
Redundancy and Novelty Between ECG Leads Based on Linear
Correlation
Utkars Jain
1 a
, Adam A. Butchy
1 b
, Michael T. Leasure
1 c
, Veronica A. Covalesky
2,3
,
Daniel McCormick
2,3
and Gary S. Mintz
4 d
1
Heart Input Output Inc., 128 N. Craig Street, Suite 406, Pittsburgh, U.S.A.
2
Cardiology Consultants of Philadelphia, Philadelphia, Pennsylvania, U.S.A.
3
Jefferson University Hospital, Philadelphia, Pennsylvania, U.S.A.
4
The Cardiovascular Research Foundation, New York, New York, U.S.A.
Keywords:
Statistics, ECGs, Cardiac, Correlation.
Abstract:
ECGs are a common diagnostic method for diagnosing cardiac pathologies. In this study, the Pearson correla-
tion coefficient is used to examine the latent linear correlations between the leads of a standard 12-lead ECG.
We utilize both the original ECG signals from the PTB-XL database and the reconstructed signal generated
by a deep learning model, ECGio. We find that leads III, aVL, V1, and V2 are, on average, the leads with the
most unique information due to their low correlation with other leads.
1 INTRODUCTION
Clinicians use a range of diagnostic techniques to de-
tect and identify heart and circulatory problems. Elec-
trocardiograms (ECGs), Troponin biomarker testing,
coronary computed tomography angiograms (CC-
TAs), magnetic resonance imaging (MRI), positron
emission tomography (PET), fractional flow reserve
(FFR), and coronary angiography are the most fre-
quently performed. These tests search for structural,
vascular, or electrical abnormalities in the cardiovas-
cular system of a patient (DeLaney et al., 2017). This
paper focuses on the noninvasive, ubiquitous, and in-
expensive electrocardiogram (ECG).
Invented in the 1870s, the ECG is a simple yet
effective monitoring device for the heart’s electrical
beats and rhythms (AlGhatrif and Lindsay, 2012).
The standard 12-lead ECG consists of nine electrodes
connected to specified locations on the patient’s body
and one electrode serving as an electrical ground. The
ECG machine then measures the voltage difference
between specific electrodes on the body to generate
waveforms that represent the building, release, and
refractory phases of the heart’s electrical cycle (Yang
a
https://orcid.org/0000-0002-1800-0768
b
https://orcid.org/0000-0002-0096-0031
c
https://orcid.org/0000-0002-1488-712X
d
https://orcid.org/0000-0003-3296-8705
et al., 2015). A bipolar electrode is created by sub-
tracting voltage of the ground electrode from the volt-
age of another electrode. Unipolar have a single pos-
itive electrode and utilize a combination of the other
electrodes to serve as the negative electrode (Derganc
and Gomi
ˇ
s
ˇ
cek, 2021). ECGs can be configured in a
variety of ways - all configurations differ based on the
placement, location, and number of leads.
The 12-lead resting ECG is the most common
(Harris, 2016) but several other types exist. Holter
ECGs may contain 2, 3, 6, or even 12 leads and are
used for continuous patient monitoring (DiMarco and
Philbrick, 1990). There exist more complicated de-
vices that use far more than 12 leads, and may even
take a full-body reading with a vest-like device (Wang
et al., 2019). Recent advancements in personal elec-
tronics have enabled the integration of 1-lead ECG
devices into smartwatches (Samol et al., 2019). In
table 1, we have a description of the 12 leads of the
normal resting ECG system.
In this paper, we analyze the linear correlation be-
tween ECG leads as a measure of information redun-
dancy using the standard 12-lead ECG and its asso-
ciated leads. We determine if there are specific leads
that carry less redundant information when compared
to others. Prior research investigated the use of AI to
more thoroughly analyze ECGs and how to use ECGs
with fewer than 12 leads to improve ECG signals (Jain
Jain, U., Butchy, A., Leasure, M., Covalesky, V., McCormick, D. and Mintz, G.
Redundancy and Novelty Between ECG Leads Based on Linear Correlation.
DOI: 10.5220/0011815700003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 4: BIOSIGNALS, pages 359-365
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
359

Table 1: The 12 leads, the location of the positive and neg-
ative electrodes, and which heart surface they are thought
to represent. “N” refers to neutral or electric ground. +
Location refers to the positive electrode location, while -
Location refers to the negative electrode location.
Lead + Location - Location Surface
I Left Arm Right Arm Lateral
II Left Leg Right Arm Inferior
III Left Leg Left Arm Inferior
aVR Right Arm N None
aVL Left Arm N Lateral
aVF Left Leg N Inferior
V1
Right side of sternum,
4th intercostal space
N Septum
V2
Left side of sternum,
4th intercostal space
N Septum
V3 Between V2 & V4 N Anterior
V4
Left midclavicular line,
5th intercostal place
N Anterior
V5 Left anterior axillary line N Lateral
V6 Left midaxillary line N Lateral
et al., 2022), we will use the methods of reconstruc-
tion to conclude if lead relationships are carried over.
The correlation between distinct sets of leads will de-
termine the redundancy and novelty of leads. Corre-
lation will be defined by the Pearson correlation coef-
ficient, also known as Pearson’s r.
2 RELATED WORKS
There have been other works that look into the cal-
culation or use of inter-lead relationships or corre-
lations between the lead of an ECG signals. Zhang
et. al set out to investigate a possible reason as to
why deep learning networks generalize pretty well in
the detection of left bundle brach block (LBBB) even
with small data sizes. They discovered that the cor-
relation between lead V1 and other leads was close
to 0, and a set correlation matrix can be indicative of
LBBB (Zhang et al., 2022). Jekova et. al used inter-
lead relationships in order to identify misplaced leads
or cable reversals resulting in a flipped signal. They
were able to build a robust algorithm that was able to
identify cable reversals based on inter-lead relation-
ships (Jekova et al., 2016).
3 METHODS
The Pearson correlation coefficient quantifies the lin-
ear correlation between two sets of data, providing a
measure of how closely a linear model would fit to
the data sets. An r value closer to 1 indicates a strong
positive correlation while a value closer to -1 indi-
cates a strong negative correlation. An r value of 0
indicates no correlation between the two datasets. A
generally accepted interpretation of the correlation is
that if there is a strong positive or negative correla-
tion that there is a redundancy in data (Schober et al.,
2018). (I.e one dataset is a scaled version of the other.)
If there is no correlation then there is novel informa-
tion contained between the two different datasets. A
visual example of the pearson correlation coefficient
based on scatter plot data is seen in figure 1. A vi-
sual example using ECG data from patient file 06275
is shown in 2.
The Pearson correlation coefficient, or r is defined
as:
r(y, ˆy) =
E[(y − µ
y
)( ˆy −µ
ˆy
)]
σ
y
σ
ˆy
(1)
Non-linear correlation cannot be measured by the
Pearson correlation coefficient. This indicates that
something may be highly uncorrelated in the linear
sense but correlated in the nonlinear sense. To main-
tain a solid basis for interpretation, nonlinear correla-
tions between leads fall outside the scope of this anal-
ysis.
We used 250 randomly selected patients from
PTB-XL, a large public electrocardiography dataset,
to conduct this study (Wagner et al., 2020). We chose
250 patients because this population served as the test
set for our reconstruction experiments and will now
serve as a good baseline to compare original signals
and reconstructed signals. These were a random sam-
ple of healthy and pathological ECGs. One in which
the original signal is present and one in which the sig-
nal has been reconstructed by a deep learning model
called ECGio (Leasure et al., 2021). In a previous
paper, the performance of ECGio in recreating ECGs
using the same 250 patients was demonstrated (Jain
et al., 2022). Maintaining patient consistency will
prevent bias and preserve the same testing set. Since
the deep learning model is expected to correct awry
signals, we will determine whether these correlations
are inherent to the ECGs or a result of noise.
The ECG signal input that was used to analyze
correlation was standardized. Each ECG signal was
clipped to represent 10 seconds of ECG time, with
the form NxM, where M was the number of leads
and N was the number of samples. Given that each
signal lasted just 10 seconds, N was also equal to
10 times the sampling rate. We resampled the sig-
nal using Fast Fourier Transform (FFT) to reduce the
sampling rate to 100Hz. This was done through the
scipy.signal.resample method
1
which assumes a pe-
riodic signal and transforms the signals to the fre-
1
docs.scipy.org/doc/scipy/reference/generated/scipy.sig
nal.resample.html
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
360
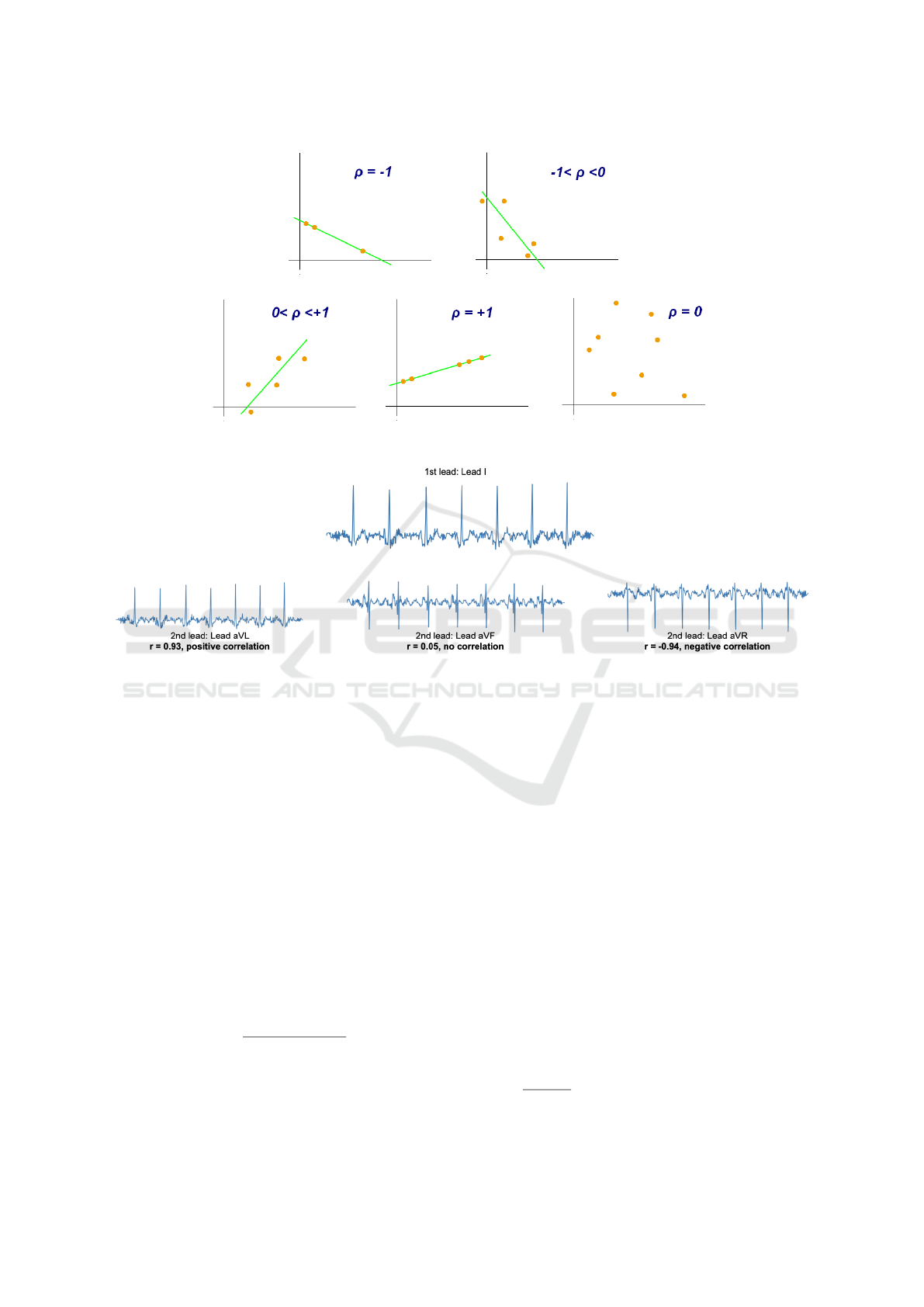
Figure 1: Visual example of the Pearson correlation coefficient based on scatter plot data. Illustration was provided by user
Kiatdd from Wikimedia Commons, under the Creative Commons Attribution-Share Alike 3.0 Unported license.
Figure 2: Visual example of the Pearson correlation coefficient based on ECG data. The initial comparative lead is lead I, and
it is compared to the unipolar leads. lead I is shown to have a strong positive correlation to lead aVR, no correlation with lead
aVF, and a strong negative correlation with lead aVL.
quency domain and removes unnecessary parts of the
domain and transforms it back. We employed a Band-
pass Butterworth filter with a passband beginning at
2Hz and ending at 40Hz to ensure that only informa-
tion within this band of frequencies was kept while
all others were discarded. Frequencies below 2Hz are
where breathing and muscle noise may interfere with
the signal (Li et al., 2017) and 40Hz is shown to be an
adequate cutoff (Ricciardi et al., 2016).
The databased ECG signals were obtained with
non-standardized minimum and maximum values in
millivolts (mV). Conforming to best practices in ma-
chine learning (Jo, 2019), we scaled each ten-second
ECG segment to values between [-1, +1] using equa-
tion 2, as is customary in deep learning to normalize
input signals:
f (x) = 2
x − min(x)
max(x) − min(x)
− 1 (2)
If x represented an ECG array in millivolts (mV),
max(x) represented the largest value along x, and
min(x) represented the minimum value along x. All
null values were transformed into zeros. Each signal
was eventually detrended to the point where the iso-
electric sections of the ECG were equal to zero.
The 12-lead standardized data served as the ba-
sis for calculating performance metrics. We did not
employ raw voltage difference values as the reference
to avoid a huge potential variance in the presence of
muscle, movement, or electric noise creating signal-
to-signal deviations. Mathematically, standardized
signals would provide a more equitable comparison,
whereas a direct comparison of performance indica-
tors between our outcomes and those of others may
not be prudent.
There are two primary methods to calculate the
Pearson correlation coefficient. One is that we loop
over every combination of the two leads in the ECG
(e.g. lead I against itself, lead I against lead II, etc.)
and apply the formula. This method would require
us to go through every combination for every ECG.
Meaning that in total there would be 66 calculations
12
2
=
12!
2!(12−2)!
for a 12-lead ECG. This is not an in-
surmountable number of calculations, but in the case
Redundancy and Novelty Between ECG Leads Based on Linear Correlation
361

for ECGs with many more leads, we would like a
more scalable solution. The second method is a vec-
torized approach, i.e. creating a correlation matrix, in
which there is a series of vector and matrix based cal-
culations, resulting in many less overall calculations.
A correlation matrix was established for each of
the 250 patients original and reconstructed ECGs.
Each of these matrices were layered to generate a
three dimensional tensor (size of 250x12x12). We did
three alternative sets of operations of the first axis of
this tensor: obtaining the mean, maximum, and mini-
mum. After these processes were finished, there were
6 separate correlation matrices. A mean, maximum,
and minimum correlation matrix for the original and
reconstructed signals. The reason why maximum and
minimum were selected was because we were under
the assumption that there would high levels of corre-
lations between any 2 leads for at least one patient -
we were looking to determine whether there were any
combinations in which their extremes broke the trend.
Algorithm 1: Algorithm for calculating correlation matrix.
Ensure: X− > Mx N ▷ X is the ECG matrix
X
∗
= X − µ(X) ▷ Subtract X by mean value in lead
axis
X
∗∗
= X
∗
· X
∗
▷ Matrix multiplication of X
∗
Z = σ(
p
diag(X
∗∗
)) ▷ Scaling based on
covariance
R =
X
∗∗
Z
▷ Correlation Matrix
4 RESULTS
The results of our two experiments are shown. In
figures 3, 4, 5, 6, 7, and 8 we have heatmaps show-
ing the cross correlation between all of the standard
ECG leads for the original patient ECGs and the re-
Figure 3: The figure below shows a heatmap of the mean
correlation matrix over the population of patients examined
with the original signals.
Figure 4: The figure below shows a heatmap of the maxi-
mum correlation matrix over the population of patients ex-
amined with the original signals.
Figure 5: The figure below shows a heatmap of the mini-
mum correlation matrix over the population of patients ex-
amined with the original signals.
constructed ones. Those that are more correlated have
a darker color while the ones that are more uncorre-
lated are a lighter color. In tables 2 and 3 we see the
mean, maximum, and minimum values for each lead
when analyzing their correlations with other leads for
the original ECGs and their reconstructions. In ta-
ble 4 we examine the differences between the maxi-
mum and minimum correlation matrices for the orig-
inal ECGs and their reconstructions.
5 DISCUSSION
The Pearson correlation coefficient is a prevalent ap-
proach for analyzing the linear relationship between
continuous variables (Benesty et al., 2009). It can
provide a great deal of broad information about vari-
ables with a single value. However, it should be high-
lighted that this is not a foolproof method for deter-
mining the correlation between signals or variables.
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
362

Figure 6: The figure below shows a heatmap of the mean
correlation matrix over the population of patients examined
with the reconstructed signals.
Figure 7: The figure above shows a heatmap of the maxi-
mum correlation matrix over the population of patients ex-
amined with the reconstructed signals.
Figure 8: The figure above shows a heatmap of the mini-
mum correlation matrix over the population of patients ex-
amined with the reconstructed signals.
We are simply evaluating the linear correlation of the
signal. Essentially, we wish to determine whether
the signals fluctuate synchronously. Do they rise or
Table 2: The table below shows the mean, maximum, and
minimum correlations per lead when compared to other
leads (sans the lead itself), when analyzing the mean cor-
relation matrix from the 250 patients. Leads that shown to
have significant degrees of uncorrelation are denoted with
an *.
Lead Mean r Max r Min r
I 0.205 0.832 -0.645
II 0.290 0.846 -0.507
III* 0.033 0.738 -0.272
aVR -0.447 0.636 -0.903
aVL* 0.057 0.787 -0.516
aVF 0.233 0.828 -0.245
V1* -0.128 0.714 -0.645
V2* 0.104 0.734 -0.163
V3 0.299 0.749 0.047
V4 0.359 0.862 -0.183
V5 0.319 0.920 -0.483
V6 0.254 0.920 -0.658
Table 3: The table below shows the mean, maximum, and
minimum correlations per lead when compared to other
leads (sans the lead itself), when analyzing the mean cor-
relation matrix from reconstructions of the 250 patients.
Leads that shown to have significant degrees of uncorrela-
tion are denoted with an *.
Lead Mean r Max r Min r
I 0.220 0.859 -0.863
II 0.291 0.867 -0.884
III* 0.050 0.687 -0.620
aVR -0.450 0.649 -0.937
aVL* 0.065 0.732 -0.620
aVF 0.241 0.804 -0.579
V1* -0.152 0.710 -0.707
V2* 0.102 0.739 -0.250
V3 0.299 0.749 -0.326
V4 0.362 0.850 -0.712
V5 0.318 0.930 -0.899
V6 0.256 0.930 -0.937
decrease simultaneously? All other connections are
considered uncorrelated.
The Pearson correlation coefficient does not ac-
count for the magnitude of differences and is highly
prone to outliers (Kim et al., 2015). For instance,
there may be a large number of points centered about
the origin and only a few along the x-axis. The Pear-
son correlation coefficient will depend heavily on how
these few points behave. We attempted to preprocess
and standardize the data in order to make fair com-
parisons, but this does not ensure the elimination of
outliers.
In a typical 12-lead ECG setup there are 2-3 limb
leads, leads attached to the arms or legs, and pre-
cordial (or chest) leads, leads attached to the chest.
Redundancy and Novelty Between ECG Leads Based on Linear Correlation
363

Table 4: The table below shows the mean correlations per
lead when compared to other leads (sans the lead itself),
when analyzing the maximum and minimum correlation
matrices from both the original 250 patients and the recon-
structed ECGs.
Lead
Original
Min
Original
Max
Reconstructed
Min
Reconstructed
Max
I -0.857 0.926 -0.904 0.940
II -0.786 0.866 -0.816 0.901
III -0.897 0.948 -0.940 0.961
aVR -0.954 0.695 -0.969 0.710
aVL -0.917 0.939 -0.957 0.964
aVF -0.827 0.954 -0.859 0.964
V1 -0.929 0.903 -0.950 0.921
V2 -0.913 0.942 -0.924 0.960
V3 -0.895 0.974 -0.929 0.982
V4 -0.890 0.984 -0.937 0.985
V5 -0.828 0.939 -0.810 0.932
V6 -0.759 0.856 -0.773 0.855
The idea behind this setup is that limb leads provided
information about the electrical propagation along a
longer axis, while the chest leads provide information
that is closer to the heart (Dower et al., 1990). These
2 sets of information combined can lead to a more
holistic interpretation of the heart’s electrical activity.
The unipolar leads (aVL, aVF, aVR) are constructed
through a linear combination of the limb leads (Ma-
dias, 2008).
Lead III, which can occasionally be created from
leads I and II (lead III = lead II - lead I), appears to
be linearly uncorrelated with the mean of the original
patients. This is unexpected because it can be viewed
as a linear combination of leads I and II, yet there ap-
pears to be novel information in terms of correlation.
Because lead I, II, and III are based upon a trigono-
metric relationship, it’s possible that correlations be-
tween them are not fully captured looking at linear
relationships.
As seen in figures 3 and 6, the precordial leads, on
average, seemed to be relatively uncorrelated when
compared against the limb leads and the unipolar
leads. This would be in line with the thought process
that precordial leads are assessing different surfaces
of the heart, and thus yield different information.
An intriguing phenomena in the maximum and
minimum correlation of the original patients is that
there are several correlations between leads that are
close to +1 or -1. Even more unexpectedly, in our
dataset, some correlations remained near 0 even when
searching for the maximum or minimum value.
For instance, when searching for the maximal cor-
relations, V6 has a correlation value of -0.14 when
compared to aVR. This implies that, across the tested
population, there was little correlation between these
two leads, but V6 showed the strong correlation with
other unipolar leads. aVR is a linear combination of
leads I and II, but its combination may contain infor-
mation that differs significantly from what was pre-
viously believed (Williamson et al., 2006). Another
similar case is between lead V6 and lead II, when
looking for the minimum correlation case the value is
-0.05. There may be an obvious explanation for unex-
pectedly significant correlations, namely that the lead
was improperly positioned and hence captured signals
that heavily overlapped with the domain of another
lead for a particular ECG of a patient.
According to tables 2 and 3, leads III, aVL, V1,
and V2 had the lowest average correlation. Using a
threshold of 0.2, it was possible to determine which
leads were uncorrelated. These leads offer informa-
tion regarding the inferior, lateral, and septal sur-
faces of the heart (both lead V1, V2 offer informa-
tion about the septal surface), which represent three
of the heart’s four primary surfaces (lateral, inferior,
septum, and anterior). Possible interpretation: these
leads contain the most novel information regarding
these surfaces. In the future, it may be necessary to
determine if the novel information contained in these
leads facilitates better levels of ECG reconstruction
performance.
Our analysis uncovered the intriguing conclusion
that the ECG reconstruction data closely resemble the
original data. The goal of comparing the original cor-
relation to that of the reconstruction was to determine
if these correlations were intrinsic to the ECG itself
and not the result of noise or lead placement, which
the deep learning model is designed to correct.
Based on the similar levels of correlation, it may
be concluded that a 12-lead ECG contains a latent
structure that is known during model training. It is
feasible that the model is just learning to match the
output as closely as possible, without learning any in-
trinsic structure. In the future, it may be necessary
to determine whether or not the model attempts to
comprehend the ECG’s underlying structure when re-
building the input.
6 CONCLUSION
In this study, we examined the average lead corre-
lations of 250 patient ECG signals from the phys-
ionet PTB-XL database. We studied the original sig-
nals while they were being normalized and when they
were being reconstructed using a novel deep learning
technique. The mean, maximum, and minimum cor-
relations between the leads of a 12-lead ECG were
studied. Leads III, aVL, V1, and V2 were identified
as the most uncorrelated in the ECG system.
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
364

REFERENCES
AlGhatrif, M. and Lindsay, J. (2012). A brief review: his-
tory to understand fundamentals of electrocardiogra-
phy. Journal of community hospital internal medicine
perspectives, 2(1):14383.
Benesty, J., Chen, J., Huang, Y., and Cohen, I. (2009).
Pearson correlation coefficient. In Noise reduction in
speech processing, pages 1–4. Springer.
DeLaney, M. C., Neth, M., and Thomas, J. J. (2017). Chest
pain triage: Current trends in the emergency depart-
ments in the united states. Journal of Nuclear Cardi-
ology, 24(6):2004–2011.
Derganc, J. and Gomi
ˇ
s
ˇ
cek, G. (2021). Teaching the basic
principles of electrocardiography experimentally. Ad-
vances in Physiology Education, 45(1):5–9.
DiMarco, J. P. and Philbrick, J. T. (1990). Use of ambula-
tory electrocardiographic (holter) monitoring. Annals
of internal medicine, 113(1):53–68.
Dower, G., Nazzal, S., Bullington, D., Pahlm, O.,
Haistey Jr, W., Marriott, H., and Bullington, R.
(1990). Limb leads of the electrocardiogram: se-
quencing revisited. Clinical cardiology, 13(5):346–
348.
Harris, P. R. (2016). The normal electrocardiogram: Rest-
ing 12-lead and electrocardiogram monitoring in the
hospital. Critical Care Nursing Clinics, 28(3):281–
296.
Jain, U., Butchy, A. A., Leasure, M., Covalesky, V. A., Mc-
Cormick, D., and Mintz, G. H. (2022). 12-lead ecg re-
construction via combinatoric inclusion of fewer stan-
dard ecg leads with implications for lead information
and significance. In BIOSIGNALS.
Jekova, I., Krasteva, V., Leber, R., Schmid, R., Tweren-
bold, R., M
¨
uller, C., Reichlin, T., and Ab
¨
acherli, R.
(2016). Inter-lead correlation analysis for automated
detection of cable reversals in 12/16-lead ECG. Com-
puter Methods and Programs in Biomedicine, 134:31–
41.
Jo, J.-M. (2019). Effectiveness of normalization pre-
processing of big data to the machine learning perfor-
mance. The Journal of the Korea institute of electronic
communication sciences, 14(3):547–552.
Kim, Y., Kim, T.-H., and Erg
¨
un, T. (2015). The instabil-
ity of the pearson correlation coefficient in the pres-
ence of coincidental outliers. Finance Research Let-
ters, 13:243–257.
Leasure, M., Jain, U., Butchy, A., Otten, J., Covalesky,
V. A., McCormick, D., and Mintz, G. S. (2021).
Deep learning algorithm predicts angiographic coro-
nary artery disease in stable patients using only a stan-
dard 12-lead electrocardiogram. Canadian Journal of
Cardiology.
Li, J., Deng, G., Wei, W., Wang, H., and Ming, Z. (2017).
Design of a real-time ECG filter for portable mobile
medical systems. IEEE Access, 5:696–704.
Madias, J. E. (2008). On recording the unipolar ecg limb
leads via the wilson’s vs the goldberger’s terminals:
avr, avl, and avf revisited. Indian Pacing and Electro-
physiology Journal, 8(4):292.
Ricciardi, D., Cavallari, I., Creta, A., Giovanni, G. D., Cal-
abrese, V., Belardino, N. D., Mega, S., Colaiori, I.,
Ragni, L., Proscia, C., Nenna, A., and Sciascio, G. D.
(2016). Impact of the high-frequency cutoff of band-
pass filtering on ECG quality and clinical interpreta-
tion: A comparison between 40hz and 150hz cutoff
in a surgical preoperative adult outpatient population.
Journal of Electrocardiology, 49(5):691–695.
Samol, A., Bischof, K., Luani, B., Pascut, D., Wiemer, M.,
and Kaese, S. (2019). Single-lead ecg recordings in-
cluding einthoven and wilson leads by a smartwatch:
a new era of patient directed early ecg differential di-
agnosis of cardiac diseases? Sensors, 19(20):4377.
Schober, P., Boer, C., and Schwarte, L. A. (2018). Corre-
lation coefficients: appropriate use and interpretation.
Anesthesia & Analgesia, 126(5):1763–1768.
Wagner, P., Strodthoff, N., Bousseljot, R.-D., Samek, W.,
and Schaeffter, T. (2020). Ptb-xl, a large publicly
available electrocardiography dataset.
Wang, L.-H., Zhang, W., Guan, M.-H., Jiang, S.-Y.,
Fan, M.-H., Abu, P. A. R., Chen, C.-A., and Chen,
S.-L. (2019). A low-power high-data-transmission
multi-lead ecg acquisition sensor system. Sensors,
19(22):4996.
Williamson, K., Mattu, A., Plautz, C. U., Binder, A., and
Brady, W. J. (2006). Electrocardiographic applica-
tions of lead avr. The American journal of emergency
medicine, 24(7):864–874.
Yang, X.-L., Liu, G.-Z., Tong, Y.-H., Yan, H., Xu, Z., Chen,
Q., Liu, X., Zhang, H.-H., Wang, H.-B., and Tan, S.-
H. (2015). The history, hotspots, and trends of elec-
trocardiogram. Journal of geriatric cardiology: JGC,
12(4):448.
Zhang, C., Li, J., Pang, S., Xu, F., and Zhou, S. (2022).
A 12-lead ECG correlation network model explor-
ing the inter-lead relationships. Europhysics Letters,
140(3):31001.
Redundancy and Novelty Between ECG Leads Based on Linear Correlation
365
