
Campos, D. G. (2011). On the distinction between peirce’s
abduction and lipton’s inference to the best explana-
tion. Synthese, 180(3):419–442.
Clark, K., Luong, M.-T., Le, Q. V., and Manning, C. D.
(2020). Electra: Pre-training text encoders as dis-
criminators rather than generators. arXiv preprint
arXiv:2003.10555.
Cyras, K., Rago, A., Albini, E., Baroni, P., and Toni,
F. (2021). Argumentative xai: A survey. ArXiv,
abs/2105.11266.
Deka, P., Jurek-Loughrey, A., and Deepak, P. (2022). Im-
proved methods to aid unsupervised evidence-based
fact checking for online health news. Journal of Data
Intelligence, 3(4):474–504.
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2019). BERT: Pre-training of deep bidirectional
transformers for language understanding. In Proceed-
ings of NAACL-HLT 2019, pages 4171–4186.
Natural Language Explanatory Arguments for Correct and Incorrect Diagnoses of Clinical Cases
447

Do
˘
gan, R. I., Leaman, R., and Lu, Z. (2014). Ncbi disease
corpus: a resource for disease name recognition and
concept normalization. Journal of biomedical infor-
matics, 47:1–10.
Dragulinescu, S. (2016). Inference to the best explanation
and mechanisms in medicine. Theoretical medicine
and bioethics, 37:211–232.
Eyre, H., Chapman, A. B., Peterson, K. S., Shi, J., Alba,
P. R., Jones, M. M., Box, T. L., DuVall, S. L., and
Patterson, O. V. (2021). Launching into clinical space
with medspacy: a new clinical text processing toolkit
in python. In AMIA Annual Symposium Proceedings,
volume 2021, page 438. American Medical Informat-
ics Association.
Fleiss, J. L. (1971). Measuring nominal scale agree-
ment among many raters. Psychological bulletin,
76(5):378.
Gu, Y., Tinn, R., Cheng, H., Lucas, M., Usuyama,
N., Liu, X., Naumann, T., Gao, J., and Poon, H.
(2020). Domain-specific language model pretraining
for biomedical natural language processing.
Gururangan, S., Marasovi
´
c, A., Swayamdipta, S., Lo, K.,
Beltagy, I., Downey, D., and Smith, N. A. (2020).
Don’t stop pretraining: Adapt language models to do-
mains and tasks. In Proceedings of ACL.
Honnibal, M. and Montani, I. (2017). spaCy 2: Natural lan-
guage understanding with Bloom embeddings, convo-
lutional neural networks and incremental parsing. To
appear.
Jin, D., Pan, E., Oufattole, N., Weng, W.-H., Fang, H., and
Szolovits, P. (2021). What disease does this patient
have? a large-scale open domain question answer-
ing dataset from medical exams. Applied Sciences,
11(14):6421.
Johnson, R. H. (2000). Manifest Rationality: A Pragmatic
Theory of Argument. Lawrence Earlbaum Associates.
Josephson, J. R. and Josephson, S. G. (1994). Abductive in-
ference: Computation, Philosophy, Technology. Cam-
bridge University Press.
Kim, J.-D., Ohta, T., Tsuruoka, Y., Tateisi, Y., and Col-
lier, N. (2004). Introduction to the bio-entity recog-
nition task at jnlpba. In Proceedings of the interna-
tional joint workshop on natural language process-
ing in biomedicine and its applications, pages 70–75.
Citeseer.
K
¨
ohler, S., Gargano, M., Matentzoglu, N., Carmody, L. C.,
Lewis-Smith, D., Vasilevsky, N. A., Danis, D., Bal-
agura, G., Baynam, G., Brower, A. M., et al. (2021).
The human phenotype ontology in 2021. Nucleic
acids research, 49(D1):D1207–D1217.
Krallinger, M., Rabal, O., Leitner, F., Vazquez, M., Salgado,
D., Lu, Z., Leaman, R., Lu, Y., Ji, D., Lowe, D. M.,
et al. (2015). The chemdner corpus of chemicals and
drugs and its annotation principles. Journal of chem-
informatics, 7(1):1–17.
Kumar, S. and Talukdar, P. (2020). Nile: Natural language
inference with faithful natural language explanations.
arXiv preprint arXiv:2005.12116.
Lan, Z., Chen, M., Goodman, S., Gimpel, K., Sharma,
P., and Soricut, R. (2019). Albert: A lite bert for
self-supervised learning of language representations.
arXiv preprint arXiv:1909.11942.
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H.,
and Kang, J. (2020). Biobert: a pre-trained biomedi-
cal language representation model for biomedical text
mining. Bioinformatics, 36(4):1234–1240.
Li, J., Sun, Y., Johnson, R. J., Sciaky, D., Wei, C.-H.,
Leaman, R., Davis, A. P., Mattingly, C. J., Wiegers,
T. C., and Lu, Z. (2016). Biocreative v cdr task cor-
pus: a resource for chemical disease relation extrac-
tion. Database, 2016.
Liu, Y., Ott, M., Goyal, N., Du, J., Joshi, M., Chen, D.,
Levy, O., Lewis, M., Zettlemoyer, L., and Stoyanov,
V. (2019). Roberta: A robustly optimized bert pre-
training approach. arXiv preprint arXiv:1907.11692.
Manzini, E., Garrido-Aguirre, J., Fonollosa, J., and Perera-
Lluna, A. (2022). Mapping layperson medical termi-
nology into the human phenotype ontology using neu-
ral machine translation models. Expert Systems with
Applications, 204:117446.
Michalopoulos, G., Wang, Y., Kaka, H., Chen, H., and
Wong, A. (2020). Umlsbert: Clinical domain knowl-
edge augmentation of contextual embeddings using
the unified medical language system metathesaurus.
arXiv preprint arXiv:2010.10391.
Miller, T. (2019). Explanation in artificial intelligence: In-
sights from the social sciences. Artif. Intell., 267:1–
38.
Mohan, S. and Li, D. (2019). Medmentions: A large
biomedical corpus annotated with umls concepts.
arXiv preprint arXiv:1902.09476.
Narang, S., Raffel, C., Lee, K., Roberts, A., Fiedel, N.,
and Malkan, K. (2020). Wt5?! training text-to-text
models to explain their predictions. arXiv preprint
arXiv:2004.14546.
Naseem, U., Khushi, M., Reddy, V. B., Rajendran, S., Raz-
zak, I., and Kim, J. (2021). Bioalbert: A simple and
effective pre-trained language model for biomedical
named entity recognition. 2021 International Joint
Conference on Neural Networks (IJCNN), pages 1–7.
Ngai, H. and Rudzicz, F. (2022). Doctor XAvIer: Explain-
able diagnosis on physician-patient dialogues and
XAI evaluation. In Proceedings of the 21st Workshop
on Biomedical Language Processing, pages 337–344,
Dublin, Ireland. Association for Computational Lin-
guistics.
Pennington, J., Socher, R., and Manning, C. D. (2014).
Glove: Global vectors for word representation. In
Proceedings of EMNLP 2014, pages 1532–1543.
Radford, A., Wu, J., Child, R., Luan, D., Amodei, D.,
Sutskever, I., et al. (2019). Language models are un-
supervised multitask learners. OpenAI blog, 1(8):9.
Raffel, C., Shazeer, N., Roberts, A., Lee, K., Narang,
S., Matena, M., Zhou, Y., Li, W., and Liu, P. J.
(2019). Exploring the limits of transfer learning
with a unified text-to-text transformer. arXiv preprint
arXiv:1910.10683.
raj Kanakarajan, K., Kundumani, B., and Sankarasubbu,
M. (2021). Bioelectra: pretrained biomedical text en-
coder using discriminators. In Proceedings of the 20th
NLPinAI 2023 - Special Session on Natural Language Processing in Artificial Intelligence
448

Workshop on Biomedical Language Processing, pages
143–154.
Reimers, N. and Gurevych, I. (2019). Sentence-bert: Sen-
tence embeddings using siamese bert-networks. arXiv
preprint arXiv:1908.10084.
Reiter, E. and Dale, R. (1997). Building applied natural
language generation systems. Natural Language En-
gineering, 3(1):57–87.
Saeed, W. and Omlin, C. W. (2021). Explainable ai (xai):
A systematic meta-survey of current challenges and
future opportunities. ArXiv, abs/2111.06420.
Smith, L., Tanabe, L. K., Kuo, C.-J., Chung, I., Hsu, C.-
N., Lin, Y.-S., Klinger, R., Friedrich, C. M., Ganchev,
K., Torii, M., et al. (2008). Overview of biocreative
ii gene mention recognition. Genome biology, 9(2):1–
19.
Soysal, E., Wang, J., Jiang, M., Wu, Y., Pakhomov, S., Liu,
H., and Xu, H. (2018). Clamp–a toolkit for efficiently
building customized clinical natural language process-
ing pipelines. Journal of the American Medical Infor-
matics Association, 25(3):331–336.
Tjoa, E. and Guan, C. (2019). A survey on explainable
artificial intelligence (XAI): towards medical XAI.
CoRR, abs/1907.07374.
Vassiliades, A., Bassiliades, N., and Patkos, T. (2021). Ar-
gumentation and explainable artificial intelligence: a
survey. The Knowledge Engineering Review, 36:e5.
Wolf, T., Debut, L., Sanh, V., Chaumond, J., Delangue,
C., Moi, A., Cistac, P., Rault, T., Louf, R., Funtow-
icz, M., Davison, J., Shleifer, S., von Platen, P., Ma,
C., Jernite, Y., Plu, J., Xu, C., Scao, T. L., Gugger,
S., Drame, M., Lhoest, Q., and Rush, A. M. (2020).
Transformers: State-of-the-art natural language pro-
cessing. In Proceedings of the 2020 Conference on
Empirical Methods in Natural Language Processing:
System Demonstrations, pages 38–45, Online. Asso-
ciation for Computational Linguistics.
Natural Language Explanatory Arguments for Correct and Incorrect Diagnoses of Clinical Cases
449
