
Knowledge Graph Based Trustworthy Medical Code Recommendations
Mutahira Khalid
1 a
, Asim Abbas
2 b
, Hassan Sajjad
3 c
, Hassan Ali Khattak
1 d
,
Tahir Hameed
4 e
and Syed Ahmad Chan Bukhari
2 f
1
School of Electrical Engineering and Computer Science, NUST, H-12, Islamabad, Pakistan
2
Division of Computer Science, Mathematics and Science, St. John’s University, Queens, NY 11439, U.S.A.
3
Faculty of Computer Science, Dalhousie University, Halifax, Canada
4
Girard School of Business, Merrimack College, North Andover, Massachusetts, U.S.A.
Keywords:
Medical Coding, Computer Assisted Coding (CAC), Deep Learning, Attention Mechanism, Symbolic AI,
Knowledge Graphs, Ontologies, Explainability.
Abstract:
Medical coding is about assigning standardized alphanumeric codes to diagnoses, procedures, and interven-
tions recorded in patients’ clinical notes. These codes are essential for correct medical claims and billing pro-
cesses, which are critical in maintaining efficient revenue cycles. Computer-Assisted-Coding (CAC) employs
AI models to automate medical coding hence cutting down human effort and errors. Despite their unrivalled
performance, these models lack ‘explainability’. Explainability opens up the inner workings and results of
black-box deep learning models. Attention mechanisms are the most common approach for ‘explainability’,
but they leave some questions unanswered, for instance, the relationship between highlighted words and pre-
dictions. Where black-box models fail to answer such questions, ‘Symbolic AI’ such as ‘Knowledge Graphs’
provide a superior alternate approach. We consolidated the attention mechanism with Symbolic AI to help
users understand the results of a deep-learning model for CAC. We evaluated its performance on the basis
of strong and weak relationships on word-to-word and word-to-code levels by employing a semantically-
enriched Knowledge Graph. We achieved 64% word-to-word and 53% word-to-code level accuracy. This
paper is among the earliest ones on knowledge graphs for explainability in medical coding. It is also the deep-
est in applying attention-based mechanisms and knowledge graphs to any medical domain.
1 INTRODUCTION
Medical coding assigns standardized alphanumeric
medical codes to patients’ diagnoses, procedures, and
other healthcare information (Aalseth, 2014). The
standardized medical billing codes include the In-
ternational Classification of Diseases (ICD), Current
procedural terminology (CPT), and the Healthcare
standard procedure coding system (HCPCS) (John-
son and Linker, 2015). The diagnosis, procedures,
and intervention codes are used for claims and billing
management with payers including insurance com-
a
https://orcid.org/0000-0001-8482-4004
b
https://orcid.org/0000-0001-6374-0397
c
https://orcid.org/0000-0002-8584-6595
d
https://orcid.org/0000-0002-8198-9265
e
https://orcid.org/0000-0002-6824-6803
f
https://orcid.org/0000-0002-6517-5261
panies, government agencies like medicare, and pa-
tients. Medical coders manually assign codes to un-
structured text in EHRs and clinical notes. Main-
taining steady cash flows and revenue cycle manage-
ment is an ongoing major challenge for healthcare
providers such as hospitals, hospices, nursing facili-
ties, and small clinics. Errors and speed of medical
coding is a major cause of lost revenues or delays in
accounts receivable for healthcare providers (Alonso
et al., 2020).
In recent years, there have been notable advance-
ments to reduce efforts and errors in medical coding.
Computer Assisted Coding (CAC) assists medical
coders by translating clinical notes to medical billing
codes with the help of machine learning and deep
learning models (Campbell and Giadresco, 2020).
The models scan the unstructured textual notes and
predict applicable medical billing codes, which saves
medical coders the time and effort required to review
Khalid, M., Abbas, A., Sajjad, H., Khattak, H., Hameed, T. and Bukhari, S.
Knowledge Graph Based Trustworthy Medical Code Recommendations.
DOI: 10.5220/0011925700003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 5: HEALTHINF, pages 627-637
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
627

long summaries and complex code databases (Catling
et al., 2018). Medical coders can assign from the pre-
dicted codes or still delve deeper to assign their own
codes. Deep Learning models were especially effec-
tive in automating this burdensome task as CAC sys-
tems keep learning from each prediction and assign-
ment of the codes (Moons et al., 2020).
Some of the common models used for CAC
are Recurrent Neural Networks (RNNs), Convolu-
tional Neural Networks (CNNs), Hierarchical At-
tention Networks (HAN), and Long-Short-Term-
Memory (LSTM), etc (Gu et al., 2021). Despite
their accurate and precise results, these deep learn-
ing models have some limitations, mainly the absence
of ‘explainability’ or ‘transparency’ about their in-
ner workings. That’s why deep learning models are
called black-box models. The black-box nature of
these models makes them less trustworthy and limits
their wide acceptance to real-world applications such
as health, finance and law (von Eschenbach, 2021).
That’s why ‘explainability’ has become an integral
need of time for healthcare information systems. AI-
based systems would not be adopted if there are sig-
nificant chances for incorrect predictions and if there
are no ways to interpret and comprehend the basis of
such life-critical and business-critical decisions.
No wonder improving transparency of deep learn-
ing models has emerged as important research for the
scientific community as well as industry (Xu et al.,
2019). Some recent advances include the incorpo-
ration of an attention mechanism (Niu et al., 2021),
in which the important words are assigned atten-
tion weights in the encoding process and later these
weights are used in the decoding process to help the
model focus on a specific part of the input. Still,
the internal working of the deep learning models re-
mains unclear. In medical coding, the attention mech-
anism incorporated into deep learning models high-
lights some particular terms. It implies that they have
some impact on the prediction of a particular medical
code. However, some issues are brought up, such as:
How are these terms related to the other highlighted
words? What link does this vocabulary have to the
predicted codes? The answer is, regrettably, none.
This limitation can be overcome by adopting sym-
bolic AI, which has given rise to a new era of neuro-
symbolic AI, also known as the third wave of artificial
intelligence (Garcez and Lamb, 2020).
Symbolic Artificial Intelligence is also known as
Classical AI or Good Old Fashioned AI (GOFAI)
which was a prominent research area from the 1950s
to 1990s (Confalonieri et al., 2021). Symbolic AI
works by training the machines the way humans learn
using symbols and symbolic representations of this
world. Knowledge Graphs (KG) or Knowledge Bases
are called the brain behind symbolic AI, which is
heterogeneous, labelled, and structured multi-graphs
(Hogan et al., 2021). Knowledge Graphs contain a
huge network of entities and their relations that could
be used as a reasoning system for causal inference.
KG could be used to open the black box of deep learn-
ing models as they are self-explainable. The consol-
idation of deep learning and KG can lead to accurate
and explainable applications (Hitzler et al., 2020).
We proposed a novel approach of ‘Explainable
Knowledge Graph Creation’ to evaluate the attention
results and provide visualization for the sake of ex-
plainability. (Dong et al., 2021) used a Hierarchi-
cal label-wise attention network (H-LAN) deep learn-
ing model for predicting Medical codes. In this pa-
per, we have customized H-LAN with KG for generic
medical code predictions with higher explainability
and transparency. H-LAN alone predicts multiple la-
bels with attention to particular words and sentences
per label. However, it does not explicate the choice
of specific words, their combinations, or their rela-
tionships with labels. Our approach predicts ICD-10
medical billing codes, labels, and words with spe-
cific attention weights. In addition, KGs evaluated
the model performance in predicted labels and high-
lighted words while providing visual connections be-
tween labels. The word-to-word and word-to-label
level explainability exactly follows human cognition
and learning patterns. As a result, medical coders see
the knowledge graphs and are more confident in mak-
ing their choices of the billing codes from the pre-
dicted labels.
This paper makes several theoretical and empiri-
cal contributions. Talking about the theoretical and
methodological contributions first, we have extended
the use of the H-LAN model in combination with
much deeper KGs for enhanced explainability of pre-
dicted labels. To that end, we have successfully
demonstrated a method to visualize connections on
word-to-word and word-to-code levels via a knowl-
edge graph at a scale not witnessed before in the med-
ical domain. On the practical side, this is the first
paper that has trained and predicted ICD-10 medi-
cal billing codes annotated using pre-trained Clinical
BERT (Surolia, 2022).
To summarise the contributions described above:
1. We fine-tuned a deep learning model from ICD-9
to ICD-10, with an enhanced problem domain.
2. An approach called ”Explainable Knowledge
Graph Creation” is proposed to make explain-
able systems more understandable and get over
the drawbacks of the attention mechanism.
3. A visualization application was made to display
CCH 2023 - Special Session on Machine Learning and Deep Learning for Preventive Healthcare and Clinical Decision Support
628
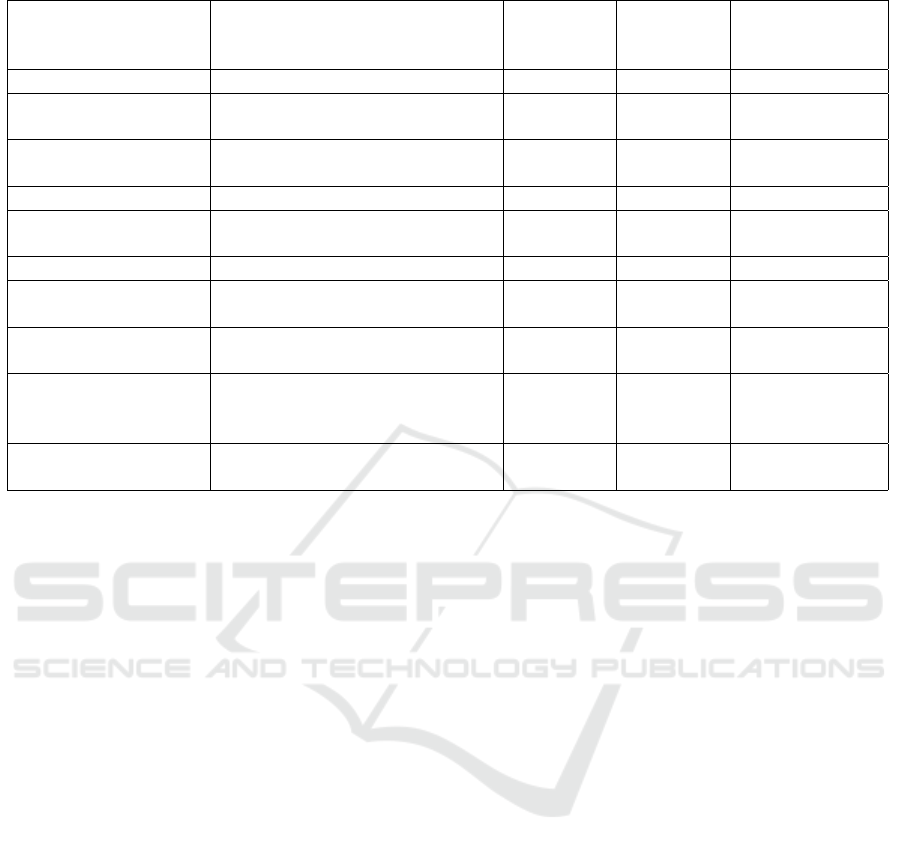
Table 1: Current Neuro-Symbolic research with the level of explainability and underlying approaches.
Related Work Neuro-Symbolic Approach Graph
Type
Deep
Learning
Model
Explainability-
Level
(Chai, 2020) KG embedding as training data KG LSTM None
(Gaur et al., 2022) Shallow Infusion KG Neural
Network
None
(Malik et al., 2020) Added Ensemble learning predic-
tions as graph nodes
KG Ensemble
Learning
Low
(Dranc
´
e, 2022) KG Embedding KG GNN Low
(Sheth et al., 2022) Knowledge Infusion KG Neural
Network
Low
(Lu et al., 2022) Graphs as input of neural network Bipartite GNN Low
(Gaur et al., 2021) Shallow and deep infusion of KG
with deep learning
KG BERT Moderate
(Wang et al., 2019) KG embedding with bidirectional
LSTM
KG LSTM High
(Teng et al., 2020) KG + Data infused to Model KG Multi-
Layer
CNN
Attention Mech-
anism
(Ahmed et al., 2022) KG embedding with Bidirectional
LSTM
HyperGraph LSTM Attention Mech-
anism
word-to-word and word-to-code level links for a
reliable and trustable medical coding application.
To the best of our knowledge, no one has consoli-
dated symbolic AI with an ‘attention mechanism’ for
explainable medical code predictions. The rest of the
paper is organized as follows. We reviewed Neuro-
Symbolic approaches with their explainability level in
section 2. Section 3 describes the materials and pro-
posed methodology. Section 4 contains the results and
analysis. Section 5 concludes the paper along with
some limitations and pointers for future research.
2 BACKGROUND
Explainability is not just a desired characteristic, it
is also a current necessity in fields where human lives
are involved e.g healthcare, finance, law, etc. Incorpo-
rating transparency in deep learning requires the ma-
nipulation of mathematical models by experts (Futia
and Vetr
`
o, 2020). An expanding field of study that has
attracted a lot of attention recently is neuro-Symbolic
AI. To accomplish both accuracy and explainability,
it integrates symbolic AI and deep learning (Sarker
et al., 2021). Interpretability and explainability are
sometimes used interchangeably, but they are funda-
mentally different concepts. Explainability is the abil-
ity of an AI model to defend its predictions, whereas
interpretability is primarily the AI model’s ability to
be transparent about its internal workings (Gaur et al.,
2021). In this section, we’ll go over the research on
the subject of neuro-symbolic AI, the methods for in-
tegrating KG with deep learning models, the level of
explainability they offer, and the methods for KG pro-
duction, their types, strengths, shortcomings, and lim-
itations.
Knowledge Graphs because of their nature are
considered a clean data source. The subject, object,
and predicate are all present in the triplets’ hub. If
these graphs are used in conjunction with deep learn-
ing models, results can be predicted more accurately.
To diagnose thyroid disease, (Chai, 2020) combined
KGs with a long-short-term memory (LSTM) model
(Hochreiter and Schmidhuber, 1997). (Gaur et al.,
2022) did a shallow infusion of knowledge graphs
with neural networks on mental health data. A
graph based on the use case of subarachnoid haem-
orrhage was constructed using an automated knowl-
edge graph-generating method (Malik et al., 2020).
A dataset of 1000 summaries was procured for graph
generation along with ensemble learning to add rup-
ture probability as nodes.
An emerging concept called ”graph embedding”
produces the vector representation of graph facts in
a manner similar to word embedding. These embed-
dings may help ensure accurate model outputs. (Lu
et al., 2022) Combined graphs and neural networks
(GNN) for predicting the risk of mental illness. For
the benefit of computer specialists, node embedding
was utilised, along with visualisation, to view the
model in action prior to applying it to the prediction
Knowledge Graph Based Trustworthy Medical Code Recommendations
629

layer. Similar to this, link prediction algorithms were
used to design a medicine repurposing strategy. The
study established the connection(s) between a chemi-
cal and a certain target while maintaining their trans-
parency and interpretability (Dranc
´
e, 2022).
The training data is what deep learning models
rely on the most. Lack of domain-specific informa-
tion results in either no forecasts or false positive re-
sults. Knowledge graphs are enriched data sources
that can assist in finding a solution to this problem.
In numerous use cases, knowledge was infused both
superficially and deeply (Gaur et al., 2021). On the
one hand, a self-supervised BERT model was given
a shallow infusion of domain knowledge on a drug-
abuse ontology (Devlin et al., 2018), mostly to help
the model comprehend the context. On the other
hand, shallow and deep knowledge graph infusions
were carried out in educational settings in an effort to
comprehend a student’s performance and identify his
or her poor domain knowledge regions. For clarity
in this situation, certain low-level visuals were cre-
ated. (Khalid et al., 2022) enriched medical sum-
maries with knowledge graphs to improve model pre-
dictions. Another comparable method was devised to
improve the accuracy of the classifiers for which they
used process-knowledge infusion. It utilised psycho-
metric questionnaires (PHQ-9) and process knowl-
edge (Sheth et al., 2022).
Some recent research has been done on knowl-
edge consolidation with inputs. The objective is to
show enriched knowledge via attention mechanisms
to enhance the level of explainability. (Teng et al.,
2020) proposed an approach named “G-coder” in
which a multi-layer CNN was employed with an at-
tention mechanism. The results included a knowl-
edge graph mapping the ICD-9 description with Free-
base ontology data that had 1560 nodes and more than
20,000 relations. The enriched knowledge graph was
combined with the attention mechanism to make the
terminologies and coding results interpretable. The
model performed well for the prediction of the top
50 codes, but the explainability remained restricted
to the attention mechanism. A graph attention em-
bedding method was employed in research on the
identification of depression symptoms. A hypergraph
was made using a psychometric questionnaire (PHQ-
9) and patient-written text, which then allowed em-
beddings to be created. An Internet-Psychological
Treatment (IDPT) was developed employing a bidi-
rectional LSTM (Graves et al., 2005) with an atten-
tion mechanism to help people deal with depression
while using fewer resources (Ahmed et al., 2022). At-
tention was applied both on the node level and on the
edge level.
Knowledge graphs were only occasionally used
in techniques to increase explainability. (Wang
et al., 2019) created a ”Knowledge-aware path recur-
rent network (KPRN)” that made suggestions using
knowledge graphs. The networks and connections
that exist between various things in the graphs can be
used to comprehend not only user preferences but also
the semantics of entities and relationships. Addition-
ally, it provided explainable predictions.
In conclusion, the publications examined in this
section combined knowledge graphs with either ma-
chine learning models or deep learning techniques.
They share a few characteristics. Prior to model train-
ing, KGs were used in the majority of these stud-
ies with the aim of either incorporating domain in-
formation to enhance model performance or display-
ing the enriched knowledge as an output in the atten-
tion layer. The limited explainability is not the true
essence or intention of knowledge graphs.
Some researchers (Wang et al., 2019; Xian et al.,
2020; Spillo et al., 2022) used graphs to explain the
outputs but they were not in the medical fields, e.g
movies and music, where ‘name entity recognition’
(NER), datasets and trained corpus are present. None
of the aforementioned studies attempted to offer ex-
plainability at the level of prediction. Neuro-symbolic
AI was not used to explain either the suggestions or
the attention mechanisms that were predicted by the
model. Table 1 summarizes the findings from our re-
view.
3 DATA AND METHODS
3.1 Data Preparation and Acquisition
‘Medical Information Mart for Intensive Care’
(MIMIC-III) is a large and freely accessible database
(Johnson et al., 2016). It contains more than 40k pa-
tient records admitted to emergency rooms (ER) units
of ‘Beth Israel Deaconess Medical Center’ between
2001 and 2012. Each admission record contains
data on demographics, diagnosis, vitals, lab measure-
ments, and survival along with discharge summaries.
The summaries do contain history, primary diagnosis,
and much more.
MIMIC-III data is annotated with ICD-9 codes.
We employed the Hugging Face model and annotated
nearly 5k discharge summaries with ICD-10 codes
since the latter is more prevalent now. Hugging Face
is an AI community containing thousands of freely
accessible datasets and models (Delangue, 2016). We
acquired a pre-trained Clinical BERT (Surolia, 2022)
for annotation. The output model was tested and ver-
CCH 2023 - Special Session on Machine Learning and Deep Learning for Preventive Healthcare and Clinical Decision Support
630

Table 2: Comparison between pretrained and fine-tuned HLAN model.
Dataset Train Validation Test Label
Count
Score cal-
culation
F-1 Score
MIMIC-III (pretrained) 8066 1573 1729 50 Top-50 64.1%
MIMIC-III (pretrained) 4574 153 322 20 Top-20 74.6%
MIMIC-III (fine-tuned) 3266 800 800 550 Top-5 67.2%
ified by the medical coder itself.
Knowledge Graphs are attractive to computer sci-
entists in all domains, especially to researchers in the
medical domain to address the need for explainability.
Ontologies are expert-created rules that have abun-
dant information which can be leveraged to create
domain-specific knowledge graphs. Freebase, Bio-
Portal, UMLS, and many other open-source ontolo-
gies are used for graph creation. We employed Bio-
Portal ontologies as a rich data source (Noy et al.,
2009) They are the world’s most comprehensible
repository of biomedical ontologies.
The BioPortal ontologies support recommenda-
tions based on dataset description, term search across
multiple ontologies, annotation of medical concepts
with ontology terms, and much more. BioPortal
is a hub of 1000+ ontologies, having 14,427,459
classes and 79,636,946 mappings. The ontologies
are present in different formats such as Resource
Description Framework (RDF), Web Ontology Lan-
guage (OWL), Extensible Markup Language (XML),
and Comma Separated Files (CSV). We mapped the
model-predicted weighted medical concepts and the
description of medical codes with the ontologies to
build a connected final medical knowledge graph.
The medical terms are matched with the ontology
classes, their definitions, synonyms, and hierarchy till
level 5. Neo4j, a graph database management system,
was used for the construction of Knowledge graphs.
BioPortal REST API was employed for information
retrieval.
3.2 Explainable Knowledge Graph for
Medical Coding
3.2.1 Problem Formulation
Medical coding is a multi-label text classification
problem in which text information is translated into
medical codes, an extremely laborious task. Accord-
ing to one estimate, four out of five generated medical
billing codes are erroneous (Tate, 2017), which has
revenue implications for both payers and providers.
AI-enabled CAC models predict medical codes with
higher precision. However, they are confronted with
acceptance challenges due to the lack of transparency
and explainability.
We propose a novel approach for an explainable
knowledge graph. Figure 1 depicts the workflow of
the proposed approach which consists of four mod-
ules; ICD-10 prediction model, semantic enrichment,
semantic knowledge consolidation, and explainable
knowledge graph creation. The last module is further
divided into two parts i.e. word-to-word and word-to-
code level connections. The following sub-sections
elaborate on these modules at some length.
3.2.2 ICD-10 Code Prediction
After testing and reproducing results from multiple
CAC models (Mullenbach et al., 2018; Desai, 2020;
Biswas et al., 2021), we selected a baseline model
titled “Hierarchical Label-Wise Attention Network”
(Dong et al., 2021) for ICD-10 billing codes predic-
tions. The architecture of HLAN and its use of the
attention mechanism for explainable code prediction
led to its selection. The model was trained for multi-
label classification on ICD-9 (9th version) on top-50
ICD-9 and top-20 ICD-10 codes. Apart from the pre-
diction of medical codes, an attention mechanism was
also applied at both word and sentence levels. We
mainly reconfigured the model for ICD-10 predic-
tions from the original ICD-9 predictions. We used
11x more labels for predictions mainly to enhance the
problem domain. We also annotated the MIMIC-III
dataset with ICD-10 codes by employing the Hug-
ging Face model, later tested and verified by an ex-
perienced professional medical coder. The reason for
not using the pre-trained model for our ‘Explainable
Knowledge Graph Creation Approach’ is a lack of at-
tention mechanism and the issue of scalability.
Around 4.8k summaries were annotated with
nearly 22k ICD-10 labels where the unique label
count was 550. An average annotation count per sum-
mary was 4 codes. We also developed the hierarchi-
cal structure for even better results. Only diagnos-
tic codes were procured for model training. We di-
vided the dataset for training, testing and validation.
3266 summaries were used for training, 800 for test-
ing, and 800 for validation. The model trained for
550 labels was somewhat less accurate but that was
expected due to an increase in the label count. Table
2 shows the comparison between the trained HLAN
model and fine-tuned HLAN model with respect to
Knowledge Graph Based Trustworthy Medical Code Recommendations
631
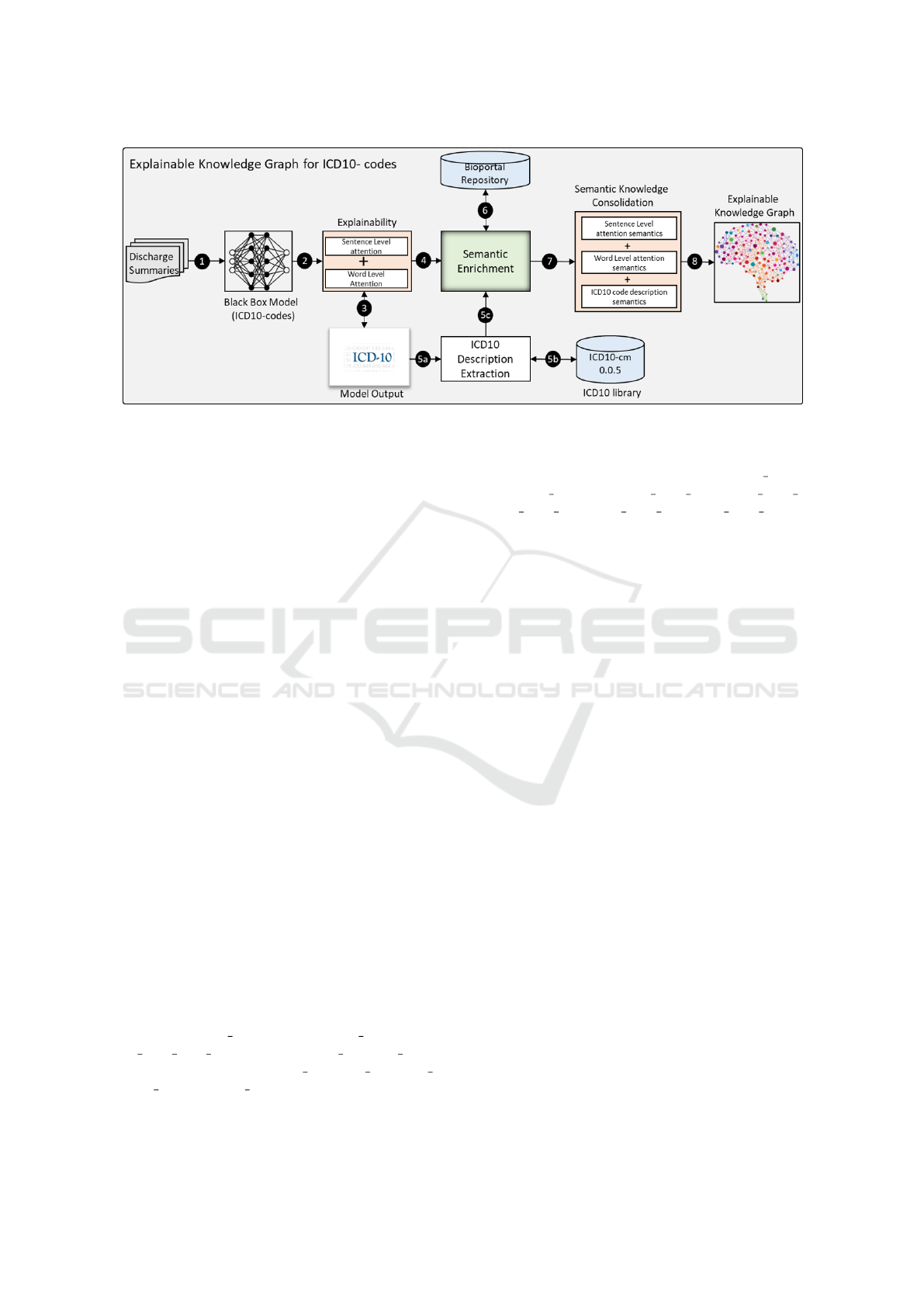
Figure 1: Explainable Knowledge Graph Creation Approach.
the dataset size and F1 score.
3.2.3 Semantic Enrichment
Semantics plays an important role in understanding
the context and boundary of particular words (Abbas
et al., 2019; Abbas et al., 2021). In our case, we need
to comprehend the meaning of particular attention
words and their connections with each other and with
the predicted labels. The highlighted words from the
patient discharge summary and the description of the
medical billing codes were the inputs for semantic en-
richment. We leveraged the ICD10-cm library to ex-
tract predicted code descriptions. Our pre-processing
involved stop-word removal (the, is, am, what, etc.),
stemming and lemmatization, and removal of dupli-
cate words. N-grams with n=4 were used to find
all possible combinations of words that were mapped
onto Bioportal Ontologies, mainly 1000+ ontologies
in Bioportal REST API, to get as much information as
possible. Words were enriched with synonyms, defi-
nitions, parent / hierarchy-level-1, parent / hierarchy-
level-2, till parent / hierarchy-level-5. Such rich in-
formation can provide a deeper level of explainability
for the end user.
3.2.4 Semantic Knowledge Consolidation
Different types of knowledge relevant to at-
tention weights and description words were
consolidated removing the repetitive synonyms,
definitions, and hierarchy descriptions. We
also separated the nodes by their types, for
instance, patient summary, medical code, med-
ical code desp words, model attention words,
synonyms, definitions, parent 1, parent 2, parent 3,
parent 4, and parent 5. Similarly, the relationships
and their types were also separated and duplica-
tions were removed. The types of relationships
were synonyms, definitions, highlighted words,
description words, parent level 1, parent level 2,
parent level 3, parent level 4, parent level 5, and
connected.
3.2.5 Explainable Knowledge Graph Creation
An automated approach was used to create the knowl-
edge graph which requires a Cypher query to be cre-
ated in the Neo4j platform. See (Khalid et al., 2022)
for similar earlier work. Knowledge graph generation
is a computation-heavy task with run-time creation
even more complex due to the diversity and complex-
ity of data. A specific graph is created for each sum-
mary as it is entered for ICD-10 code recommenda-
tions in the deep learning model. The relationships
between the graph nodes help us understand the con-
text and semantics in general, but the ‘connected re-
lationship’ specifically focuses on working of the at-
tention mechanism and model prediction. Attention
weights are assigned to words in the medical billing
codes prediction based on similarity. We have sim-
plified the graph visualization for the users by pro-
viding two different types of explainability, word-to-
word connections, and word-to-code connections.
3.2.6 Visualisation Application
Graphs are extremely useful to comprehend the pre-
dictions of deep-learning models. But explainability
is more complex and computationally intensive than
most tasks even for deep-learning models. The ac-
tual knowledge graphs created for patient discharge
summaries contain thousands of nodes and relation-
ships making it difficult to analyze just by looking
at them. Having that much visual information be-
comes information overload for a medical coder in-
CCH 2023 - Special Session on Machine Learning and Deep Learning for Preventive Healthcare and Clinical Decision Support
632
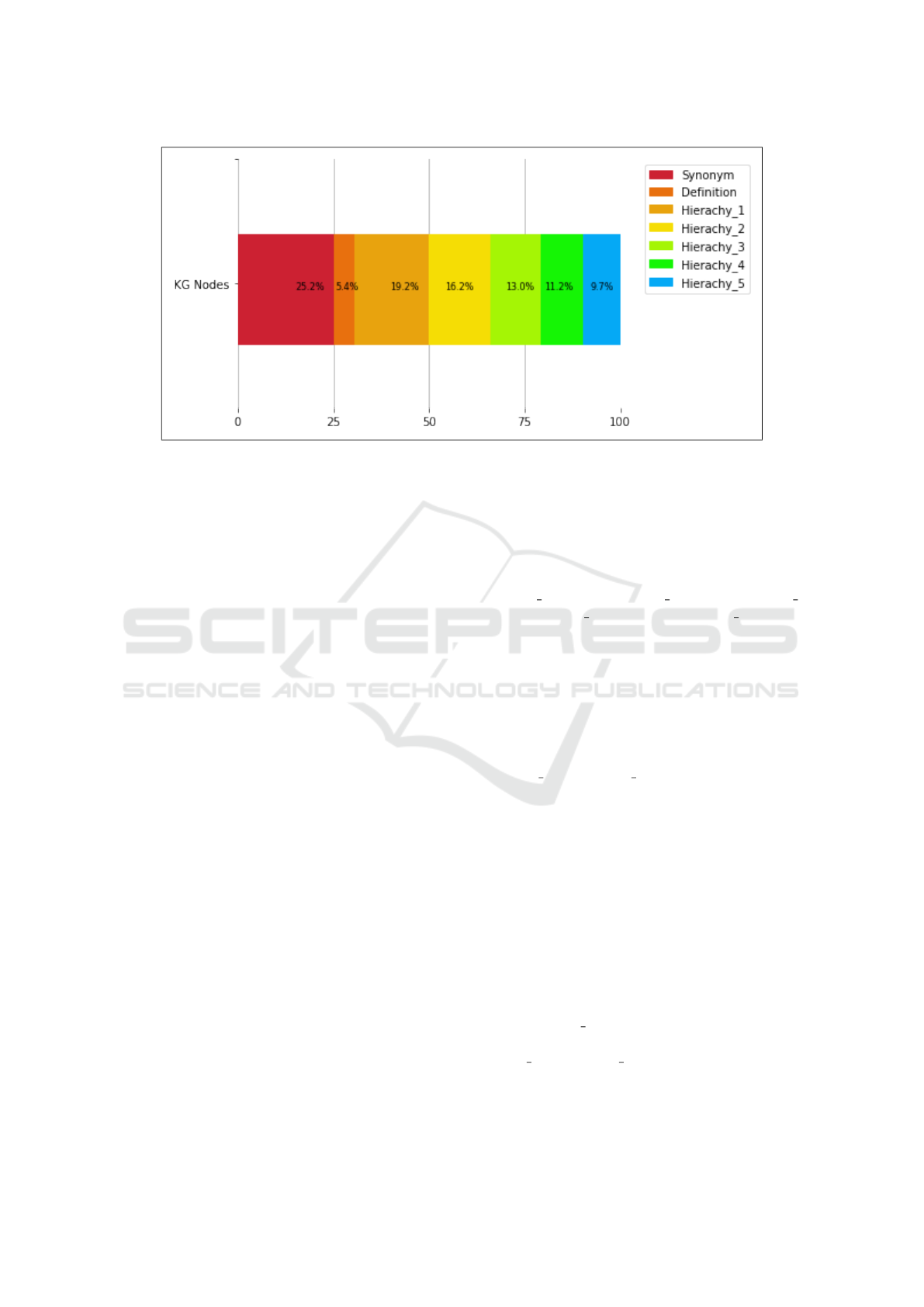
Figure 2: Semantic Enrichment Results.
stead of supporting them in the task. We have gener-
ated Knowledge Graphs on word-to-word and word-
to-code levels taking human cognitive behaviour into
account, mainly visual cues in terms of object size,
bolded shapes, colour differences, and colour attri-
bution to events, etc. The graphs can be searched
for user-entered words or codes and they can also be
restricted/filtered to visualize direct and strong con-
nections only. The actual graph was queried using
Cypher language with the help of the Trinity library.
Neo4j is an extremely good platform for visualiza-
tion but it does not offer automatic manipulation of
the size and the colour of the nodes. The simpli-
fied visualization was done using the kglab library.
Path detection algorithms such as depth-first search
and breadth-first search algorithms in Neo4j were ap-
plied to find the shortest path. However, these algo-
rithms performed poorly due to the diversity of cre-
ated graphs, so we used simple Cypher functions to
get a path between chosen words or labels.
4 RESULTS AND ANALYSIS
We achieved a 67.2% F1 score on the top 5 codes in
model training (800 summaries for testing and 800
for validation). Nearly 100 discharge summaries were
passed to the model and the predicted labels and high-
lighted words were procured. Each summary consists
of an average of 25 medical concepts comprising at-
tention words and in-code descriptions. The model
predicted an average of three medical codes (ICD-
10) for each summary. The ‘Explainable Knowledge
Graph Creation Approach’ was applied to the model
results.
The semantic enrichment module extracted the
biomedical concepts and their synonyms, definitions,
and hierarchies at level 5 from BioPortal ontolo-
gies. The semantic information was then consoli-
dated. We obtained 736 synonyms per summary on
average. There were 159 definitions, 562 nodes for
Hierachy 1, 473 for Hierachy 2, 379 for Hierachy 3,
327 for Hierachy 4, and 284 for Hierachy 5, as shown
in figure 2.
The explainable knowledge graph created for each
summary was a collection of 2900 nodes and 3340
relationships on average. A graph of this size can
be effectively used for reasoning and explainabil-
ity. To that end, the model’s performance was eval-
uated based on strength of connections. The pro-
posed semantic enrichment (synonyms, definitions,
hierarchy 1 to hierarchy 5) process plays a vital role
in identifying word-to-words and word-to-code level
connections or relations. They were made either
through string matching or based on the semantic rel-
evance of the medical concepts.
During the processing of 100 summaries, the
model highlights the words that contributed to the pre-
diction of ICD-10 codes. After scrutinizing these re-
sults for word-to-word level connections, we found
an average number of 176 connections based on syn-
onyms and 75 connections owing to the definition per
summary, as shown in Figure 3. Similarly, we ana-
lyzed that hierarchical levels of semantic information
also have a crucial role in word-to-word level connec-
tions. Hierarchy 1 produces an average of 39 connec-
tions or relations which is much more than other hi-
erarchy 2 to hierarchy 5. The reason for no or fewer
connections on some hierarchy levels is due to the na-
ture of BioPortal Ontologies, which is not critical at
the word-to-word level.
Knowledge Graph Based Trustworthy Medical Code Recommendations
633
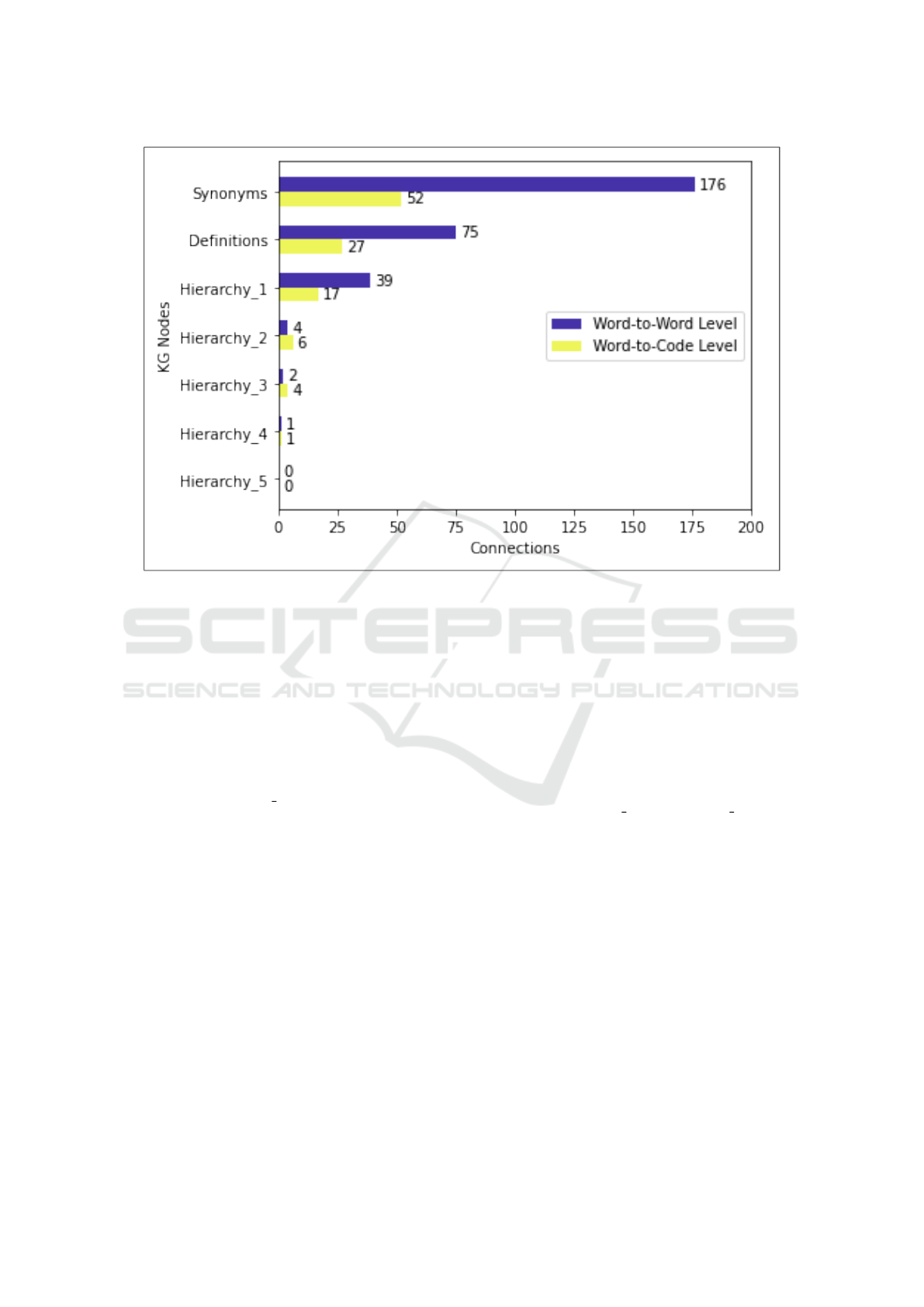
Figure 3: Word-to-Word and Word-to-Code Level Connections.
While analyzing the word-to-code level relations,
we found an average of 52 connections based on
synonyms and 27 connections based on definitions.
Moreover, we found hierarchy levels 1 and 2 add
more to the word-to-code level relations in compar-
ison to hierarchy levels 3, 4, and 5, as shown in Fig-
ure 3. Consequently, the connections are relatively
fewer when compared to the word-to-word level but
expected, due to the fact the average amount of words
per discharge summary is 21 but only 4 for code de-
scriptions. The hierarchy 5 showed zero connections
for both levels but they are included to check the dif-
ferences and reduction in the number of connections.
Subsequently, we evaluated the performance of
the deep learning model for its predictions and at-
tention mechanism results. The relationship be-
tween labels-with-words and words-with-words was
assessed based on its strength. The relationships be-
ing weak or strong were marked by experts. More
than 30 connections were taken as strong and fewer
than 10 as weak connections.
The procured 100 summaries contain nearly 2500
medical concepts. The manual assessment of them
is not possible in terms of strong and weak connec-
tions. We used a small section of randomly chosen
40 medical codes and nearly 100 words to retrieve re-
sults (Nearly 10 summaries). Out of the 100 words,
64 words had strong, weak, or no connection with re-
spective words as marked by the experts. 21 out of the
40 labels contained were also correct. Eq.1 has been
used to measure the connection accuracy at different
levels, where accuracy is equal to the correct relations
instances divided by total relationships. Hence, we
measure accuracy in terms of strong and weak con-
nections. The model achieved 64% of accuracy for
word-to-word level connections and 53% accuracy for
word-to-code level connections.
Accuracy = Correct Relations/Total Relations
(1)
It has been found that there could be direct or in-
direct relationships (or connections) between words.
For instance, the biomedical concept ‘flap’ has a di-
rect association with the word ‘graft’ but carries an in-
direct relationship with the biomedical concept ‘anas-
tomosis’. We assigned half count to the indirect re-
lationships keeping it full for the direct relationship.
For instance, if ‘flap’ and ‘anastomosis’ are connected
with 4 nodes in between, we would count it as 2 (in
terms of equivalence to direct relationships). The re-
sults of our approach were totally dependent on the
model output. As we trained our model on generic
ICD-10 codes, and not specific top-50 labels, some-
what low accuracy was expected. The average words
in the accuracy calculation were around 200.
CCH 2023 - Special Session on Machine Learning and Deep Learning for Preventive Healthcare and Clinical Decision Support
634

5 CONCLUSIONS, DISCUSSION,
AND FUTURE WORK
The black-box nature of deep learning models hin-
ders the end-users from trusting predictions and deci-
sion support offered by AI systems. It is especially
true for medical and other critical fields. The lack of
trust makes sense in the wake of risks to human lives,
health and costs. Recent advancements in ’attention
mechanisms’ based explainability are helping allevi-
ate such trust issues by elucidating the inner workings
of the black-box deep learning AI models. However,
the user still is not made aware of how the highlighted
term is related to other terms and predicted labels.
This paper has demonstrated a novel deep learn-
ing approach titled ‘Explainable Knowledge Graph
Creation’ to introduce explainability in computer-
assisted medical coding (CAC). It has not only suc-
cessfully predicted applicable medical codes in in-
patient discharge summaries, but it has also gener-
ated corresponding knowledge graphs (KG) that help
users review the basis of the predictions. The gener-
ated KGs are very broad and deep, yet they are con-
figurable in ways where the users can view the re-
lationships between different concepts found in pa-
tient summaries based on their strengths. Strong and
weak word-to-word and word-to-code level connec-
tions make it very valuable for the users in under-
standing and verifying the predictions. Visualization
brings it closer to the process of knowledge creation
and understanding. The proposed approach refers
to reliable medical ontologies and medical coding
databases. While AI-based automation finds the most
applicable medical codes, the attention mechanisms
and knowledge graphs build user trust in automati-
cally predicted codes. Finally, the proposed system
learns from previous predictions, gradually improv-
ing its performance. To the best of our knowledge, our
approach is among the earliest ones on using knowl-
edge graphs for explainability in medical coding. It
also goes the deepest so far in incorporating explain-
ability in any medical domain.
Multiple practical use cases exist for this approach
mainly in professional services using unstructured
knowledge bases and ontologies, such as medical cod-
ing, accounting, auditing and legal services. For ex-
ample, as shown in this paper, medical coders and
medical claims auditors can be provided automati-
cally predicted codes which they can accept or reject
with higher confidence due to the incorporation of ex-
plainability. Similarly, accounting, tax and legal pro-
fessions rely on extensive textual knowledge bases as
well as text documents from the client side that should
be coded with relevance to specific clauses in the ac-
counting manuals, tax codes or legal clauses. This
approach can be helpful in building trustworthy rec-
ommenders in these areas.
Despite its precise results, there are certain limi-
tations in our research. At first, an accuracy of 64%
and 53% was achieved on word-to-word and word-to-
code levels respectively. These are low but they will
be considered very good considering this is an early
paper in this direction. Even though the model will
learn and improve the accuracy of code predictions
and identifying relationships over time, an important
point is the performance of the Explainable Knowl-
edge Graph Creation approach depends on the accu-
racy of the outputs of the deep learning model. If the
model’s performance is poor, it will directly impact
the accuracy of our novel approach. Secondly, the
enriched knowledge graph is limited to certain nodes
and relationships excluding some entities and detailed
domain knowledge which could have led to even bet-
ter results and explainability. A third limitation comes
from the testing and training datasets containing pa-
tient summaries. MIMIC-III is limited to emergency
room clinical notes and patient discharge summaries
where the focus is on stabilizing the patient rather
than long-term prognosis, so comorbidities and other
issues might not be deeply focused on or addressed
by the ER physicians.
We plan to address the aforementioned limitations
in our future research. To improve model perfor-
mance, a deep infusion of knowledge graphs with
deep learning could increase the overall accuracy of
the Multi-Label Classification problem. In order to
enhance the reliability and accuracy of predictions,
future research should employ other medical ontolo-
gies with a deeper knowledge of the domain, further
improving the understanding and visualization of ex-
plainability. Using broader all-cause hospital admis-
sions datasets is also recommended. All the above
steps would go a long way in opening the black box
of deep learning CAC models.
REFERENCES
Aalseth, P. (2014). Medical Coding: What it is and how it
Works. Jones & Bartlett Publishers.
Abbas, A., Afzal, M., Hussain, J., Ali, T., Bilal, H. S. M.,
Lee, S., and Jeon, S. (2021). Clinical concept extrac-
tion with lexical semantics to support automatic an-
notation. International Journal of Environmental Re-
search and Public Health, 18(20):10564.
Abbas, A., Afzal, M., Hussain, J., and Lee, S. (2019).
Meaningful information extraction from unstructured
clinical documents. Proc. Asia Pac. Adv. Netw, 48:42–
47.
Knowledge Graph Based Trustworthy Medical Code Recommendations
635

Ahmed, U., Lin, J. C.-W., and Srivastava, G. (2022). Hyper-
graph-based attention curriculum learning using a lex-
ical algorithm for mental health. Pattern Recognition
Letters, 157:135–143.
Alonso, V., Santos, J. V., Pinto, M., Ferreira, J., Lema, I.,
Lopes, F., and Freitas, A. (2020). Problems and bar-
riers during the process of clinical coding: a focus
group study of coders’ perceptions. Journal of medi-
cal systems, 44(3):1–8.
Biswas, B., Pham, T.-H., and Zhang, P. (2021). Transicd:
Transformer based code-wise attention model for ex-
plainable icd coding. In International Conference
on Artificial Intelligence in Medicine, pages 469–478.
Springer.
Campbell, S. and Giadresco, K. (2020). Computer-assisted
clinical coding: A narrative review of the literature
on its benefits, limitations, implementation and impact
on clinical coding professionals. Health Information
Management Journal, 49(1):5–18.
Catling, F., Spithourakis, G. P., and Riedel, S. (2018). To-
wards automated clinical coding. International jour-
nal of medical informatics, 120:50–61.
Chai, X. (2020). Diagnosis method of thyroid disease com-
bining knowledge graph and deep learning. IEEE Ac-
cess, 8:149787–149795.
Confalonieri, R., Coba, L., Wagner, B., and Besold, T. R.
(2021). A historical perspective of explainable arti-
ficial intelligence. Wiley Interdisciplinary Reviews:
Data Mining and Knowledge Discovery, 11(1):e1391.
Delangue, C. (2016). Hugging face – the ai community
building the future.
Desai, G. (2020). gauravkdesai/mids-w210-
medical
insurance payment assistant.
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2018). Bert: Pre-training of deep bidirectional trans-
formers for language understanding. arXiv preprint
arXiv:1810.04805.
Dong, H., Su
´
arez-Paniagua, V., Whiteley, W., and Wu,
H. (2021). Explainable automated coding of clinical
notes using hierarchical label-wise attention networks
and label embedding initialisation. Journal of biomed-
ical informatics, 116:103728.
Dranc
´
e, M. (2022). Neuro-symbolic xai: Application to
drug repurposing for rare diseases. In International
Conference on Database Systems for Advanced Ap-
plications, pages 539–543. Springer.
Futia, G. and Vetr
`
o, A. (2020). On the integration of
knowledge graphs into deep learning models for a
more comprehensible ai—three challenges for future
research. Information, 11(2):122.
Garcez, A. d. and Lamb, L. C. (2020). Neurosymbolic ai:
the 3rd wave. arXiv preprint arXiv:2012.05876.
Gaur, M., Faldu, K., and Sheth, A. (2021). Semantics of
the black-box: Can knowledge graphs help make deep
learning systems more interpretable and explainable?
IEEE Internet Computing, 25(1):51–59.
Gaur, M., Gunaratna, K., Bhatt, S., and Sheth, A. (2022).
Knowledge-infused learning: A sweet spot in neuro-
symbolic ai. IEEE Internet Computing, 26(4):5–11.
Graves, A., Fern
´
andez, S., and Schmidhuber, J. (2005).
Bidirectional lstm networks for improved phoneme
classification and recognition. In International con-
ference on artificial neural networks, pages 799–804.
Springer.
Gu, P., Yang, S., Li, Q., and Wang, J. (2021). Disease cor-
relation enhanced attention network for icd coding.
In 2021 IEEE International Conference on Bioinfor-
matics and Biomedicine (BIBM), pages 1325–1330.
IEEE.
Hitzler, P., Bianchi, F., Ebrahimi, M., and Sarker, M. K.
(2020). Neural-symbolic integration and the semantic
web. Semantic Web, 11(1):3–11.
Hochreiter, S. and Schmidhuber, J. (1997). Long short-term
memory. Neural computation, 9(8):1735–1780.
Hogan, A., Blomqvist, E., Cochez, M., d’Amato, C., Melo,
G. D., Gutierrez, C., Kirrane, S., Gayo, J. E. L.,
Navigli, R., Neumaier, S., et al. (2021). Knowledge
graphs. ACM Computing Surveys (CSUR), 54(4):1–
37.
Johnson, A. E., Pollard, T. J., Shen, L., Lehman, L.-w. H.,
Feng, M., Ghassemi, M., Moody, B., Szolovits, P.,
Anthony Celi, L., and Mark, R. G. (2016). Mimic-
iii, a freely accessible critical care database. Scientific
data, 3(1):1–9.
Johnson, S. L. and Linker, R. (2015). Understanding medi-
cal coding: A comprehensive guide. Cengage Learn-
ing.
Khalid, M., Khattak, H. A., Ahmad, A., and Bukhari, S.
A. C. (2022). Explainable prediction of medical codes
through automated knowledge graph curation frame-
work. In Proceedings of 2022 19th International
Bhurban Conference on Applied Sciences & Technol-
ogy (IBCAST) Islamabad, Pakistan, 16th-20th August,
2022, pages 1–6. IEEE.
Lu, H., Uddin, S., Hajati, F., Khushi, M., and Moni, M. A.
(2022). Predictive risk modelling in mental health
issues using machine learning on graphs. In Aus-
tralasian Computer Science Week 2022, pages 168–
175.
Malik, K. M., Krishnamurthy, M., Alobaidi, M., Hussain,
M., Alam, F., and Malik, G. (2020). Automated
domain-specific healthcare knowledge graph curation
framework: Subarachnoid hemorrhage as phenotype.
Expert Systems with Applications, 145:113120.
Moons, E., Khanna, A., Akkasi, A., and Moens, M.-F.
(2020). A comparison of deep learning methods for
icd coding of clinical records. Applied Sciences,
10(15):5262.
Mullenbach, J., Wiegreffe, S., Duke, J., Sun, J., and
Eisenstein, J. (2018). Explainable prediction of
medical codes from clinical text. arXiv preprint
arXiv:1802.05695.
Niu, Z., Zhong, G., and Yu, H. (2021). A review on the
attention mechanism of deep learning. Neurocomput-
ing, 452:48–62.
Noy, N. F., Shah, N. H., Whetzel, P. L., Dai, B., Dorf, M.,
Griffith, N., Jonquet, C., Rubin, D. L., Storey, M.-A.,
Chute, C. G., et al. (2009). Bioportal: ontologies and
CCH 2023 - Special Session on Machine Learning and Deep Learning for Preventive Healthcare and Clinical Decision Support
636

integrated data resources at the click of a mouse. Nu-
cleic acids research, 37(suppl 2):W170–W173.
Sarker, M. K., Zhou, L., Eberhart, A., and Hitzler, P. (2021).
Neuro-symbolic artificial intelligence: Current trends.
arXiv preprint arXiv:2105.05330.
Sheth, A., Gaur, M., Roy, K., Venkataraman, R., and Khan-
delwal, V. (2022). Process knowledge-infused ai:
Toward user-level explainability, interpretability, and
safety. IEEE Internet Computing, 26(5):76–84.
Spillo, G., Musto, C., De Gemmis, M., Lops, P., and Se-
meraro, G. (2022). Knowledge-aware recommenda-
tions based on neuro-symbolic graph embeddings and
first-order logical rules. In Proceedings of the 16th
ACM Conference on Recommender Systems, pages
616–621.
Surolia, A. (2022). Akshatsurolia/icd-10-code-prediction ·
hugging face.
Tate, N. (2017). 4 in 5 medical bills contain errors: Here’s
what you can do.
Teng, F., Yang, W., Chen, L., Huang, L., and Xu, Q. (2020).
Explainable prediction of medical codes with knowl-
edge graphs. Frontiers in Bioengineering and Biotech-
nology, 8:867.
von Eschenbach, W. J. (2021). Transparency and the black
box problem: Why we do not trust ai. Philosophy &
Technology, 34(4):1607–1622.
Wang, X., Wang, D., Xu, C., He, X., Cao, Y., and Chua,
T.-S. (2019). Explainable reasoning over knowledge
graphs for recommendation. In Proceedings of the
AAAI conference on artificial intelligence, volume 33,
pages 5329–5336.
Xian, Y., Fu, Z., Zhao, H., Ge, Y., Chen, X., Huang,
Q., Geng, S., Qin, Z., De Melo, G., Muthukrish-
nan, S., et al. (2020). Cafe: Coarse-to-fine neural
symbolic reasoning for explainable recommendation.
In Proceedings of the 29th ACM International Con-
ference on Information & Knowledge Management,
pages 1645–1654.
Xu, F., Uszkoreit, H., Du, Y., Fan, W., Zhao, D., and Zhu,
J. (2019). Explainable ai: A brief survey on history,
research areas, approaches and challenges. In CCF in-
ternational conference on natural language process-
ing and Chinese computing, pages 563–574. Springer.
Knowledge Graph Based Trustworthy Medical Code Recommendations
637
