
Foreground Extraction in Histo-Pathological Image by Combining
Mathematical Morphology Operations and U-Net
Jia Li
1,3 a
, Junling He
2
, Jingmin Long
1
, Chenxu Wang
3 b
, Jesper Kers
2 c
and Fons J. Verbeek
1 d
1
Leiden Institute of Advanced Computer Science, Leiden University, Leiden, The Netherlands
2
Leiden University Medical Center, Leiden, The Netherlands
3
School of Computer Science, Xi’an Jiaotong University, Beilin, Xi’an, China
Keywords:
Tissue Segmentation, Foreground Extraction, U-Net, Whole Slide Image.
Abstract:
In recent years, computational pathology is rapidly developing. This resulted in various artificial intelligence
approaches that have been proposed and applied to images common to the pathology practice, i.e. Whole Slide
Images. It is very important to pre-process these images for a deep learning classifier because they are simply
too large to feed into such a network. In order to get useful information from these images, we propose a new
background removal method for the extracted Regions Of Interest in these images. We combine traditional
morphology image operators and a U-Net framework. Firstly, we pre-process the images by using Contrast
Limited Adaptive Histogram Equalization and thresholding. Then we predict the mask by using pre-trained
U-Net weights. Finally, we use morphological opening and propagation operators on the predicted mask to
refine the masks. The experiments based on different types of staining (H&E, PAS, and JONES silver) show
the effectiveness of our method compared to 3 state-of-the-art models.
1 INTRODUCTION
Pathological examination of biopsies is an important
method for clinical diagnosis and plays a crucial role
in the diagnostic process. Over the past decade, dig-
ital pathology has become one of the main directions
for development in pathology (Li et al., 2022). The
introduction and use of digital slide scanner systems
provide high-resolution whole slide images (WSIs)
which are obtained from the traditional pathologi-
cal slides resulting in image sizes in gigabyte order.
Digitization of pathological slides contributes a lot
to the preservation, sharing, and analysis of patho-
logical information. On the basis of WSIs, an au-
tomated or computer-assisted diagnosis comes into
reach. This requires that dedicated pattern recogni-
tion systems need to be developed and, consequently,
the WSIs need to be prepared for these pattern recog-
nition procedures. At present, pattern recognition
methods are based on, so-called, deep learning sys-
tems. So, WSIs can be used in computational ap-
a
https://orcid.org/0000-0001-8842-1042
b
https://orcid.org/0000-0002-9539-5046
c
https://orcid.org/0000-0002-2418-5279
d
https://orcid.org/0000-0003-2445-8158
proaches to recognize certain pathologies in these im-
ages (Neuner et al., 2021). Pattern recognition can as-
sure efficient and accurate pathological assessment of
diseases. Although the computer-aided diagnosis of
histo-pathological images gains critical acclaim for its
accuracy, stability and efficiency, still, the quality of
the histo-pathological images has posed various chal-
lenges to those proposed techniques. We will discuss
some of these limitations in terms of slide preparation
and computational preparation. In this paper, we em-
ploy histo-pathological images of the kidney, in par-
ticular by investigating biopsies taken from patients
with a kidney transplant.
There are several staining methods for tissues
commonly used in kidney pathology; haematoxylin
and eosin (H&E) is the most frequently used staining
technique. For kidney transplants, also the Periodic
Acid Schiff (PAS) and the JONES silver staining are
used. Examples of these three different staining meth-
ods are depicted in Figure 1. These images are typical
examples for kidney biopsies.
As mentioned, the WSIs are large and have a high
resolution, this complicates the use of the image as
a whole for pattern recognition; i.e., computer mem-
ory is still limited. Therefore, it is important to first
find where slide of the biopsy and thus on the WSI the
146
Li, J., He, J., Long, J., Wang, C., Kers, J. and Verbeek, F.
Foreground Extraction in Histo-Pathological Image by Combining Mathematical Morphology Operations and U-Net.
DOI: 10.5220/0011803500003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 2: BIOIMAGING, pages 146-153
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

relevant information is.. Once the locations of infor-
mation are found, just the tissue information on those
locations needs to be extracted. Only this part of the
WSI is considered relevant for further processing.
The relevant information on the biopsy WSI is re-
ferred to as the foreground, and this part is used to find
the patterns. Most often there are a number biopsy
sections mounted on one slide. The first step is to
identify the regions where the sections are. The empty
background, i.e. the part of the slide where no sec-
tions are mounted, is often indicated with one color,
this needs to be set to zero first; an example of this
can be seen in Figure 2a.
Existing methods for foreground extraction for
WSIs are mostly focused on H&E staining (Riasa-
tian et al., 2020). In kidney transplant biopsies, how-
ever, also the PAS staining is important for histo-
pathological analysis. Foreground extraction of PAS
stained WSI can be complicated as the staining fades
over time resulting in a low contrast image. In gen-
eral, the stained tissue in a WSI should be evaluated
for fading, artifact, dirt, and low contrast. In Figure 2b
and 2c some typical examples are depicted.
In order to analyse the information in the tissue,
all tissue areas need to be identified. The analysis
consists of building a classifier for the pathological
state of the tissue. Therefore it is important to only
use relevant, i.e. tissue, information in the classifica-
tion. Therefore, the WSI needs to be pre-processed to
that this information can be submitted to the classifi-
cation system. We focus on getting this information
from the WSI.
Once the tissue regions in the WSI are identified,
the tissues themselves need to be identified as fore-
ground. To this end, a segmentation procedure is ap-
plied. Therefore, the next step of histo-pathological
image analysis is tissue segmentation (Khened et al.,
2021).
The classification of the foreground parts is ac-
complished using a deep neural network. The com-
putational task is facilitated trough the use of graphic
processing units (GPUs). However, the memory of
GPUs has limitations. Consequently, the common ap-
proach is to divide the image into smaller patches.
These patches are then used to train the deep neural
network.
So, the preparation of the WSIs for the training
of a classifier requires constructing “patches” for the
relevant areas with tissue. Once, these areas are es-
tablished and “clean”, the patching is the last step for
the preparation. This needs to be done in such a man-
ner that the patches contain useful information for the
training of a classification. Therefore our work aims
to pre-process the WSI in such a manner that first
the regions are established where sections are on the
slide, and subsequently process the tissue area in each
of the regions such that a binary mask is obtained that
can be used for the construction of patches that con-
tain relevant tissue information.
(a) H&E (b) JONES (c) PAS
Figure 1: Different types of staining.
(a) Empty area (b) fade staining (c) dirty staining
Figure 2: Some challenging examples for tissue segmenta-
tion.
In our approach, the empty area, cf. Figure 2a, of
the WSI is set to zero through a simple thresholding
operation. Next, the tissue part of the WSI needs to be
assessed. In order to deal with low contrast in the im-
age, image enhancement is used. This is typically the
case for staining that is fading over time. We assess
the contrast distribution in the image and enhance the
contrast through a redistribution of the intensity val-
ues. For our approach we use Contrast Limited Adap-
tive Histogram Equalization (CLAHE) (Pizer et al.,
1990) for low-contrast images. This enhancement
method operates on a local assessment of the intensity
distributions and combines well with the subdivision
of patches later in the process. Artifacts and dirt on
the slides are often seen as tissue by the segmentation
procedure. In order to remove these artifacts we use
mathematical morphology operators. The combina-
tion of these procedures will result in areas that are
suitable for consistent and robust patching.
Further, based on results from the literature, we
propose a new method that combines mathematical
morphology operations with a U-Net deep learning
structure. We first assess the contrast distribution,
then we use thresholding to remove the empty parts
Foreground Extraction in Histo-Pathological Image by Combining Mathematical Morphology Operations and U-Net
147

from the analysis. Next, we use the MobileNet neu-
ral network (Howard et al., 2017) as the U-Net en-
coder backbone to predict the tissue foreground ar-
eas. This neural network was pre-trained on The Can-
cer Genome Atlas (TCGA) datasets
1
. Next, we post-
process the initial masking area through mathematical
morphology operations. The resulting mask is then
used to create the patches for the classifier.
The main contributions of this paper are: (1). A
better solution for the extraction of the relevant fore-
ground from the WSI. (2). Development of a new
generic procedure for the pre-processing of kidney
biopsy images. (3) Comparison with three state-of-
the-art methods (Otsu, MobileNet, and EfficientNet-
B3.) on 7 typical images with two binary evaluation
indexes.
The remainder of this paper is organized as fol-
lows: in section 2, several existing approaches related
to our algorithm are presented. Then, in section 3, we
will introduce our method. Section 4 provides the ex-
periment results. Finally, we present our conclusions
in section 5.
2 RELATED WORK
For the processing of WSIs There are two categories
related to our work: background removal and whole
slide image processing.
2.1 Background Removal
In tissue segmentation tasks, background removal can
refer to different approaches. One is to correct for
uneven illumination in the background. And elabo-
rate methods are available for this (Cai and Verbeek,
2015). For WSIs background correction refers to the
removal of non-object parts in the image. This of-
ten uses a segmentation method and the crux is to
find the right threshold value(s). Here we see two
categories, one based on traditional machine learn-
ing algorithms and the other based on a deep learn-
ing method. The traditional approaches entail re-
gion growing, the watershed-based method, and Otsu
thresholding (Otsu, 1979). The main idea of the
threshold-based algorithm is to compute an optimum.
In a deep learning approach, patching over the image
is applied to find local optima.
By dividing the section parts of the WSI, aka the
regions of interest (ROI), into small patches, a deep
neural network for the segmentation can be used. This
1
Pretrained model available at: https://kimialab.uwater
loo.ca/kimia/index.php/data-and-code/
works by predicting a label for each of the pixels in a
patch. The label denotes whether it is foreground or
not. Next, the segmented patches are stitched back to
the overall images. There are several neural network
models for image segmentation (Sultana et al., 2020),
such as FCN (Fully Convolutional Network) (Long
et al., 2015), U-Net (Ronneberger et al., 2015), and
Mask R-CNN (He et al., 2017). Riasatian et al. com-
pared different U-Net topologies for background re-
moval in histo-pathological images (Riasatian et al.,
2020). By training on different backbones in their
experiments, they have shown that MobileNet and
EfficientNet-B3 (Tan and Le, 2019) perform better
than the others.
2.2 Whole Slide Image Pre-Processing
Chen et al. proposed a tissue localization pipeline
to process WSIs (Chen and Yang, 2019). They use
thresholding on grayscale images followed by filling
the holes, which works well on H&E staining. Neuner
et al. developed an open-source library to process
WSIs, which helps the training and evaluation task for
classification (Neuner et al., 2021). The general pro-
cedure of their software consists of several steps: ROI
definition, tile filtering, tile extraction, and tile collec-
tion. After this procedure, we can get a batch of tiles,
which can be directly used for downstream tasks.
They employed several kinds of filters to segment the
background and foreground. Clustering-constrained
Attention Multiple Instance Learning (CLAM) is an-
other deep-learning-based method that uses attention-
based learning to classify the WSIs (Lu et al., 2021).
In CLAM, thresholding is used on the saturation
channel after blurring the image with a median filter.
In addition, morphological operators are used to fill
the small holes.
3 METHOD
Our method consists of three main steps: image pre-
processing by contrast assessment and thresholding,
MobileNet mask prediction using pretrained weights,
and mask post-processing using propagation and mor-
phological opening. The workflow of our method is
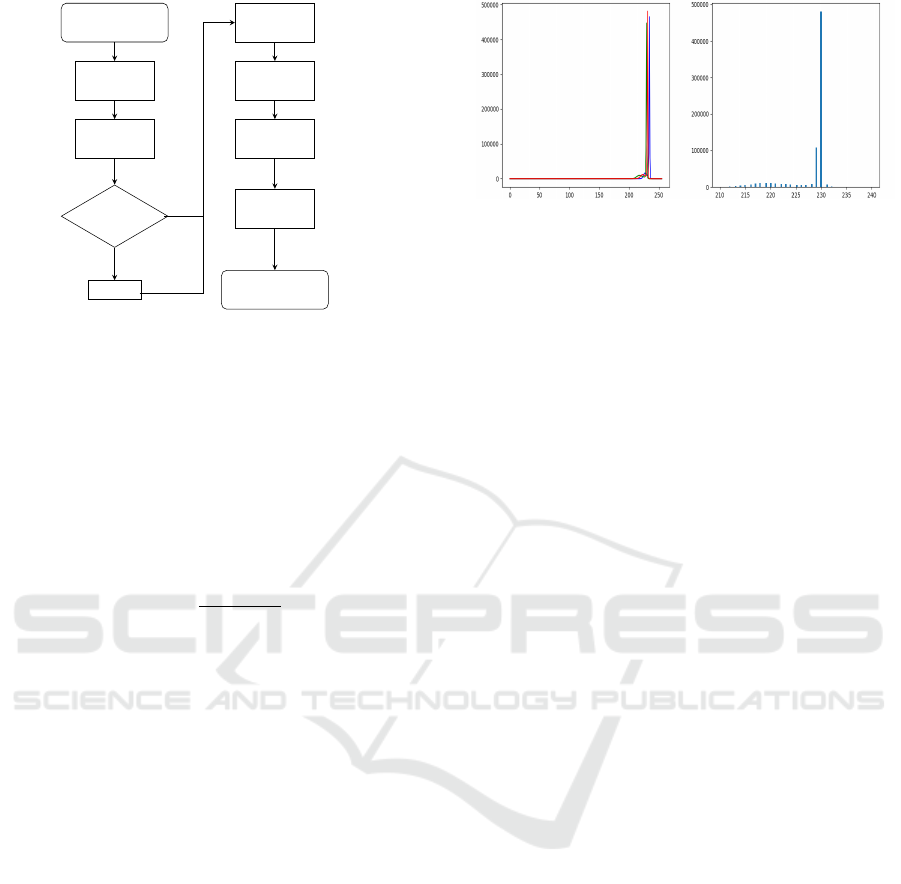
shown in Figure 3.
3.1 Image Pre-Processing
Since there is often some debris on the slide as well as
color coding of non-section areas in the WSIs, we use
thresholding to remove the very dark and very light
BIOIMAGING 2023 - 10th International Conference on Bioimaging
148
Original WSI
Extract ROI
Distribution
Assessment
Low Contrast
CLAHE
Thresholding
MobileNet
Propagation
Opening
Binary Mask
yes
no
Figure 3: Flow chart of our method.
areas. First, we convert the image to a grayscale im-
age and then assess the contrast, i.e. low or sufficient.
The grayscale image is calculated using this conver-
sion:
Y = 0.299 ∗ R + 0.587 ∗ G + 0.114 ∗ B (1)
Where R,G,B is the intensity value of red, green, and
blue channels. From the resulting intensity image, we
calculate the contrast by using:
C
M
=
I
max
− I
min
I
f ull
(2)
where I
max
and I
min
is the maximum and minimum
intensity value in the image respectively. I
f ull
repre-
sents the dynamic range for the given image type; typ-
ical, for an 8-bit image, this is [0, 255]. If the image
contrast is lower than a given value, it is considered
a low-contrast image (see Figure 4 for a histogram
example from the source image in Figure 2(b)). For
this image, the max gray value is 241, and the min
gray value is 80. As we can see, the value above 231
and below 214 are no more than 1%. In this case, the
I
max
is 231, and I
min
is 214. I
f ull
is 256. So the contrast
value would be 0.066; meaning that only 6.6% of the
dynamic range is used. We consider this image to be
of low contrast and we employ CLAHE as an image
enhancement method on this image. If it is not the
low-contrast image, we directly proceed to threshold-
ing.
In the thresholding step, we set the pixels whose
intensity values are below 70 and above 230 to be
255 (from empirical assessments). By doing this, the
pixels with values beyond this interval are viewed as
background by the neural network. Through image
enhancement and thresholding, some of the noise is
removed. After thresholding, we will get an image
that keeps almost all of the tissues and contains less
background.
(a) (b)
Figure 4: Histogram of low contrast image (Figure 2(b)) (a)
RGB histogram of the image. (b) Zoom in to the highest
frequency gray values.
3.2 U-Net Architecture
Image segmentation tasks can be accomplished in
many ways. In order to further classify the image con-
tent, we need to create masks over the regions where
the tissues are found. Following recent accomplish-
ments in WSI segmentation, we invoke a deep learn-
ing strategy for the segmentation. As a widely used
convolutional neural network model, U-Net works
well for image segmentation tasks, especially for
biomedical microscopy images (Ronneberger et al.,
2015). There are several different backbones that we
can choose for the tissue segmentation task. Accord-
ing to experiments in previous work (Riasatian et al.,
2020), the MobileNet works better than the others.
The main idea of MobileNet is the depth-wise separa-
ble convolution, with which the number of parameters
can be reduced. Therefore, we choose the MobileNet
as the backbone of U-Net, and we use the default set-
tings (the patch size is 400) to produce the (initial)
mask.
3.3 Post-Processing
After the initial prediction of U-Net, we get the mask
with small holes both in the tissue are and on the bor-
der of the tissue area. To fill the holes in the tissue,
we process the mask using image propagation. Image
propagation is to fill the holes in the overall mask. As
for small concavities on the border of the tissue, we
use an opening with a rectangular structuring element,
size 20x20, to fill in the small concavities. Finally, in
this manner. we obtain a binary mask for each of the
ROIs.
Given the generated mask, we could keep only tis-
sues by combining it with the original image using
logic AND operation. Subsequently, we generate the
image patches for the neural network classifier from
the masked area. A simple patch example is shown in
Figure 5. The green line represents the overall mask
outline. We generate 256*256 sized patches. As an
Foreground Extraction in Histo-Pathological Image by Combining Mathematical Morphology Operations and U-Net
149

Table 1: Similarity measure between the masks from our method and the others.
Image
binary correlation binary overlap
EfficientNet MobileNet Otsu EfficientNet MobileNet Otsu
I
1
0.9864 0.9817 0.8592 0.9919 0.9890 0.9055
I
2
0.7178 0.7967 0.4940 0.7476 0.8258 0.5733
I
3
0.9060 0.6395 0.8753 0.9184 0.6215 0.8915
I
4
0.9217 0.3483 0.1447 0.9269 0.3363 0.0387
I
5
0.9661 0.9782 0.9108 0.9703 0.9810 0.9199
I
6
0.9881 0.9923 0.0335 0.9900 0.9935 0.0230
I
7
0.9902 0.9860 0.6770 0.9919 0.9885 0.7365
extra heuristic, we establish if there is sufficient infor-
mation in the patch for the training. This is to prevent
the classifier to train on the background. In a patch,
the tissue should have at least an area (256*256)/2
pixels, i.e. 50%, for it to be relevant and kept to feed
it into the classifier (cf. shown in red square in Fig-
ure 5).
Figure 5: A simple example of the resulting patches.
4 EXPERIMENT
4.1 Dataset Preparation
We have selected 7 WSIs from kidney transplants on
different types of staining, which are difficult for the
neural network to predict nice masks. The WSIs we
used are scanned with a Philips DP v1.0. The Au-
tomated Slide Analysis Platform (ASAP) is used to
annotate the best quality ROIs. From the annotation
generated by ASAP (XML file), we extract the infor-
mation on the ROIs. A WSI is acquired at different
resolutions, aka levels. These levels are ranged from
0 to 9. On average 1 ROI per WSI, which leads in
total to 500 useful patches for training. To speed up
the prediction of neural networks and include as much
information as possible, we extract the level 5 ROIs
corresponds with a magnification of 1.25×. The orig-
inal images are shown in Figure 6 (a). The images
from top to down are denoted as I
1
to I
7
. I
1
is the
H&E staining. I
3
is the PAS staining. The others are
all JONES staining. I
1
, I
2
, and I
3
are from the same
kidney but with different types of staining.
4.2 Experimental Settings
We have implemented our algorithms in Python and
use the OpenCV and Diplib library
2
to process the
image. The experiments were run on a Windows 11
system with Intel 3.4GHz Processor and 16GB mem-
ory. We compare our method with Otsu threshold seg-
mentation in OpenCV, MobileNet, and EfficientNet-
B3
3
. The mask results are shown in Figure 6.
4.3 Performance Evaluation
To be able to compare the masks generated by our
method with other approaches, we employ binary cor-
relation and binary overlap to measure the correlation
between two binary images (Verbeek, 1995). In this
calculation, the two images should be binary images
of the same size. The total pixels in the image are
denoted as N
tot
. The binary correlation could be cal-
culated as:
bc(I
1
,I
2
) =
|N
I
1
∩I
2
∗ N
tot
− N
I
1
∗ N
I
2
|
h
(N
I
1
∗ N
tot
− N
2
I
1
) ∗ (N
I
2
∗ N
tot
− N
2
I
2
)
i
1/2
(3)
2
https://diplib.org/.
3
Code and pretrained weights available at: https://kimia
lab.uwaterloo.ca/kimia/index.php/ijcnn-2020-u-net-based-
background-removal-in-histopathology/.
BIOIMAGING 2023 - 10th International Conference on Bioimaging
150

(a) Original (b) Otsu (c) MobileNet (d) EfficientNet (e) Our method
Figure 6: The results of the different methods.
where I
1
denotes the mask generated by our method,
and I
2
denotes the mask generated by another method.
N
I
1
is the number of object pixels in I
1
. N
I
2
is the num-
ber of object pixels in I
2
. N
I
1
∩I
2
denotes the number
Foreground Extraction in Histo-Pathological Image by Combining Mathematical Morphology Operations and U-Net
151

of the patterns that result from the logical AND op-
eration of I
1
and I
2
. And the binary overlap could be
calculated as:
bo(I
1
,I
2
) =
2 ∗ N
I
1
∩I
2
N
I
1
+ N
I
2
(4)
4.4 Results Analysis
The binary correlation and binary overlap between the
masks from our method and the others are shown in
Table 1. The measures indicate the discrepancies be-
tween our methods compared to the other approaches.
The results show that all the methods work well for
H&E staining. Our method can, however, remove all
the background in the image resulting from a dirty
staining I
2
. For images resulting from a weak staining
I
3
, MobileNet predicts fewer tissues and EfficientNet
could find more tissues. Otsu and MobileNet view
the empty area in I
4
as foreground, EfficientNet and
our method can recognize the empty area and only
consider the tissue as foreground. Due to the empty
area and the white part inside the empty area, the Otsu
missed most tissues in I
4
and I
6
. Our method removes
the small holes predicted by MobileNet in I
5
, I
6
, and
I
7
.
5 CONCLUSIONS
In this paper, we have proposed a solution for the con-
struction of a tissue mask as a pre-processing step
for tissue classification. The masking is based on a
tissue segmentation task, which uses a combination
of mathematical morphology processing on results
from the U-Net architecture. Several experiments of
our method on different types of staining show the
method performs well and leads to better results for
the patching. For the PAS staining, there are still a few
parts of tissue missing. So, here we need to do further
filter and parameter optimization to be as complete
as possible in identifying the tissue parts. Further-
more, we aim to automatically extract the parameters
from the images. This will require further analysis of
a larger number of images.
ACKNOWLEDGEMENTS
This work is partially supported by the Chinese
Scholarship Council (CSC No.202106280008). We
would like to thank the LUMC (Leiden University
Medical Center) to provide the research data.
REFERENCES
Cai, F. and Verbeek, F. J. (2015). Dam-based rolling ball
with fuzzy-rough constraints, a new background sub-
traction algorithm for image analysis in microscopy.
In 2015 International Conference on Image Process-
ing Theory, Tools and Applications (IPTA), pages
298–303. IEEE.
Chen, P. and Yang, L. (2019). Tissueloc: Whole slide digital
pathology image tissue localization. J. Open Source
Software, 4(33):1148.
He, K., Gkioxari, G., Doll
´
ar, P., and Girshick, R. (2017).
Mask r-cnn. In Proceedings of the IEEE international
conference on computer vision, pages 2961–2969.
Howard, A. G., Zhu, M., Chen, B., Kalenichenko, D.,
Wang, W., Weyand, T., Andreetto, M., and Adam,
H. (2017). Mobilenets: Efficient convolutional neu-
ral networks for mobile vision applications. arXiv
preprint arXiv:1704.04861.
Khened, M., Kori, A., Rajkumar, H., Krishnamurthi, G.,
and Srinivasan, B. (2021). A generalized deep learn-
ing framework for whole-slide image segmentation
and analysis. Scientific reports, 11(1):1–14.
Li, X., Li, C., Rahaman, M. M., Sun, H., Li, X., Wu, J.,
Yao, Y., and Grzegorzek, M. (2022). A comprehensive
review of computer-aided whole-slide image analy-
sis: from datasets to feature extraction, segmentation,
classification and detection approaches. Artificial In-
telligence Review, pages 1–70.
Long, J., Shelhamer, E., and Darrell, T. (2015). Fully con-
volutional networks for semantic segmentation. In
Proceedings of the IEEE conference on computer vi-
sion and pattern recognition, pages 3431–3440.
Lu, M. Y., Williamson, D. F., Chen, T. Y., Chen, R. J., Bar-
bieri, M., and Mahmood, F. (2021). Data-efficient
and weakly supervised computational pathology on
whole-slide images. Nature biomedical engineering,
5(6):555–570.
Neuner, C., Coras, R., Bl
¨
umcke, I., Popp, A., Schlaf-
fer, S. M., Wirries, A., Buchfelder, M., and Jabari,
S. (2021). A whole-slide image managing library
based on fastai for deep learning in the context of
histopathology: Two use-cases explained. Applied
Sciences, 12(1):13.
Otsu, N. (1979). A threshold selection method from gray-
level histograms. IEEE transactions on systems, man,
and cybernetics, 9(1):62–66.
Pizer, S., Johnston, R., Ericksen, J., Yankaskas, B., and
Muller, K. (1990). Contrast-limited adaptive his-
togram equalization: speed and effectiveness. In
[1990] Proceedings of the First Conference on Visu-
alization in Biomedical Computing, pages 337–345.
Riasatian, A., Rasoolijaberi, M., Babaei, M., and Tizhoosh,
H. R. (2020). A comparative study of u-net topologies
for background removal in histopathology images. In
2020 International Joint Conference on Neural Net-
works (IJCNN), pages 1–8. IEEE.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-Net :
Convolutional Networks for Biomedical. In MICCAI,
pages 234–241.
BIOIMAGING 2023 - 10th International Conference on Bioimaging
152

Sultana, F., Sufian, A., and Dutta, P. (2020). Evolution
of image segmentation using deep convolutional neu-
ral network: a survey. Knowledge-Based Systems,
201:106062.
Tan, M. and Le, Q. (2019). Efficientnet: Rethinking model
scaling for convolutional neural networks. In Interna-
tional conference on machine learning, pages 6105–
6114. PMLR.
Verbeek, F. J. (1995). Three-Dimensional Reconstruction
of Biological Objects from Serial Sections Including
Deformation Correction., chapter 4, pages 83–84.
Foreground Extraction in Histo-Pathological Image by Combining Mathematical Morphology Operations and U-Net
153
