
MR to CT Synthesis Using GANs: A Practical Guide Applied to Thoracic
Imaging
Arthur Longuefosse
1
, Baudouin Denis De Senneville
2
, Ga
¨
el Dournes
3
, Ilyes Benlala
3
,
Franc¸ois Laurent
3
, Pascal Desbarats
1
and Fabien Baldacci
1
1
LaBRI, Universit
´
e de Bordeaux, Talence, France
2
Institut de Math
´
ematiques de Bordeaux, Universit
´
e de Bordeaux, Talence, France
3
Service d’Imagerie M
´
edicale Radiologie Diagnostique et Th
´
erapeutique, CHU de Bordeaux, France
Keywords:
Generative Adversarial Networks, CT Synthesis, Lung.
Abstract:
In medical imaging, MR-to-CT synthesis has been extensively studied. The primary motivation is to benefit
from the quality of the CT signal, i.e. excellent spatial resolution, high contrast, and sharpness, while avoiding
patient exposure to CT ionizing radiation, by relying on the safe and non-invasive nature of MRI. Recent
studies have successfully used deep learning methods for cross-modality synthesis, notably with the use of
conditional Generative Adversarial Networks (cGAN), due to their ability to create realistic images in a target
domain from an input in a source domain. In this study, we examine in detail the different steps required
for cross-modality translation using GANs applied to MR-to-CT lung synthesis, from data representation and
pre-processing to the type of method and loss function selection. The different alternatives for each step were
evaluated using a quantitative comparison of intensities inside the lungs, as well as bronchial segmentations
between synthetic and ground truth CTs. Finally, a general guideline for cross-modality medical synthesis is
proposed, bringing together best practices from generation to evaluation.
1 INTRODUCTION
In clinical practice, computed tomography (CT) is
typically used to diagnose lung conditions. However,
this modality exposes patients to ionizing radiation,
which may have negative effects on their health. Re-
cently, lung MRI with ultrashort or zero echo-time
(UTE/ZTE) has shown promise for high-resolution
structural imaging of the lung (Dournes et al., 2015;
Dournes et al., 2018). However, the appearance of
images obtained using this technique is substantially
different from those obtained using CT, notably imag-
ing texture, blurring, and noise which has limited its
adoption in clinical practice (cf Figure 1). The gen-
eration of CT images from MRI may be a good alter-
native and could improve patient diagnosis by provid-
ing high quality images to radiologists based solely on
the safe and non-invasive nature of MRI. Over recent
years, deep learning approaches, particularly genera-
tive adversarial networks (GANs) (Goodfellow et al.,
2014), have been extensively studied for image syn-
thesis in medical imaging. This type of network is
made of a generator and a discriminator, and is able to
produce high quality synthetic data similar to a given
dataset by learning a complex non-linear relationship
between MR and CT. Previous research on cross-
modality synthesis has used GANs to synthesize im-
ages in several different regions of the body, such as
the brain (Wolterink et al., 2017; Nie et al., 2017),
pelvic region (Lei et al., 2019), and also in the lungs
using Dixon MRI (Baydoun et al., 2020). Many stud-
ies have been conducted on the development of spe-
cific GAN models, including unpaired methods based
on cycleGAN (Zhu et al., 2017) and paired methods
based on pix2pix (Isola et al., 2017). In addition,
research also focused on the development of various
loss functions, such as cycle consistency (Zhu et al.,
2017), feature-matching (Wang et al., 2018), percep-
tual (Johnson et al., 2016), and contrastive loss (An-
donian et al., 2021). However, most state-of-the-art
studies are limited to these developments and do not
properly address the full range of steps involved in
medical translation tasks, such as preprocessing and
robust evaluation.
In this paper, we present a general guideline for
image-to-image translation applied to thoracic MR
to CT synthesis, covering key topics such as pre-
processing steps, data normalization and quantiza-
tion, and the importance of an adapted resampling
before registering the input. We review the differ-
ent types of GANs and losses and compare their per-
formances in thoracic image-to-image translation. A
268
Longuefosse, A., Denis De Senneville, B., Dournes, G., Benlala, I., Laurent, F., Desbarats, P. and Baldacci, F.
MR to CT Synthesis Using GANs: A Practical Guide Applied to Thoracic Imaging.
DOI: 10.5220/0011895700003417
In Proceedings of the 18th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2023) - Volume 3: IVAPP, pages
268-274
ISBN: 978-989-758-634-7; ISSN: 2184-4321
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

quantitative evaluation of the different models and
parameters is presented, using traditional metrics as
well as a comparison of the segmentations of air-
ways in synthesized CT images versus ground truth
CT images, to help identify the factors that have the
biggest impact on the performance of medical image-
to-image translation. Overall, our evaluation helps to
provide a better understanding of the different mod-
els and parameters used for medical image-to-image
translation and can serve as a useful reference for re-
searchers and practitioners in this field.
(a) UTE MR. (b) CT.
Figure 1: Visual comparison of thoracic UTE MR and CT
modalities of the same patient at a corresponding axial slice.
The CT scan shows a higher signal quality, greater contrast
and sharpness, and fewer artifacts compared to the MRI.
2 METHODS
2.1 Data Acquisition
The dataset used in this study consists of UTE MR
and CT thoracic images of 110 patients. Both modal-
ities were acquired on the same day, from 2018 to
2022. CT images were obtained using a Siemens
SOMATOM Force, in end-expiration, with sharp fil-
ters. The parameters used were a DLP of 10 mGy.cm
and a SAFIRE iterative reconstruction. UTE MR im-
ages were acquired using the SpiralVibe sequence on
a SIEMENS Aera scanner, with the following param-
eters: TR/TE/flip angle=4.1ms/0.07ms/5°. Since the
slice plane is encoded in Cartesian mode, the native
acquisition was performed in the coronal plane with
field-of-view outside the anterior and posterior chest
edges to prevent aliasing. It should be noted that res-
olutions, voxel spacings, and fields of view are not
identical in CT and MR images. In addition, modal-
ities may have been taken at different points in the
respiratory cycle. To obtain a paired dataset, an ad-
equate resampling and a deformable registration will
thus be required between CT and MR volumes.
2.2 Preprocessing
2.2.1 Resampling
In multimodal registration, it is typically advised to
use the image with the highest resolution as the fixed
image and the image with the lower resolution as the
moving image, since a higher level of detail and ac-
curacy in the fixed image can help improve the per-
formance of the registration process. In our case, we
have to register the CT volume, with a voxel size of
0.6 × 0.6 × 0.6 mm
3
, on the MRI, with a voxel size
of 1 × 1 × 1 mm
3
, which implies a resampling of the
CT to the MRI resolution, and thus a loss of infor-
mation, as shown in Figure 2. To avoid this issue,
we propose to upsample the MRI voxel size to the
CT voxel size, allowing to keep the initial resolution
of the CT, which implies a better convergence of the
registration algorithm as well as better performances
for the GAN. The two modalities are therefore resam-
pled on a common grid of 0.6 × 0.6 × 0.6 mm
3
using
tricubic interpolation. For comparison purposes, CT
and MR volumes were also resampled on a 1 × 1 × 1
mm
3
grid.
2.2.2 Multimodal Registration
Accurate alignment of images from different modali-
ties often requires non-rigid registration, especially in
parts of the body subject to severe periodic deforma-
tions, such as cardiac and respiratory motions. Edge-
alignment methods seem particularly well suited for
multimodal medical registration since they don’t rely
on input landmarks and can overcome differences in
intensity and contrast between modalities, by focus-
ing on boundary information. In our dataset, a rigid
translation is estimated to ease convergence, before
the EVolution algorithm (Denis de Senneville et al.,
2016) is employed to estimate the elastic deformation,
a patch-based approach that includes a diffusion reg-
ularization term and a similarity term that favors edge
alignments. To prevent physically implausible folding
of the volumes during the registration process, a dif-
feomorphic transformation is ensured by minimizing
the inverse consistency error ((Christensen and John-
son, 2001), (Heinrich et al., 2012)).
2.2.3 Intensity Normalization
CT and MR modalities have fundamental differences
that must be taken into account when normalizing
intensity values. CT intensity values are defined in
Hounsfield units (HU) and have a physical meaning,
whereas MR intensity values strongly depend on ac-
quisition parameters. Therefore, methods used for in-
tensity normalization must be tailored to the specific
MR to CT Synthesis Using GANs: A Practical Guide Applied to Thoracic Imaging
269
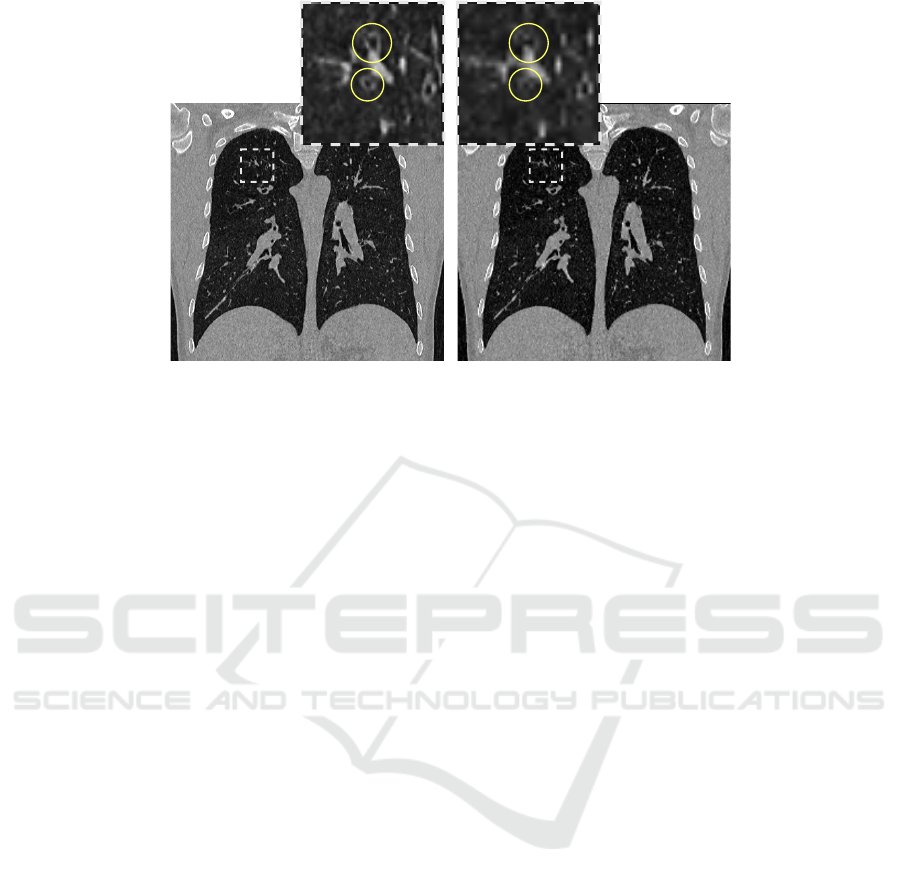
(a) CT 0.6 × 0.6 × 0.6 mm
3
. (b) CT 1 × 1 × 1 mm
3
.
Figure 2: Visual comparison of identical CT slices at different voxel spacing, with zoomed regions that highlight bronchi,
circled in yellow. Pulmonary bronchi are nearly indistinguishable in the 1 mm
3
version due to the lower resolution of the
image.
characteristics of each modality. In our study, CT in-
tensities are cropped to [-1000; 2000] HU window to
remove irrelevant values from the table or background
and rescaled to [-1; 1] using the same window lim-
its. MR intensity inhomogeneities, also known as bias
field, are first corrected using the popular N4 bias field
correction algorithm (Tustison et al., 2010). MR val-
ues are then normalized using z-score, i.e. zero mean
and unit variance, cropped to [−3σ ; 3σ] to remove
outliers, σ being the standard deviation, and rescaled
to [-1; 1] based on minimum and maximum inten-
sities. Nyul histogram matching (Ny
´
ul et al., 2000)
was also considered, but the findings of Reinhold et
al. (Reinhold et al., 2019) indicated that the synthesis
process was robust to the choice of MR normalization
method used. As a result, we opted for a traditional
Z-score normalization approach.
2.2.4 Field of View Standardization
As shown in Figure 1, modalities may have different
fields of view (FOV). Due to the use of narrow beams
of X-rays to produce images, CT field of view is typi-
cally limited to a small area of the body, whereas MRI
allows capturing a wider field of view. In our dataset,
patients may also be in a different position depending
on the modality. This is reflected by the visibility of
the arms on the MRI, as opposed to the CT image. To
uniformize FOV, which can be useful to speed up cal-
culations and guide training, one common approach
is to identify a region of interest (ROI) using segmen-
tation methods. Few methods for lung segmentation
in MRI have been developed due to the lower sig-
nal and contrast, as well as the lack of data. On the
other hand, many effective methods are available for
CT, such as the U-Net R-231 convolutional network
(Hofmanninger et al., 2020). The CT volume being
registered on the MRI, it is then possible to apply the
segmentation of the lungs from the CT on the MRI,
allowing to obtain the same FOV on both modalities.
All axial slices are then either cropped or zero-padded
to 512 × 512, depending on the CT lung mask size.
2.2.5 Impact of Intensity Quantization
In this study, we also investigate the impact of the bit
depth of input medical images on the performance of
a GAN in lung MR to CT translation. We create two
datasets, one in line with most of the current state-
of-the-art papers in the field with 8-bit images, and
another dataset with 16-bit images, and evaluate the
GAN’s performance on each dataset. This allows us
to determine whether using higher bit-depth images
can improve the performance of the GAN for thoracic
CT synthesis.
2.3 Image-to-Image Translation
Conditional generative adversarial networks
(cGANs), are a variant of GANs trained with
additional constraints on a specific input image and
have demonstrated significant potential for image-to-
image translation tasks. CGANs are typically divided
into two main categories: unpaired methods, often
based on the CycleGAN model (Zhu et al., 2017),
designed for image-to-image translation without
the need for corresponding pairs of images, and
IVAPP 2023 - 14th International Conference on Information Visualization Theory and Applications
270

paired methods, based on the pix2pix model, using
corresponding pairs of images.
Since the introduction of the cycle consistency
loss in CycleGAN, many unpaired methods have been
developed, including NICE-GAN (Chen et al., 2020),
a decoupled network training method that uses the
discriminator to encode the image of the target do-
main. As for the paired methods, pix2pix improve-
ments are described in the method pix2pixHD (Wang
et al., 2018), which is no longer dependent on the
pixel-wise loss, but on a new feature matching loss,
as well as a multi-scale discriminator and a percep-
tual loss. SPADE (Park et al., 2019) also enhanced
the performance of paired methods by injecting class-
specific information into the generator network. This
model introduces a spatially adaptive normalization
based on the inputs, that improves the performance
and reliability of the generator, allowing synthesized
images that are conditioned on the input class. The
SPADE architecture can be integrated in other mod-
els, such as pix2pixHD, to apply additional con-
straints on the inputs and guide training.
Finally, recent works on paired image-to-image
translation developed a new type of bidirectional con-
trastive loss, called PatchNCE loss (Andonian et al.,
2021), that assesses the similarity between two im-
ages based on the mutual information from embedded
patches, unlike GANs discriminator that only evalu-
ates the realism of a synthesized image. This con-
trastive loss produces a smooth and interpretable loss
trajectory, which makes it easier to evaluate the con-
vergence of the training process and determine the
number of epochs needed. This is a common chal-
lenge with GANs since their traditional loss functions
tend to be noisy and provide no clear indication of
training progress.
2.4 Assessment of the Synthesis
In order to evaluate the performance of generative
models, past research has proposed several extrin-
sic evaluation measures, most notably Inception Dis-
tances (FID (Heusel et al., 2017), KID (Binkowski
et al., 2018)), which compare the generated images to
a set of real images and assess their quality and sim-
ilarity. Such measures have been proven to be insen-
sitive to global structural problems (Tsitsulin et al.,
2020), and may not be sufficient for the evaluation of
medical image translation.
Traditional image processing metrics, such as
MSE, PSNR, SSIM, are the state-of-the-art reference
metrics for evaluating synthetic images. They can
provide information on how well the model preserves
spatial structure and content of the original images,
but are still highly sensitive to noise and distortion,
and may not accurately reflect the visual quality of an
image with low-level artifacts (Toderici et al., 2017).
Our assumption is that task-specific metrics are re-
quired to accurately quantify synthesized images and
evaluate the performance of a model, by taking into
account the structure and semantics of the images.
In our dataset, we defined them as Dice score, preci-
sion, and sensitivity between synthesized and ground
truth bronchial tree segmentations, by using NaviAir-
way (Wang et al., 2022), a bronchiole-sensitive air-
way segmentation pipeline designed for CT data. This
allows us to accurately quantify false positives and
false negatives at the bronchial level for each syn-
thetic CT image. A qualitative evaluation conducted
by radiologists or other medical experts can also be
valuable, to ensure that the translation has preserved
overall fidelity with the ground truth and diagnostic
information.
3 EXPERIMENTS AND RESULTS
(a) UTE-MR input. (b) Ground truth CT.
(c) SPADE 1. (d) SPADE 0.6.
Figure 3: Comparison between MR, synthetic CT from
SPADE 1 × 1 × 1 mm
3
and SPADE 0.6 × 0.6 × 0.6 mm
3
and ground truth CT axial slices.
The initial dataset of 110 MR-CT thoracic images is
split in a training set of 82 patients, and testing set
of 28 patients. Although 3D GANs allow perception
of volumetric and neighborhood spatial information,
they involve an excessive computational cost and a re-
duction of the number of samples, which can be chal-
lenging to implement for some datasets. Therefore,
we choose to train the models on the 2D axial slices
of the CT and MR volumes and define datasets that
will allow us to assess the impact of each preprocess-
ing step:
• unpaired + unregistered 0.6 × 0.6 × 0.6 mm
3
CT,
MR to CT Synthesis Using GANs: A Practical Guide Applied to Thoracic Imaging
271

Table 1: Mean squared error (MSE), cross-correlation (CC) and structural similarity index (SSIM) between synthesized CT
and real CT inside the lungs.
Model MSE CC SSIM
NICE-GAN 88,45 ± 9,91 0,9283 ± 0,023 0,9725 ± 0,024
NICE-GAN registered 82,41 ± 9,74 0,9406 ± 0,015 0,9776 ± 0,022
pix2pixHD 78,46 ± 13,03 0,9499 ± 0,011 0,9834 ± 0,032
pix2pixHD w/ contrast 75,51 ± 12,05 0,9557 ± 0,010 0,9900 ± 0,030
SPADE 67,82 ± 8,18 0,9635 ± 0,083 0,9915 ± 0,016
SPADE w/ contrast 67,53 ± 7,31 0,9646 ± 0,088 0,9927 ± 0,017
SPADE 8-bit w/ contrast 67,76 ± 7,70 0,9630 ± 0,096 0,9932 ± 0,018
SPADE 1mm
3
w/ contrast 76,36 ± 8,79 0,9505 ± 0,016 0,9830 ± 0,026
Table 2: Dice, precision and sensitivity between synthesized and ground truth airways segmentations.
Model Dice Precision Sensitivity
NICE-GAN 0,590 ± 0,088 0,636 ± 0,0752 0,583 ± 0,1298
NICE-GAN-registered 0,640 ± 0,071 0,665 ± 0,069 0,642 ± 0,104
pix2pixHD 0,707 ± 0,054 0,796 ± 0,060 0,660 ± 0,102
pix2pixHD w/ contrast 0,741 ± 0,031 0,787 ± 0,052 0,715 ± 0,088
SPADE 0,733 ± 0,068 0,829 ± 0,060 0,681 ± 0,108
SPADE w/ contrast 0,743 ± 0,060 0,819 ± 0,054 0,706 ± 0,104
SPADE 8-bit w/ contrast 0,742 ± 0,055 0,802 ± 0,057 0,719 ± 0,098
SPADE 1mm w/ contrast 0,687 ± 0,078 0,766 ± 0,068 0,652 ± 0,120
8bit
• unpaired + registered 0.6× 0.6× 0.6 mm
3
CT, 8bit
• paired + registered 0.6 × 0.6 × 0.6 mm
3
CT, 8bit
• paired + registered 0.6 × 0.6 × 0.6 mm
3
CT, 16bit
• paired + registered 1 × 1 × 1 mm
3
CT, 16bit
The NICE-GAN model was trained using un-
paired datasets, while the pix2pixHD and SPADE
models were trained using paired datasets. We also
evaluated the performance gain of the contrastive loss
when applied to these paired methods.
All models are trained using the same proce-
dure and architecture defined in the respective papers,
apart from pi2pixHD/SPADE dataloader and infer-
ence parts, which have been adapted to support 16-
bit input and output arrays. Table 1 lists the quan-
titative evaluation using mean squared error, cross-
correlation and structural similarity index between
synthesized CT images and ground truth CT. Calcula-
tions are constrained within the intersection between
CT and synthesized CT lung masks, to avoid compar-
ing the backgrounds and narrow the results inside the
lungs. Figure 3 shows an example axial slice between
input MR, ground truth CT and synthetic CT from
SPADE model with different samplings. SPADE re-
sults based on CT with a voxel size of 0.6× 0.6 × 0.6
mm
3
present enhanced contrast and sharpness, and
therefore allow a more accurate distinction of vessels
and bronchi inside the lungs.
To validate these assumptions, we performed
the airways segmentation of synthesized and ground
truth CT using NaviAirway (Wang et al., 2022),
a bronchiole-sensitive airway segmentation pipeline
designed for CT data, and computed dice score, preci-
sion, and sensitivity (Table 2). To enable comparison,
the SPADE 1× 1 × 1 mm
3
was resampled to the same
resolution as the ground truth CT before calculating
the airways segmentation. Figure 4 shows a com-
parison of ground truth and synthetic CT bronchial
trees, and illustrates the ability of the SPADE method
to produce high quality airways, with levels of depth
generation that closely approximate those of ground
truth CT.
4 DISCUSSION
Results from Table 1 based on image processing met-
rics and Table 2 based on the evaluation of airways
segmentation are strongly correlated, with identical
trends. Unpaired methods seem to benefit from the
elastic registration but produce less satisfactory re-
sults than paired methods, which is in agreement with
statements in state-of-the-art (Kaji and Kida, 2019).
Paired pix2pixHD method combined with the con-
ditional normalization layer SPADE provides better
performances than the pix2pixHD method alone since
it can overcome false positives and false negatives by
adding constraints on the inputs. The introduction
IVAPP 2023 - 14th International Conference on Information Visualization Theory and Applications
272
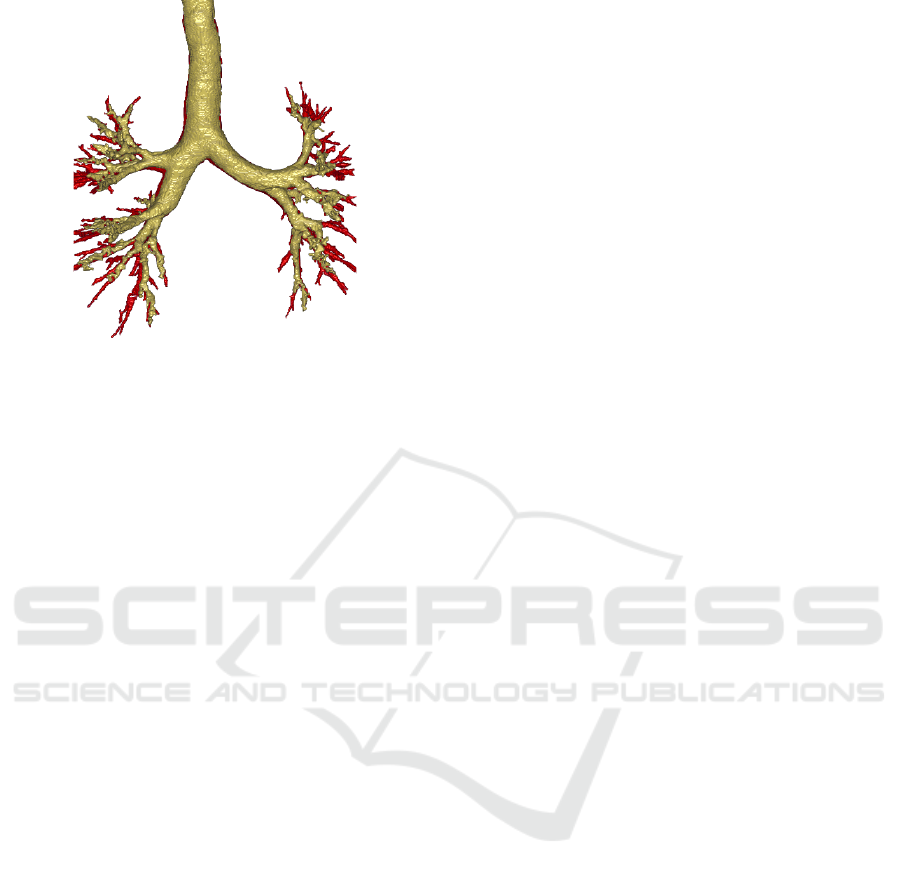
Figure 4: Airways segmentation example based on SPADE
with contrastive loss (yellow) and real CT (red) using the
NaviAirway pipeline (Wang et al., 2022).
of the PatchNCE (Andonian et al., 2021) contrastive
loss has improved the performance of paired meth-
ods, particularly for the pix2pixHD model that tends
to diverge. This addition had only a minor impact
on the SPADE model, but still provided better control
over convergence during training and a more accu-
rate way to differentiate epochs. The performance of
the SPADE model with a voxel size of 0.6× 0.6 × 0.6
mm
3
is significantly superior to that of the model with
a voxel size of 1 × 1 × 1 mm
3
, both in terms of sig-
nal quality and bronchi reconstruction. These results
confirm our initial hypothesis that input data should
be registered based on the voxel size of the modal-
ity with the highest resolution, since downsampling
the ground truth reference leads to a loss of informa-
tion, especially in fine structures such as vessels and
bronchi. Surprisingly, the intensity quantization in the
input dataset images does not appear to have a signif-
icant impact on GAN performances; both SPADE 16-
bit and SPADE 8-bit models performed similarly. The
reason for this could be that our dataset is composed
of highly contrasted information, such as vessels and
bronchi in the lungs, and the representation in 8-bit
instead of the initial 12-bit would barely impact the
reconstruction using GANs. Future works will aim to
confirm this hypothesis by conducting similar exper-
iments using different medical datasets in other parts
of the body.
5 CONCLUSION
In this paper, we present a comprehensive guide for
medical image translation using GANs. We focus on
the importance of data preprocessing, and its impact
on performance; the benefits of using a resampling
based on the modality with the highest resolution,
as opposed to state-of-the-art statements, have been
demonstrated. We advocate the use of contrastive
loss methods, such as PatchNCE, to address one of
the most significant challenges of GANs, which is
assessing convergence and stability during training.
In addition, we argue that traditional GAN metrics
commonly used in the field, such as FID and KID,
as well as standard image processing metrics, do not
provide sufficient information to adequately evaluate
GAN performances in medical image-to-image trans-
lation tasks. We recommend defining task-specific
quantitative evaluation methods, ideally in conjunc-
tion with a qualitative evaluation by experts, in order
to robustly assess the performance of a model in this
context. In future work, we plan to investigate the
validity of our assumptions on different datasets for
other parts of the body and provide guidance on in-
corporating 3D information into the training process
for medical image-to-image translation.
REFERENCES
Andonian, A., Park, T., Russell, B., Isola, P., Zhu, J.-Y., and
Zhang, R. (2021). Contrastive feature loss for image
prediction. In Proceedings of the IEEE/CVF Interna-
tional Conference on Computer Vision, pages 1934–
1943.
Baydoun, A., Xu, K., Yang, H., Zhou, F., Heo, J., Jones,
R., Avril, N., Traughber, M., Traughber, B., Qian,
P., and Muzic, R. (2020). Dixon-based thorax syn-
thetic ct generation using generative adversarial net-
work. Intelligence-Based Medicine, 3-4.
Binkowski, M., Sutherland, D. J., Arbel, M., and Gretton,
A. (2018). Demystifying mmd gans. International
Conference on Learning Representations.
Chen, R., Huang, W., Huang, B., Sun, F., and Fang, B.
(2020). Reusing discriminators for encoding: Towards
unsupervised image-to-image translation. In Proceed-
ings of the IEEE/CVF Conference on Computer Vision
and Pattern Recognition (CVPR).
Christensen, G. E. and Johnson, H. J. (2001). Consis-
tent image registration. IEEE Trans Med Imaging,
20(7):568–582.
Denis de Senneville, B., Zachiu, C., Ries, M., and Moo-
nen, C. (2016). EVolution: an edge-based variational
method for non-rigid multi-modal image registration.
Phys Med Biol, 61(20):7377–7396.
Dournes, G., Grodzki, D., Macey, J., Girodet, P. O., Fayon,
M., Chateil, J. F., Montaudon, M., Berger, P., and Lau-
rent, F. (2015). Quiet Submillimeter MR Imaging of
the Lung Is Feasible with a PETRA Sequence at 1.5
T. Radiology, 276(1):258–265.
Dournes, G., Yazbek, J., Benhassen, W., Benlala, I., Blan-
chard, E., Truchetet, M. E., Macey, J., Berger, P.,
MR to CT Synthesis Using GANs: A Practical Guide Applied to Thoracic Imaging
273

and Laurent, F. (2018). 3D ultrashort echo time
MRI of the lung using stack-of-spirals and spherical
k-Space coverages: Evaluation in healthy volunteers
and parenchymal diseases. J Magn Reson Imaging,
48(6):1489–1497.
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B.,
Warde-Farley, D., Ozair, S., Courville, A., and Ben-
gio, Y. (2014). Generative adversarial nets. In Ad-
vances in Neural Information Processing Systems,
volume 27.
Heinrich, M. P., Jenkinson, M., Bhushan, M., Matin, T.,
Gleeson, F. V., Brady, S. M., and Schnabel, J. A.
(2012). MIND: modality independent neighbourhood
descriptor for multi-modal deformable registration.
Med Image Anal, 16(7):1423–1435.
Heusel, M., Ramsauer, H., Unterthiner, T., Nessler, B., and
Hochreiter, S. (2017). Gans trained by a two time-
scale update rule converge to a local nash equilibrium.
In Proceedings of the 31st International Conference
on Neural Information Processing Systems, NIPS’17,
page 6629–6640.
Hofmanninger, J., Prayer, F., and et al., J. P. (2020). Au-
tomatic lung segmentation in routine imaging is pri-
marily a data diversity problem, not a methodology
problem. Eur Radiol Exp, 50.
Isola, P., Zhu, J.-Y., Zhou, T., and Efros, A. A. (2017).
Image-to-image translation with conditional adversar-
ial networks. CVPR.
Johnson, J., Alahi, A., and Fei-Fei, L. (2016). Perceptual
losses for real-time style transfer and super-resolution.
In Computer Vision – ECCV 2016, pages 694–711.
Kaji, S. and Kida, S. (2019). Overview of image-to-image
translation by use of deep neural networks: denois-
ing, super-resolution, modality conversion, and recon-
struction in medical imaging. Radiol Phys Technol,
12(3):235–248.
Lei, Y., Harms, J., Wang, T., Liu, Y., Shu, H. K., Jani, A. B.,
Curran, W. J., Mao, H., Liu, T., and Yang, X. (2019).
MRI-only based synthetic CT generation using dense
cycle consistent generative adversarial networks. Med
Phys, 46(8):3565–3581.
Nie, D., Trullo, R., Lian, J., Petitjean, C., Ruan, S., Wang,
Q., and Shen, D. (2017). Medical Image Synthe-
sis with Context-Aware Generative Adversarial Net-
works. Med Image Comput Comput Assist Interv,
10435:417–425.
Ny
´
ul, L. G., Udupa, J. K., and Zhang, X. (2000). New vari-
ants of a method of MRI scale standardization. IEEE
Trans Med Imaging, 19(2):143–150.
Park, T., Liu, M.-Y., Wang, T.-C., and Zhu, J.-Y. (2019).
Semantic image synthesis with spatially-adaptive nor-
malization. In Proceedings of the IEEE Conference
on Computer Vision and Pattern Recognition.
Reinhold, J. C., Dewey, B. E., Carass, A., and Prince, J. L.
(2019). Evaluating the Impact of Intensity Normaliza-
tion on MR Image Synthesis. Proc SPIE Int Soc Opt
Eng, 10949.
Toderici, G., Vincent, D., Johnston, N., Jin Hwang, S.,
Minnen, D., Shor, J., and Covell, M. (2017). Full
resolution image compression with recurrent neural
networks. In Proceedings of the IEEE conference
on Computer Vision and Pattern Recognition, pages
5306–5314.
Tsitsulin, A., Munkhoeva, M., Mottin, D., Karras, P., Bron-
stein, A., Oseledets, I., and Mueller, E. (2020). The
shape of data: Intrinsic distance for data distributions.
In International Conference on Learning Representa-
tions.
Tustison, N. J., Avants, B. B., Cook, P. A., Zheng, Y., Egan,
A., Yushkevich, P. A., and Gee, J. C. (2010). N4ITK:
improved N3 bias correction. IEEE Trans Med Imag-
ing, 29(6):1310–1320.
Wang, A., Tam, T. C. C., Poon, H. M., Yu, K.-C., and Lee,
W.-N. (2022). Naviairway: a bronchiole-sensitive
deep learning-based airway segmentation pipeline for
planning of navigation bronchoscopy. arXiv preprint
arXiv:2203.04294.
Wang, T.-C., Liu, M.-Y., Zhu, J.-Y., Tao, A., Kautz, J., and
Catanzaro, B. (2018). High-resolution image synthe-
sis and semantic manipulation with conditional gans.
In Proceedings of the IEEE Conference on Computer
Vision and Pattern Recognition.
Wolterink, J., Dinkla, A., Savenije, M., Seevinck, P., van
den Berg, C., and I
ˇ
sgum, I. (2017). Deep mr to ct
synthesis using unpaired data. In Simulation and Syn-
thesis in Medical Imaging, Lecture Notes in Computer
Science, pages 14–23.
Zhu, J.-Y., Park, T., Isola, P., and Efros, A. A. (2017).
Unpaired image-to-image translation using cycle-
consistent adversarial networks. In Computer Vision
(ICCV), 2017 IEEE International Conference on.
IVAPP 2023 - 14th International Conference on Information Visualization Theory and Applications
274
