
Agnostic eXplainable Artificial Intelligence (XAI) Method Based on
Volterra Series
Jhonatan Contreras
1,2,∗,† a
and Thomas Bocklitz
1,2,∗,† b
1
Leibniz Institute of Photonic Technology, Albert Einstein Straße 9, 07745 Jena, Germany
2
Institute of Physical Chemistry (IPC) and Abbe Center of Photonics (ACP), Friedrich Schiller University,
Helmholtzweg 4, 07743 Jena, Germany
Keywords:
Explainabe Artificial Intelligence, Volterra Series, Model Approximation, Model Interpretation.
Abstract:
Convolutional Neural Networks (CNN) have shown remarkable results in several fields in recent years. Tradi-
tional performance metrics assess model performance but fail to detect biases in datasets and models. Explain-
able artificial intelligence (XAI) methods aim to evaluate models, identify biases, and clarify model decisions.
We propose an agnostic XAI method based on the Volterra series that approximates models. Our model ar-
chitecture is composed of three second-order Volterra layers. Relevant information can be extracted from the
model to be approximated and used to generate relevance maps that explain the contribution of the input ele-
ments to the prediction. Our Volterra-XAI learns its Volterra kernels comprehensively and is trained using a
target model outcome. Therefore, no labels are required, and even when training data is unavailable, it is still
possible to generate an approximation utilizing similar data. The trustworthiness of our method can be mea-
sured by considering the reliability of the Volterra approximation in comparison with the original model. We
evaluate our XAI method for the classification task on 1D Raman spectra and 2D images using two common
CNN architectures without hyperparameter tuning. We present relevance maps indicating higher and lower
contributions to the approximation prediction (logit).
1 INTRODUCTION
The remarkable rise of convolutional neural networks
(CNNs) makes them attractive for application in var-
ious fields, including the medical domain, where
CNNs have been used to classify medical data suc-
cessfully, such as Raman spectra, cystoscopy, and
histological images (Lin et al., 2019)(Halicek et al.,
2020)(Rodner et al., 2019) (Niioka et al., 2018).
However, traditional accuracy metrics fail to detect
(or can hide) biases in both datasets and models,
which is critical in this sector. The models must be
reliable and transparent. Therefore, explainable arti-
ficial intelligence (XAI) methods seek to describe the
model behaviour using relevance maps, the notion of
that class, and heat maps. Relevance maps (R-Map)
specify the input elements that contribute the most to
the classification output. The notion of that class in-
a
https://orcid.org/0000-0002-0491-9896
b
https://orcid.org/0000-0003-2778-6624
∗
Member of Leibniz Health Technologies
†
Member of the Leibniz Centre for Photonics in Infec-
tion Research (LPI)
dicates what the model expects as input to maximize
a particular class. Heat maps (H-Map) show the fea-
tures extracted by the model at different stages. The
most popular XAI methods can be divided into three
groups: perturbation-based (Mishra et al., 2017),
which computes a set of duplicate inputs with per-
turbations (removing pixels or spectra) and evalu-
ates how the prediction changes. Deconvolution-
based (Lundberg and Lee, 2017), which generates
salience maps using convolution transpose opera-
tions. Gradient-based (Simonyan et al., 2013), which
uses backpropagation to calculate logit gradients to
visualize the notion of the class.
Some XAI methods require access to model
architecture and parameters, such as Integrated
Gradient (Sundararajan et al., 2017), which utilizes
an integral approximation by averaging gradients
over a set of perturbed versions of the input image.
Similarly, Taylor-based (Montavon et al., 2017)
(TD) and Layer-wise Relevance Propagation (Bach
et al., 2015) (LRP) produce relevance maps using
partial derivatives of the model weights, requiring
the model definition to backpropagate gradients.
Contreras, J. and Bocklitz, T.
Agnostic eXplainable Artificial Intelligence (XAI) Method Based on Volterra Series.
DOI: 10.5220/0011889700003411
In Proceedings of the 12th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2023), pages 597-606
ISBN: 978-989-758-626-2; ISSN: 2184-4313
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
597

Alternatively, XAI methods can be agnostic, in-
dicating that they consider models a black box. For
instance, a series of polynomial models derived from
a Taylor expansion can approximate a non-linear
model’s output function (Bocklitz, 2019). Among
the advantages of agnostic methods is their flexibility,
which allows them to be applied to any AI model. A
simpler and more explainable model increases the ex-
plainability. At the same time, it becomes more flex-
ible by being able to choose a feature representation
that may be different from the original model.
We propose an agnostic XAI method based on the
Volterra series to approximate a target model (Koren-
berg and Hunter, 1996). In this manner, relevant infor-
mation can be extracted to generate a relevance map
that explains the contribution of the pixels to the pre-
diction. Our Volterra-XAI learns its Volterra kernels
comprehensively and is trained using the target model
outcome. Therefore, no labels are required, and even
when training data is unavailable, it is still possible
to generate an approximation employing similar data.
The relevance map is extracted from the second to
last layer of the Volterra-XAI, and its trustworthiness
can be measured by considering the reliability of the
Volterra approximation. This value can be obtained
from the difference between the outcome of the target
model and the approximation model.
Section 2 and Section 3 introduce the Volterra se-
ries and Volterra Network. Section 4 presents the
experiment protocol. Section 4.1 shows the result
on a 1D dataset. Section 4.2 evaluates 2D models
on the CIFAR 10 and the histology dataset. In all
cases, our method compares some trained methods.
Finally, Section 5 summarizes the contribution and set
together the conclusions.
2 VOLTERRA SERIES
Consider a single-input, single-output (SISO) system
with an input time function, x(t), and output time
function y(t). This system can be, for example, an
artificial intelligence method, such as a Linear Dis-
criminant Analysis (LDA) model or a neural network.
It can be extended in an infinite Volterra series as
y(t) =
∞
∑
i=0
V
i
[h
i
, x] (1)
Where the zero-order Volterra term is a constant
called the impulse response.
V
0
[h
0
, x] = h
0
(2)
and for i ≥ 1, the i-th-order Volterra term is
Figure 1: Visualization of the second-order Volterra series
as block operation.
V
i
[h
i
, x] =
Z
∞
−∞
· ··
Z
∞
−∞
h
i
(τ
1
, · · · , τ
i
)x(t − τ
1
)
· · · x(t − τ
i
)d
τ
1
· · · d
τ
i
(3)
Equations 1, 2 and 3 represent the Volterra series ex-
pansion, where the kernels h
i
are the Volterra ker-
nels (Stegmayer et al., 2004), (Korenberg and Hunter,
1996). Although the calculation of the Volterra ker-
nels is a complicated and time-consuming task, sev-
eral methods have been proposed (Stegmayer, 2004),
(Azpicueta-Ruiz et al., 2010), (Franz and Sch
¨
olkopf,
2006), (Orcioni, 2014) (Orcioni et al., 2018).
In this work, we propose an agnostic explainable
artificial technique that approximates non-linear sys-
tems employing the second-order Volterra series, as
shown in Equation 4 and Figure 1.
y(t) = h
0
+
Z
∞
−∞
h
1
(τ)x(t − τ)d
τ
+
Z
∞
−∞
Z
∞
−∞
h
2
(τ
1
, τ
2
)x(t − τ
1
)x(t − τ
2
)d
τ
1
d
τ
2
(4)
3 VOLTERRA NETWORK
Finding the kernels that satisfy the equation 4 is com-
putationally expensive, and it is a problem similar to
that faced by neural networks. Although it is theoreti-
cally possible to represent any possible function with
a single Volterra expansion, we reduce the complexity
by decomposing it into cascading layers, where each
layer is a simpler Volterra series. Our Volterra layers
are also second-order Volterra expansions, where one
layer’s output serves as the next layer’s input. It re-
duces kernel sizes and helps learn more complex and
abstract relationships in data.
Figure 2 shows the architecture of our feed-
forward Volterra network. The base model has three
Volterra layers, a tanh activation layer, and a dense
layer with the c neurons corresponding to the number
of classes of the respective task. The number of layers
and kernels can be increased or decreased depending
on the complexity of the data, the target model, and
the task. In this paper, we present the results for 1D
and 2D data. We do not perform hyperparameter op-
timization and use the same architecture to compare
ICPRAM 2023 - 12th International Conference on Pattern Recognition Applications and Methods
598
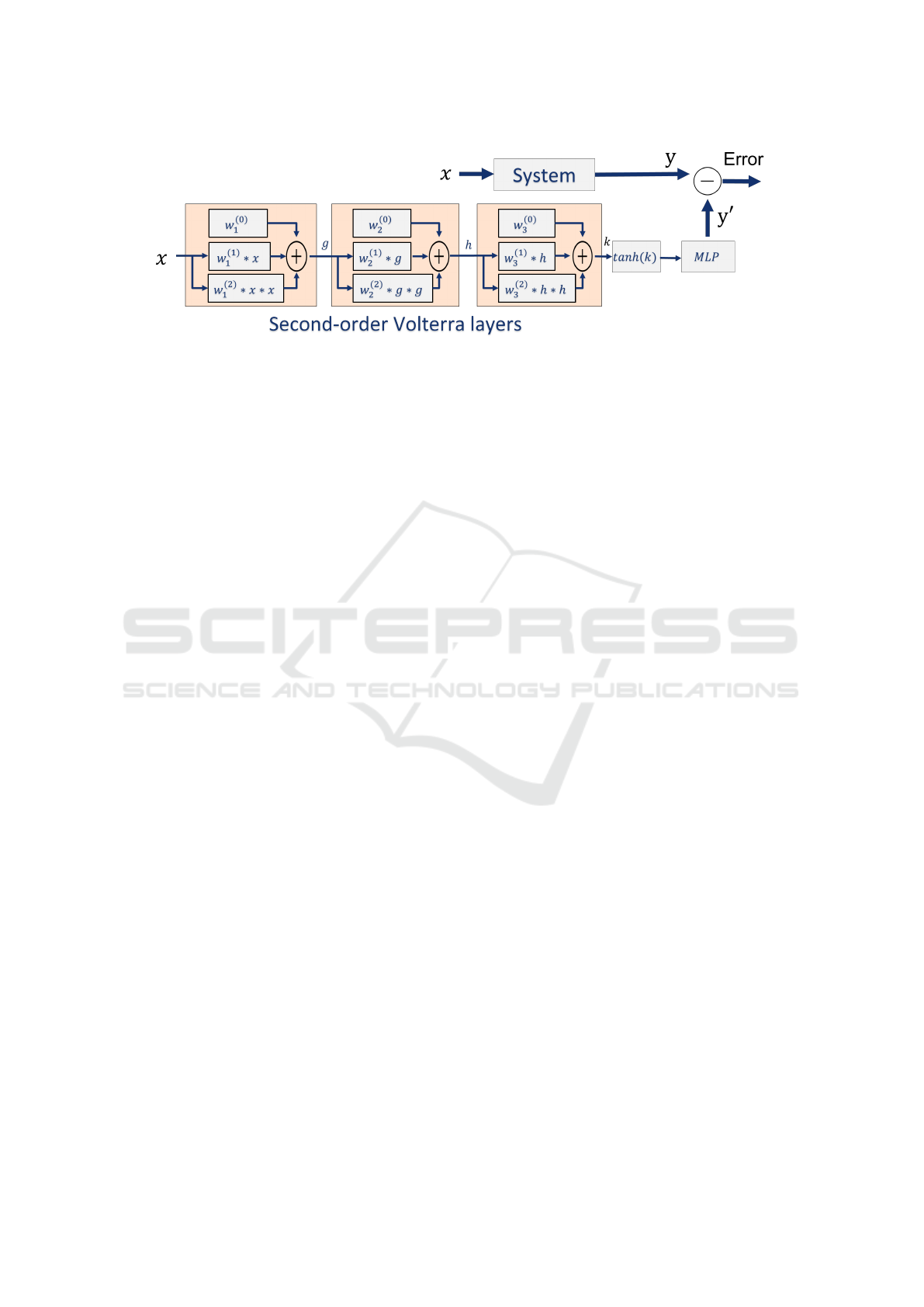
Figure 2: Architecture of our Volterra-XAI Network, composed of three Volterra layers, a tanh activation layer, and a dense
layer with the c neurons corresponding to the number of classes of the respective task.
models. The following formulation can be extrapo-
lated for the analysis of 2D images.
Let’s define our 1D input vector x and a set of
Volterra kernels w = [w
(0)
, w
(1)
, w
(2)
] , and say that
x has length n, w
(0)
has length 1, w
(1)
has length m,
and w
(2)
has length mxm. The second-order Volterra
approximation layer in the discrete domain can be de-
fined as
y[n] = w
(0)
+
∑
m
i=1
w
(1)
i
x[n − i] +
∑
m
i=1
∑
m
j=1
w
(2)
i j
x[n − i]x[n − j]
(5)
Therefore, the zero-order kernel w
(0)
corresponds
to a learnable 1D tensor in our implementation, and
the first-order kernel w
(1)
can be implemented as a
discrete convolution operation, according to Equation
5. The second-order kernel w
(2)
requires the multipli-
cation of the input signal, which can be implemented
by defining a local multiplication in a sliding window.
The multiplication of the input signal can be effi-
ciently implemented by local multiplications of slid-
ing windows. Therefore, the input signal can be de-
fined as a set of patches in the form x = [x
1
, x
2
, ..., x
m
].
Therefore, we can reformulate Equation 5 as follow-
ing:
y(x) = w
(0)
+
m
∑
i=1
w
(1)
i
x
i
+
m
∑
i=1
m
∑
j=1
w
(2)
i j
x
i
x
j
(6)
In matrix notations, the kernel w
(2)
has a dimen-
sion of (mxm, 1). The second term of Equation 6 can
be replaced by the Khatri-Rao product (Khatri and
Rao, 1968), (Seber, 2008), which is the column-wise
Kronecker product of two matrices. Given a M × N
matrix A and a P × Q matrix B, Khatri-Rao product
A ⊙ B has size MP × N is given by
A ⊙ B = [a
1
⊗ b
1
a
2
⊗ b
2
· · · a
N
⊗ b
N
] (7)
Where the Kronecker product (Seber, 2008) is de-
noted by ⊗, given the matrices M × N matrix A and a
P × Q matrix B, the Kronecker product A ⊗ B has size
MP × NQ is given by
A ⊗ B =
a
11
B a
12
B · · · a
1N
B
a
21
B a
22
B · · · a
2N
B
.
.
.
.
.
.
.
.
.
.
.
.
a
M1
B a
M2
B · · · a
MN
B
(8)
Therefore, the Equation 6 can be rewrite as
y(x) = w
(0)
+ (w
(1)
)
T
x + (w
(2)
)
T
(x ⊗ x) (9)
4 EXPERIMENTS
We evaluate our XAI method for the classification
task, in this particular case, for Raman spectra and
2D images. For the experiments, we present mod-
els that obtain training, validation, and test accu-
racy relatively close to state-of-the-art without hyper-
parameter tuning. Parameter selection and accuracy
could be improved with a more sophisticated hyper-
parameter search, a learning rate program, a different
optimizer, or even more modern models. However,
our goal is not to train the best model but to agnosti-
cally evaluate the trained models and provide insights
to scientists with extensive knowledge in areas other
than data science, such as medicine, chemistry, and
physics.
4.1 Raman Spectra
Dataset. This Raman-spectral data set contains six
bacterial species, including Escherichia coli DSM
423, Klebsiella terrigena DSM 2687, Pseudomonas
stutzeri DSM 5190, Listeria innocua DSM 20649,
Staphylococcus warneri DSM 20316, and Staphylo-
coccus cohnii DSM 20261, from Deutsche Samm-
lung von Mikroorganismen and Zellkulturen GmbH
(DSMZ) (Ali et al., 2018). The dataset contains 5420
Agnostic eXplainable Artificial Intelligence (XAI) Method Based on Volterra Series
599
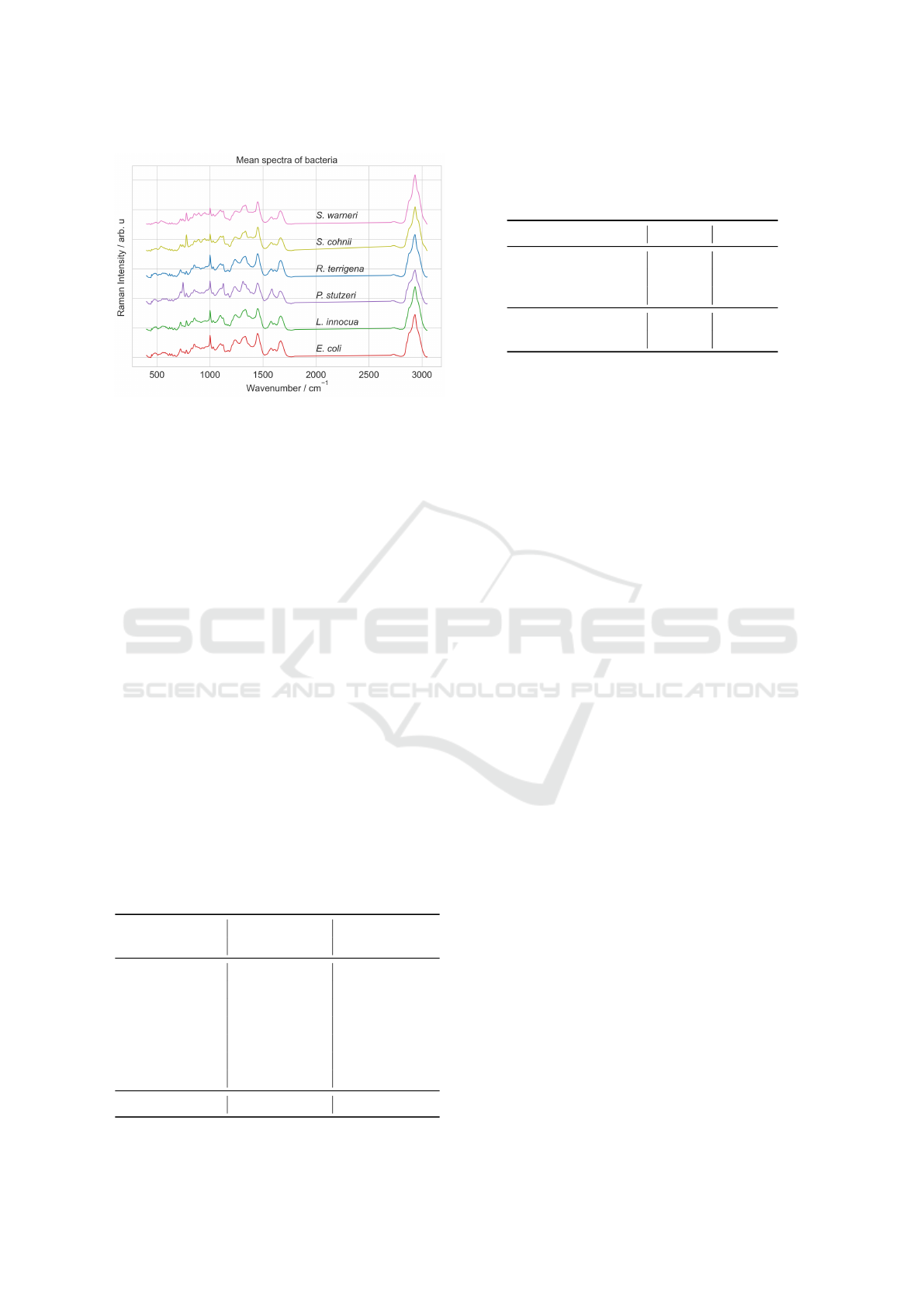
Figure 3: Mean spectra of bacteria dataset.
preprocessed spectra with 584 wavenumbers, Figure
3 shows the mean spectra for each class.
The preprocessing consists of cosmic spikes re-
moval, wavenumber calibration, spectra aligned be-
tween 240–3190 cm–1, and baseline correction. All
species were cultivated in nine independent biologi-
cal replicates (batches). Accordingly, we evaluated
by cross-validation, where the test set is composed of
two batches, and the remaining (7 batches) are used
for training (70%) and validation (30%).
We compare two CNN models trained from
scratch, a traditional 1D CNN and a transformer base
model (TrCNN) (Vaswani et al., 2017). The CNN
comprises a feature extraction part (convolutional lay-
ers) and the classification part (dense layers). The Tr-
CNN comprises multi-head attention layers for fea-
ture extraction and dense layers for classification.
Table 1 presents both models’ mean sensitivity ob-
tained for cross-validation on a total of 36 models.
The CNN model has a mean sensitivity of 83.05%
with a standard deviation of 4.08%, while the mean
sensitivity of the TrCNN was 82.06% with a standard
deviation of 5.34%.
Table 1: Sensitivities of all classes for the first fold in Per-
cent (%). Cross-validation mean sensitivity for the CNN
and Transformer (TrCNN) model.
CNN TrCNN
Class Test Train Test Train
E. coli 68.87 100 51.53 92.0
L. innocua 97.57 100 96.60 90.0
P. stutzeri 88.00 100 80.00 100.0
R. terrigena 57.84 100 88.23 94.12
S. cohnii 93.93 100 84.84 98.11
S. warneri 89.10 100 89.60 97.10
AVG Sens 82.55 100 81.80 95.22
CV-Mean Sens 83.05± 4.08 82.06±5.34
Table 2: Mean absolute error (MAE) of the logits expresses
the difference between a target model and the Volterra ap-
proximation on the bacteria spectra dataset. The number of
parameters for the target models and Volterra network.
Model CNN TrCNN
MAE Train 0.5040 0.2174
MAE Validation 1.3211 0.4119
MAE Test 1.2521 0.3874
Model parameters 19.5M 104.4K
Volterra parameters 175.2K 175.2K
Table 1 also reports the average and the sensitivity
of all classes for the first fold. The average sensitiv-
ity of batch 0 is close to the mean cross-validation
sensitivity, indicating that it is a successfully trained
model. The traditional 1D CNN performs slightly
better. Nevertheless, note that the number of CNN
parameters used is significantly higher (19.5 M) than
the TrCNN model parameters (104.4 K), as shown in
table 2.
4.1.1 1D Volterra Approximation
We evaluate the models trained on the first fold using
our Volterra method. Although the models have dif-
ferent architectures and parameters, we used the same
architecture in our Volterra network to approximate
both models. A better approximation can be found
by parameter selection. The tanh activation function
at the last Volterra layer transforms the output values
from −1 to 1. These values are combined linearly us-
ing a dense layer to approximate the logit values of
the original models. This linear combination can be
visualized as a relevance map or saliency map.
Table 2 exhibits the approximation error, which is
the difference between the target model output and the
Volterra approximation. As expected, the errors of the
transformer model (Tr-CNN) are much lower. This
model has fewer parameters and has a slightly lower
performance than the CNN model, with 104.4K pa-
rameters, while the CNN has 19.5M parameters. The
two Volterra models are identical. The approximation
error can be reduced by increasing the number of pa-
rameters, layers, or training strategies.
Figure 4 shows examples of two saliency maps for
the CNN model generated using our Volterra approx-
imation and a Taylor-based (Montavon et al., 2017)
method. Each saliency map is divided into two sec-
tions. The top part corresponds to the classifier pre-
diction, and the bottom corresponds to the ground
truth class.
The spectra in Figure 4a correspond to the E. coli
class, but is wrongly classified as R. terrigena. Cor-
rectly classified spectra have equal saliency maps, as
ICPRAM 2023 - 12th International Conference on Pattern Recognition Applications and Methods
600

a. Wrongly classified spectra.
b. Correctly classified spectra.
Figure 4: Saliency Map for the CNN model generated using
the Volterra approximation (top) and a Taylor-based (Mon-
tavon et al., 2017) method (bottom). Each saliency map
is divided into two sections. The top part corresponds to
the classifier prediction, and the bottom corresponds to the
ground truth class.
shown in Figure 4b. For the predicted class, the areas
with dark red are the areas that influence the most,
while the areas in yellow are less important for the
classification. Simultaneously, for the correct class,
the bands that influence the most are magenta, and
the bands that influence the least are cyan.
The Volterra approximation error is an indicator of
the saliency map quality. A small error indicates that
the input is close to the expected spectra for that class.
The information provided for saliency maps is diffi-
cult to read, especially for the Taylor method, which
displays excessive noise. We can see that the result
does not focus on the areas as our method does.
We consider that a better manner to examine
this output is to focus only on the k most relevant
wavenumbers. Figure 5 shows the top 15 spectra
(wavenumbers) for the CNN model obtained using
our Volterra method (top) and the Taylor method (bot-
tom). Figure 5a shows an example of a misclassified
spectrum. The spectra in Figure 5b were correctly
classified, and the difference between the target model
output and the Volterra approximation is 0.07, which
corresponds to 2.25%.
Figure 6 shows histograms constructed using the
training data, which accumulate the k most important
wavenumbers for two classes (E. coli and L. innocua).
a. Wrongly classified spectra.
b. Correctly classified spectra.
Figure 5: K most relevant wavenumber postions (variables)
for the CNN model obtained using our Volterra method
(top) and the Taylor method (bottom). Each saliency map
is divided into two sections. The top part corresponds to
the classifier prediction, and the bottom corresponds to the
ground truth class.
The red areas indicate the most frequently used bands
by the classifier according to the Volterra and Taylor-
based methods.
a
b
Figure 6: Histograms accumulate the kth most important
wavenumbers for the CNN model. (a) Volterra approxima-
tion, (b) Taylor-based method. The red areas indicate the
most frequently used bands by the CNN model.
Agnostic eXplainable Artificial Intelligence (XAI) Method Based on Volterra Series
601

The bands shown by the Volterra model are more
concentrated, while the bands used by the Taylor
method are highly distributed. Taylor-based models
should theoretically select a root point value; by ap-
proximating this value, the results are only guaran-
teed stable for some cases. Some recent variations
of the Taylor method, such as layer-wise backpropa-
gation, employ rules to select a reference point and
to omit negative gradients or enhance positive ones.
However, there needs to be a defined way to establish
whether the generated map is reliable. On the other
hand, in this work, we use the Volterra approximation
to state directly the cases in which our model deviates
too much from the original model, so the result should
not be considered.
4.2 2D Images
We assess our 2D Volterra model on two datasets,
CIFAR-10 (Krizhevsky et al., 2009), and a public
histology dataset (Kather et al., 2016). Hundreds of
methods are used for image classification. In this pa-
per, we select two models, a popular transfer learning
strategy and a transformer-based network in the top
5 state-of-the-art. Both models can be improved by
changing the architecture, increasing the number of
layers, by parameters such as the learning rate sched-
ule, optimizer, weight decay, or data augmentation.
However, our interest is not to train the best network,
but to evaluate previously trained networks.
Transfer learning. This CNN network performs
transfer learning based on one of the most com-
mon pre-trained networks, the ResNet50, available in
Keras, where the weights are pre-trained in Imagenet.
All layers are retained except the final classification
layers, replaced by two dense layers with non-linear
activation ReLU and the last layer with Softmax acti-
vation.
Transformer-based network. We implemented Tr-
CNN, a version of the Vision Transformer (ViT)
model proposed by (Dosovitskiy et al., 2020) for
image classification. TrCNN uses the self-attentive
Transformer architecture, which requires images to be
transformed into patch sequences.
4.2.1 CIFAR-10
CIFAR-10 is a benchmark dataset consisting of 60000
32 × 32 color images in 10 classes, with 6000 im-
ages per class, including airplanes, cars, birds, cats,
deer, dogs, frogs, horses, ships, and trucks. There are
50000 training images and 10000 test images. The
training dataset is divided into validation (20%) and
training (80%) splits in our experiments. The test split
contains 1000 images from each class.
a
b
c
d
Figure 7: Relevance maps and logit values for the airplane
class. (a-b) correct classification, the error is small (rele-
vance maps can be trusted). (c) correct classification. How-
ever, the pixels used for the estimation are incorrect. (d)
incorrect classification, the error is large (the relevance map
cannot be trusted).
Table 3: Classification results for CIFAR 10 dataset, mean
accuracy, top-5 accuracy. The number of parameters for the
target models and Volterra network.
Model CNN TrCNN
Mean Accuracy 82.95 64.18
Top-5 Accuracy 99.12 96.26
Model Parameters 26.1M 507.2K
Volterra Parameters 1.4M 1.4M
Table 3 shows the classification mean accuracy
and top-5 accuracies. The accuracy for CNN (transfer
learning, Resnet-50) is lower than the typical value of
around 90%, obtained through a better learning rate
ICPRAM 2023 - 12th International Conference on Pattern Recognition Applications and Methods
602

a
b
c
d
Figure 8: Relevance maps and logit values for CNN model
and Volterra method for the CIFAR 10 dataset.
selection and a robust data augmentation strategy.
On the other hand, Alexey Dosovitskiy et al.
(Dosovitskiy et al., 2020) reported an accuracy of
99.5% achieved by pre-training the ViT model using
the JFT-300M dataset, then fine-tuning it on CIFAR
10, in our case, we trained the network from scratch,
obtaining only accuracy of 64.18% and top 5 accura-
cies of 96.26% as shown in Table 3.
Table 4 summarizes the logit errors.
Table 4: Mean absolute error of the logits on CIFAR 10
dataset, which expresses the difference between the models
and the Volterra approximation.
Model Train Validation Test
CNN 0.5040 1.3211 1.2521
TrCNN 0.2174 0.4119 0.3874
a
b
c
Figure 9: Relevance maps and logit values for the CNN
model and Volterra method for the class dog.
We employ the mean absolute error to measure the
difference between the models and the Volterra ap-
proximation logits, which have a dynamic range.
The CNN error is greater than the error for the Tr-
CNN, because it is more complex, with 26.1M param-
eters, while TrCNN has just 500K, and our Volterra
network uses only 1.4M.
Figure 7 shows relevance maps obtained using our
Volterra method for images of the airplane class. The
logit for the CNN, TrCNN, and the Volterra approxi-
mation method. The larger the logit value for the orig-
inal model, the more it indicates that the class is an
airplane. The prediction of the original model for Fig-
ure 7a and Figure 7b are correct. We can observe that
the pixels that belong to the planes are shown with
more intensity. Consequently, the model is paying at-
tention to the right place. Additionally, the Volterra
approximation error is small. Therefore, the relevance
map can be trusted. The classification of Figure 7c is
correct. However, the relevance map shows that the
classification is based on the shape of the black pixels
at the edge of the image, which are artifacts and possi-
bly correspond to the window from which the picture
was taken. The original model’s prediction in Figure
7d is incorrect. The logit corresponding to the class
airplane is negative, and the model classified it as a
Agnostic eXplainable Artificial Intelligence (XAI) Method Based on Volterra Series
603

Figure 10: Images of every tissue class from the stained
colorectal cancer histology dataset (Kather et al., 2016). (a)
tumour epithelium, (b) simple stroma, (c) complex stroma,
(d) immune cell conglomerates, (e) debris and mucus, (f)
mucosal glands, (g) adipose tissue, and (h) background.
ship. In this case, the highlighted pixels belong to the
background (sky and mountains). Fortunately, we can
use this large Volterra approximation error to indicate
that this relevance map should not be trusted.
Figure 8 shows additional relevance maps for cor-
rect predictions of images for the classes bird and
horse. Figure 8c and Figure 8d show that humans are
the most relevant pixels to classify these images as
horses. However, humans are not present in CIFAR-
10 as a class, but there are several images where they
ride horses, showing a clear example of bias.
Figure 9 shows tested images not from the dataset,
with humans and dogs. Dogs are correctly classified
if humans are next to them, as shown in Figure 9a.
However, when the humans are on top, the classifier
gets confused, and some are classified as horses. Fig-
ure 9b is correctly classified. However, the model is
almost equally confident that the image is a dog or a
horse with logit values equal to 4.08 and 3.10. Fig-
ure 9c is classified as a horse, which shows that the
relevance maps in Figure 9 classify were correct.
4.2.2 Colorectal Dataset
The H&E stained colorectal cancer histology dataset
(Kather et al., 2016) has eight different classes: tumor
epithelium, simple stroma, complex stroma (stroma
containing single tumor cells and/or single immune
cells), immune cell conglomerates, debris and mucus,
mucosal glands, adipose tissue, and background. The
images are tissue tiles at different scales ranging from
individual cells, with an approximate size of 10 µm,
e.g., Figure 10 (d) to larger structures such as mu-
Table 5: Accuracy on stained histology dataset (in Percent),
number of parameters for the target models and Volterra
network.
Model CNN TrCNN
Accuracy 92.4 79.20
Model Parameters 26.2M 703.6K
Volterra Parameters 6.4M 6.4M
Table 6: Mean absolute error (MAE) of the logits expresses
the difference between a target model and the Volterra ap-
proximation on the stained histology dataset.
Model Train Validation Test
CNN 0.0108 0.0636 0.0617
TrCNN 0.0114 0.1013 0.0992
cosal glands ≥ 50 µm, e.g., Figure 10 (f). The dataset
has 5000 images, 3200 were used for training, 800 for
validation, and 1000 for testing. The size of the RGB
images is 224 × 224. The models were trained from
scratch without data augmentation during 200 epochs.
Table 5 shows the mean accuracy and the number
of parameters for the CNN, TrCNN, and Volterra net-
work. The CNN accuracy is significantly higher than
the TrCNN model. Although TrCNN is a newer and
state-of-the-art model, this network is trained from
scratch and has no pre-training. The results for both
models can be improved using different strategies.
However, our interest is to analyze trained models.
Table 6 presents the approximation mean abso-
lute error, which measures the difference between the
models and their Volterra approximation. The errors
are low, and the validation and testing errors are in the
same range and not too far from the training error. We
use identical Volterra models even though the models
are different. The Volterra model can be more or less
complex if a more precise approximation is required.
However, the approximation found is satisfactory.
Figure 11 displays relevance maps obtained by
our method on the CNN and TrCNN. In most cases,
Volterra’s logit is remarkably close to the target
model. In the relevance maps, dark blue indicates
that the pixels are less influential, and red indicates
greater relevance. Background class, and some tis-
sues such as debris and mucus (see Figure 11d), con-
tain homogeneous texture, so the relevance is dis-
tributed throughout the image, except for some areas
in red that break homogeneity. Figure 11b and Fig-
ure 11c are opposite, while Figure 11b should high-
light only the stroma (note that the relevance map in-
dicates in blue that cells are being ignored). Figure
11c should omit the stroma and look at the immune
cell conglomerates.
ICPRAM 2023 - 12th International Conference on Pattern Recognition Applications and Methods
604
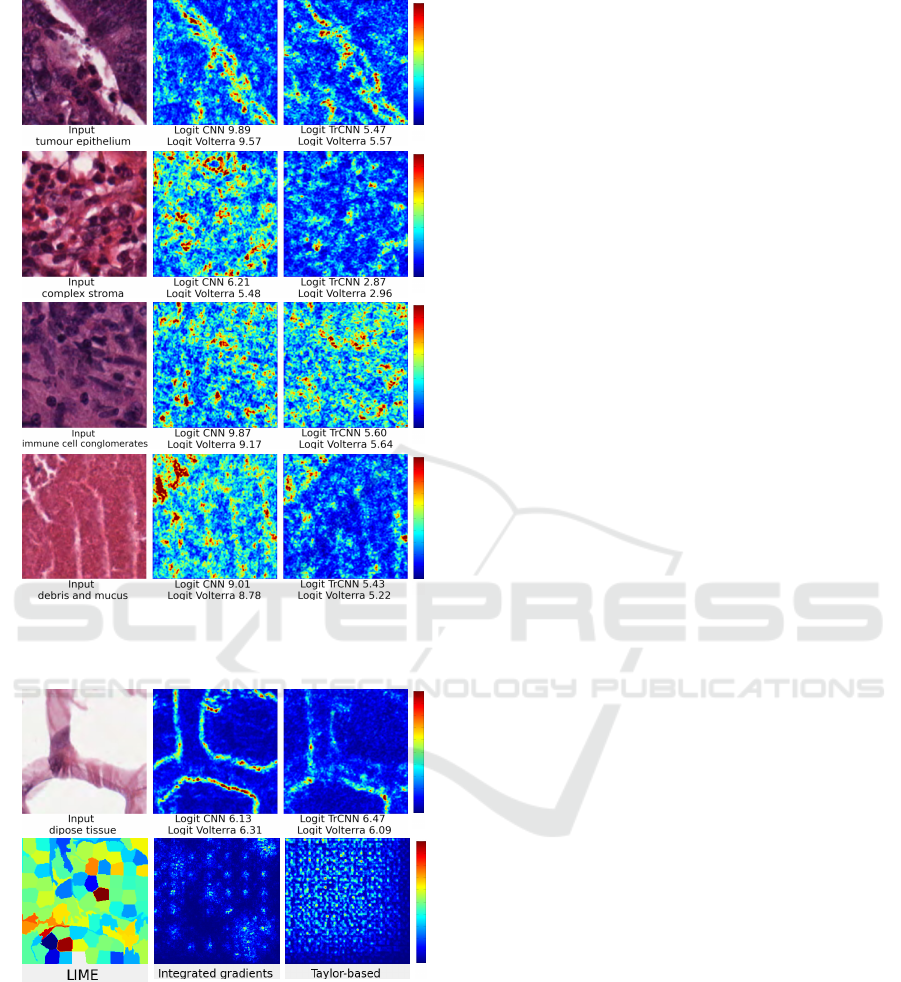
a
b
c
d
Figure 11: Histology dataset, relevance maps and logits (a)
tumour epithelium, (b) complex stroma, (c) immune cell
conglomerates, and (d) debris and mucus.
a
b
Figure 12: Histology dataset, explanations for adipose tis-
sue. (a) Input and Volterra relevance maps, (b) Comparison
with existing approaches.
Figure 12 compares the relevance map obtained
for our Volterra XAI method and other existing ap-
proaches for adipose tissue. Texture and color are
not relevant to our method for the lipid class (see Fig-
ure 12a). The edges of the lipids that form structures
and lines allow this class to be classified. In contrast,
LIME (Mishra et al., 2017) exhibits an output with
few details because it is based on the search for rele-
vant regions. The results obtained by integrated gra-
dients (Sundararajan et al., 2017) and Taylor-based
(Montavon et al., 2017) method are hardly inter-
pretable. Gradient methods describe pixel changes in
the model’s prediction and do not fully explain the
model prediction. Alternatively, Volterra relevance
maps can be used directly by more experienced users
(physicians) who can check the models for biases.
5 CONCLUSIONS
We propose an agnostic explainable artificial intelli-
gence method based on the Volterra series to approxi-
mate models, identify biases, and clarify model deci-
sions. The model architecture is composed of second-
order Volterra layers. To make fair comparisons from
our point of view, we used identical Volterra models
even though the target models were different. How-
ever, the Volterra model can be more or less com-
plex if a more precise approximation is required. Our
Volterra network allows us to create a simpler model
by emulating a target model. We evaluate the perfor-
mance of the emulation numerically by comparing a
target model’s prediction and the Volterra approxima-
tion. Therefore, no labels are required, and compa-
rable data can be employed even when training data
is unavailable. We generate relevance maps for Ra-
man spectra and 2D images. They explain the contri-
bution of the input elements to the prediction. The
trustworthiness of our method can be measured by
considering the error of the Volterra approximation.
We obtain low training errors for most of the models.
The validation and testing errors are in the same range
and not too far from the training error for the bacte-
ria dataset (TrCNN), histology dataset (CNN and Tr-
CNN), and CIFAR 10 (TrCNN). We present relevance
maps indicating higher and lower contributions to the
approximation prediction (logit) for commonly used
models. We identify biases in the models trained on
the CIFAR 10 dataset, which allows us to eliminate
them. Despite this does not seem transcendental for
the classification of simple classes, bias identification
is critical in the medial area.
ACKNOWLEDGEMENTS
This work is supported by the Ministry for Eco-
nomics, Sciences and Digital Society of Thuringia
(TMWWDG), under the framework of the Lan-
desprogramm ProDigital (DigLeben-5575/10-9) and
the Federal Ministry of Education and Research
Agnostic eXplainable Artificial Intelligence (XAI) Method Based on Volterra Series
605

of Germany (BMBF), funding program Photonics
Research Germany (FKZ: 13N15466, 13N15710,
13N15708) and is integrated into the Leibniz Cen-
ter for Photonics in Infection Research (LPI). The
LPI initiated by Leibniz-IPHT, Leibniz-HKI, UKJ
and FSU Jena is part of the BMBF national roadmap
for research infrastructures.
REFERENCES
Ali, N., Girnus, S., R
¨
osch, P., Popp, J., and Bocklitz, T.
(2018). Sample-size planning for multivariate data: a
raman-spectroscopy-based example. Analytical chem-
istry, 90(21):12485–12492.
Azpicueta-Ruiz, L. A., Zeller, M., Figueiras-Vidal, A. R.,
Arenas-Garcia, J., and Kellermann, W. (2010). Adap-
tive combination of volterra kernels and its application
to nonlinear acoustic echo cancellation. IEEE Trans-
actions on Audio, Speech, and Language Processing,
19(1):97–110.
Bach, S., Binder, A., Montavon, G., Klauschen, F., M
¨
uller,
K.-R., and Samek, W. (2015). On pixel-wise explana-
tions for non-linear classifier decisions by layer-wise
relevance propagation. PloS one, 10(7):e0130140.
Bocklitz, T. (2019). Understanding of non-linear parametric
regression and classification models: A taylor series
based approach. In ICPRAM, pages 874–880.
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn,
D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer,
M., Heigold, G., Gelly, S., et al. (2020). An image is
worth 16x16 words: Transformers for image recogni-
tion at scale. arXiv preprint arXiv:2010.11929.
Franz, M. O. and Sch
¨
olkopf, B. (2006). A unifying view
of wiener and volterra theory and polynomial kernel
regression. Neural computation, 18(12):3097–3118.
Halicek, M., Dormer, J. D., Little, J. V., Chen, A. Y., and
Fei, B. (2020). Tumor detection of the thyroid and
salivary glands using hyperspectral imaging and deep
learning. Biomedical Optics Express, 11(3):1383–
1400.
Kather, J. N., Weis, C.-A., Bianconi, F., Melchers, S. M.,
Schad, L. R., Gaiser, T., Marx, A., and Z
¨
ollner, F. G.
(2016). Multi-class texture analysis in colorectal can-
cer histology. Scientific reports, 6(1):1–11.
Khatri, C. and Rao, C. R. (1968). Solutions to some func-
tional equations and their applications to characteriza-
tion of probability distributions. Sankhy
¯
a: The Indian
Journal of Statistics, Series A, pages 167–180.
Korenberg, M. J. and Hunter, I. W. (1996). The identifi-
cation of nonlinear biological systems: Volterra ker-
nel approaches. Annals of biomedical engineering,
24(2):250–268.
Krizhevsky, A., Hinton, G., et al. (2009). Learning multiple
layers of features from tiny images.
Lin, H., Deng, F., Zhang, C., Zong, C., and Cheng, J.-X.
(2019). Deep learning spectroscopic stimulated raman
scattering microscopy. In Multiphoton Microscopy in
the Biomedical Sciences XIX, volume 10882, pages
207–214. SPIE.
Lundberg, S. M. and Lee, S.-I. (2017). A unified approach
to interpreting model predictions. Advances in neural
information processing systems, 30.
Mishra, S., Sturm, B. L., and Dixon, S. (2017). Local inter-
pretable model-agnostic explanations for music con-
tent analysis. In ISMIR, volume 53, pages 537–543.
Montavon, G., Lapuschkin, S., Binder, A., Samek, W., and
M
¨
uller, K.-R. (2017). Explaining nonlinear classifica-
tion decisions with deep taylor decomposition. Pat-
tern recognition, 65:211–222.
Niioka, H., Asatani, S., Yoshimura, A., Ohigashi, H.,
Tagawa, S., and Miyake, J. (2018). Classification of
c2c12 cells at differentiation by convolutional neural
network of deep learning using phase contrast images.
Human cell, 31(1):87–93.
Orcioni, S. (2014). Improving the approximation ability
of volterra series identified with a cross-correlation
method. Nonlinear Dynamics, 78(4):2861–2869.
Orcioni, S., Terenzi, A., Cecchi, S., Piazza, F., and Carini,
A. (2018). Identification of volterra models of tube au-
dio devices using multiple-variance method. Journal
of the Audio Engineering Society, 66(10):823–838.
Rodner, E., Bocklitz, T., von Eggeling, F., Ernst, G., Cher-
navskaia, O., Popp, J., Denzler, J., and Guntinas-
Lichius, O. (2019). Fully convolutional networks in
multimodal nonlinear microscopy images for auto-
mated detection of head and neck carcinoma: Pilot
study. Head & neck, 41(1):116–121.
Seber, G. A. (2008). A matrix handbook for statisticians.
John Wiley & Sons.
Simonyan, K., Vedaldi, A., and Zisserman, A. (2013).
Deep inside convolutional networks: Visualising im-
age classification models and saliency maps. arXiv
preprint arXiv:1312.6034.
Stegmayer, G. (2004). Volterra series and neural networks
to model an electronic device nonlinear behavior. In
2004 IEEE International Joint Conference on Neu-
ral Networks (IEEE Cat. No. 04CH37541), volume 4,
pages 2907–2910. IEEE.
Stegmayer, G., Pirola, M., Orengo, G., and Chiotti, O.
(2004). Towards a volterra series representation from
a neural network model. WSEAS Transactions on Sys-
tems, 3(2):432–437.
Sundararajan, M., Taly, A., and Yan, Q. (2017). Axiomatic
attribution for deep networks. In International confer-
ence on machine learning, pages 3319–3328. PMLR.
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones,
L., Gomez, A. N., Kaiser, Ł., and Polosukhin, I.
(2017). Attention is all you need. Advances in neural
information processing systems, 30.
ICPRAM 2023 - 12th International Conference on Pattern Recognition Applications and Methods
606
