
A Rank Aggregation Algorithm for Performance Evaluation in Modern
Sports Medicine with NMR-based Metabolomics
∗
V. Vigneron
a
and H. Maaref
b
Univ. Evry, Université Paris-Saclay, IBISC EA 4526, Evry, France
Keywords:
Deep Learning, Pooling Function, Rank Aggregation, LBP, Segmentation, Contour Extraction.
Abstract:
In most research studies, much of the gathered information is qualitative in nature. This article focuses on
items for which there are multiple rankings that should be optimally combined. More specifically, it describes
a supervised stochastic approach, driven by a Boltzmann machine capable of ranking elements related to
each other by order of importance. Unlike classic statistical ranking techniques, the algorithm does not need
a voting rule for decision-making. The experimental results indicate that the proposed model outperforms
two reference rank aggregation algorithms, ELECTRE IV and VIKOR, and it behaves more stable when
encountering noisy data.
1 INTRODUCTION AND
RELATED WORKS
In the last decades, the field of multiple criteria
decision-making (MCDM) has received considerable
attention in engineering, sciences, and humanities as
they are extremely efficient in situations where pol-
icymakers need to decide priorities (Yazdani et al.,
2017).
There are optimal resolution procedures like lin-
ear programming or nonlinear optimization for solv-
ing problems governed by single criteria. But real-life
situations demand the evaluation of a set of alterna-
tives against multiple criteria and are typically struc-
tured as MCDM problems (Thakkar, 2021; Rahman
et al., 2017). When a decision needs to be made - like
choosing a movie, buying a car, selecting a stock port-
folio, etc. - the choice should not be random or biased
by someone’s suggestion. MCDM algorithms often
produce conflicting results when compared together
because of the choice of the function to optimize.
This comes from the unavoidable trade-off be-
a
https://orcid.org/0000-0001-5917-6041
b
https://orcid.org/ 0000-0002-1192-7333
∗
This research was supported by the program Cátedras
Franco-Brasileiras no Estado de São Paulo, an initiative of
the French consulate and the state of São Paulo (Brazil).
We thank our colleagues Rémi Souriau for his helpful com-
ments and Laurence Le-Moyec who supervise the data ac-
quisition with the Institut national du sport, de l’expertise et
de la performance (INSEP).
tween conflicting objectives as well as constraints. As
a result, the optimal solution is not unique and cor-
responds to a so-called Pareto solution (Freund and
Williamson, 2015).
On the opposite, learning to rank (LTR) is a class
of approaches that apply supervised machine learn-
ing (ML) to resolve ranking problems. The training
data for a LTR model consists of a list of samples and
a "ground truth" score for each of those samples, man-
ually labeled by experts, see (Li et al., 2017). The
set of ranked data ("ground truth") becomes the data
set that the system "trains" by minimizing some loss
function to learn how best to rank automatically these
items (Chaudhuri and Tewari, 2015).
The most common application of LTR is search
engine ranking (Sharma et al., 2022). We propose to
use them in the context of sports medicine because
performance evaluation is a kind of fuzzy task.
Existing LTR algorithms may be divided into
3 main classes: (a) pointwise methods which re-
duce the rating on each item to regression or clas-
sification (Blackburn and Ukhov, 2013) (b) pairwise
methods which essentially formulate ranking on each
document pair as a classification problem (Burges
et al., 2007) (c) list-wise methods which optimize a
measure-specific loss function, on all available items.
See Chavhan et al.(Chavhan et al., 2021) for a review.
The pros and cons of using LTR vs. MCDM
are (a) LTR is essentially a black box in terms of
explainability. It’s hard to explain what exact ef-
fect specific inputs have on the outcome (b) LTR is
332
Vigneron, V. and Maaref, H.
A Rank Aggregation Algorithm for Performance Evaluation in Modern Sports Medicine with NMR-based Metabolomics.
DOI: 10.5220/0011798000003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 4: BIOSIGNALS, pages 332-339
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

greedy (c) result relevance is metric-dependent. This
work presents a new extension of the LTR based on
a continuous restricted Boltzmann machine (CRBM)
(Hinton, 2002). The CRBM is a generative stochastic
artificial neural networks (ANN) that can learn a prob-
ability distribution over (all possible permutations of)
its set of inputs. CRBMs have found applications in
dimensionality reduction (Vrábel et al., 2020), clas-
sification (Yin et al., 2018) and collaborative filtering
(Verma et al., 2019). They are excellent generative
learning models for latent space extraction. Specif-
ically, they can be trained into excellent ranking de-
vices because of their flexible loss function and the
associative memory captured in the transfer matrix W
between the visible and the hidden layers, which is a
promising advantage over other standard MCDM al-
gorithms.
The paper is organized as follows: section 2 pro-
vides a detailed description of the problem and the
metrics used for measuring aggregation of ranks. Sec-
tion 3 presents the generative model. The experimen-
tal results are presented in Section 4. The last section
concludes and outlines the way for future work.
Notations. Throughout this paper small Latin let-
ters a, b,.. . represent integers. Small bold letters a, b
are put for vectors, and capital letters A, B for ma-
trices or tensors depending on the context. The dot
product between two vectors is denoted < a, b >. We
denote by ∥a∥ =
√
< a, a >, the ℓ
2
norm of a vector.
X
1
,. . ., X
n
are non ordered variates, x
1
,. . ., x
n
non or-
dered observations. "Ordered statistics" means either
p
(1)
≤. . . ≤ p
(n)
(ordered variates) or p
(1)
≤. . . ≤ p
(n)
(ordered observations). The p
(i)
are necessarily de-
pendent because of the inequality relations among
them.
Definition 1 ((Savage, 1956)). The rank order cor-
responding to the n distinct numbers x
1
,. . ., x
n
is the
vector t = (t
1
,. . .,t
n
)
T
where t
i
is the number of x
j
’s≤
x
i
and i ̸= j.
The rank order t is always unambiguously defined
as a permutation of the first n integers.
2 GENERAL FRAMEWORK
2.1 Rank-Aggregation
Let A = {a
1
,a
2
,. . ., a
n
} be a set of alternatives, candi-
dates, individuals, etc. with cardinality |A|= n and let
V be a set of voters, judges, criteria, etc. with |V |= m.
The data is collected in a (n ×m) table T of general
term {t
i j
} crossing the sets A and V (Figure 1). t
i j
can be marks (t
i j
∈ N), value scales (t
i j
∈ R), ranks
(such that a voter can give ex-aequo positions) or bi-
nary numbers (t
i j
∈ {0, 1} such as opinion yes/no).
T represents the ranking of the n alternatives under
the form (see (Brüggemann and Patil, 2011) for a re-
minder on rank-aggregation). For ease of writing, in
the following, t
i j
= t
( j)
i
.
T =
v
(1)
v
(2)
... v
(k)
... v
(m)
a
1
a
2
.
.
.
.
.
.
a
i
. . . t
i j
.
.
.
a
n
(a) Data matrix T (t
i j
≥ 0).
→
Y
(k)
=
a
1
a
2
. . . a
j
. . . a
n
a
1
0
a
2
0
.
.
.
.
.
.
a
i
. . . y
(k)
i j
.
.
.
a
n
0
(b) kth pairwise comparison matrix between the
alternatives a
i
and a
j
.
Figure 1: The data are collected in a (n ×m) table T .
Solving a rank-aggregation problem means find-
ing a distribution of values x
∗
attributed by a virtual
judge to the n alternatives by minimizing the dis-
agreements of opinions between the m judges (Ben-
son, 2016), i.e.
x
∗
= arg min
t
m
∑
k=1
d(t,t
(k)
), s.t. t ≥ 0, (1)
where d(t,t
(k)
) is a metric measuring the proximity
between t and t
(k)
, chosen a priori, and t
(k)
is the kth
column of the table T . Depending on the properties of
d(·), we will deal with a nonlinear optimization pro-
gram with an explicit or implicit solution.
One could also stand the dual problem of the
previous one, i.e., is there a distribution of rank-
ings/marks that the m voters could have attributed to
a virtual alternative ’a’ summarizing the behavior of
the set of individuals A (Yadav and Kumar, 2015)?
The first problem is linked to the idea of aggregating
points of view, and the second to the concept of sum-
marizing behaviors.
A Rank Aggregation Algorithm for Performance Evaluation in Modern Sports Medicine with NMR-based Metabolomics
333

2.2 Explicit or Implicit Resolution
Eq. (1) defines a nonlinear optimization program
whose solution is x
∗
(Yadav and Kumar, 2015). The
distance d(t
(k)
,t
(k
′
)
) between the ranking of voters k
and k
′
can be chosen for instance as the Euclidean
distance
∑
n
i=1
(t
ik
−t
ik
′
)
2
, the disagreement distance
(Condorcet)
∑
n
i=1
sgn|t
ik
−t
ik
′
| or the order disagree-
ment distance
∑
i
∑
j
|y
(k)
i j
−y
(k
′
)
i j
|as t
(k)
can be replaced
by its permutation matrix Y
(k)
(Figure 1). In the lat-
ter, y
(k)
i j
=
i< j
denotes the indicator matrix for which
y
(k)
i j
= 1 if the rank of the alternative a
i
is less than the
alternative a
j
and 0 otherwise (Gehrlein and Lepelley,
2011). Note that y
(k)
ii
= 0 and y
(k)
i j
= 0 if i and j are
ex-aequos.
In using matrix Y
(k)
,
1
2
∑
i
∑
j
|y
(k)
i j
− y
(k
′
)
i j
| =
1
2
∑
i
∑
j
(y
(k)
ik
− y
(k
′
)
ik
)
2
since since the expressions
|y
(k)
i j
−y
(k
′
)
i j
| are 0 or 1.
As y
2
i j
= y
i j
= y
(k)
i j
2
= y
(k)
i j
= 0 or 1, the function
associated to order disagreement distance d is given
by
1
2
"
n
∑
i=1
n
∑
j=1
my
i j
+
n
∑
i=1
n
∑
j=1
m
∑
k=1
y
i j
!
−2
n
∑
i=1
n
∑
j=1
y
i j
m
∑
k=1
y
(k)
i j
#
.
(2)
Let α
i j
=
∑
m
k=1
y
(k)
i j
the total number of voters prefer-
ring alternative a
i
to a
j
and define a matrix A = {α
i j
},
summing the m matrices Y
(k)
associated to the rank-
ings t
(k)
of the voters V
(k)
, Eq. (2) becomes:
1
2
"
n
∑
i=1
n
∑
j=1
my
i j
+
n
∑
i=1
n
∑
j=1
α
i j
−2
n
∑
i=1
n
∑
j=1
α
i j
y
i j
#
. (3)
Finally, the search for a total order given by a matrix
Y is the solution of the linear program
max
Y
n
∑
i=1
n
∑
j=1
my
i j
+
n
∑
i=1
n
∑
j=1
α
i j
−2
n
∑
i=1
n
∑
j=1
α
i j
y
i j
!
s.t. α
i j
=
p
∑
k=1
y
(k)
i j
y
i j
+ y
ji
= 1, i < j,
y
ii
= 0 y
i j
+ y
ji
−y
ik
≤ 1, i ̸= j ̸= k, y
i j
∈ {0, 1}.
(4)
If the chi-2 metric is chosen, then the dependent
variables cannot be separated as in Eq. (4): The reso-
lution follows an implicit gradient-descent procedure
as in (Vigneron and Tomazeli Duarte, 2018). Section
2.3 detailed how chi-2 distance is used for solving ag-
gregation problems.
2.3 Chi-2 Metric
The distance between x and t
(k)
is given by
d(x,t
(k)
) =
n
∑
i
1
f
i·
f
ix
f
·x
−
f
ik
f
·k
2
, (5)
where
f
i·
=
∑
k
t
ik
+ x
i
∑
ik
t
ik
+
∑
i
x
i
f
ik
=
x
i
∑
ik
t
ik
+
∑
i
x
i
f
·x
=
∑
i
x
i
∑
ik
t
ik
+
∑
i
x
i
f
ik
=
t
ik
∑
ik
t
ik
+
∑
i
x
i
f
·k
=
∑
i
t
ik
∑
ik
t
ik
+
∑
i
x
i
.
(6)
Let n¯r =
∑
ik
t
ik
,n ¯x =
∑
i
x
i
,t
·k
=
∑
i
t
ik
and t
i·
=
∑
k
t
ik
.
Then, after some calculus, the optimal ranking x
∗
minimizes
∑
k
d(x,t
(k)
):
n
m
∑
k=1
n
∑
i=1
¯r + ¯x
t
i·
+ x
i
x
i
n ¯x
−
t
ik
t
·k
2
= n(¯r + ¯x)
n
∑
i=1
m
∑
k=1
x
i
√
t
i·
n ¯x
−
t
ik
√
t
i·
t
·k
2
,
(7)
assuming t
i·
≫ x
i
and
√
t
i·
+ x
i
≈
√
t
i·
.
According to Eq. (7), the ranking is performed
on the row profiles or column-profiles of the matrix T
(see Fig. 1), each row being weighted by
√
t
i·
.
So it is equivalent to compute profile matrix C
whose entry is
t
ik
√
t
i·
t
·k
, to consider the Euclidean dis-
tance between its rows and to heighten each row by
√
t
i·
. A remark has to be made at this stage: two al-
ternatives will be close if a large proportion of judges
choose them simultaneously. For example, if there is
a considerable amount of individuals chosen prefer-
ably by judges ’A’ and ’B,’ then we will say that
judges A and B are close and that they "attract" each
other. Eq. (7) is the well-known expression used in
testing for independence in contingency tables.
Eq. (7) derives from the Bhattacharyya directed
divergence between two discrete probability distri-
butions P = {p
i
} and Q = {q
i
} defined as BD =
−ln(
∑
i
√
p
i
q
i
) (Nielsen, 2022) if p
i
=
x
2
i
t
i·
n
2
¯x
2
and q
i
=
t
2
ik
t
i·
t
2
·k
. Note that n
2
is useless in the ratio and will be
removed in the entries of the continuous restricted
Boltzmann machine. Implicit methods are natural for
LTR algorithms that are usually fed by an incoming
data stream, n constantly varying. Section 3 proposes
a learning model in which the rank probabilities take
the form of a Boltzmann distribution.
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
334
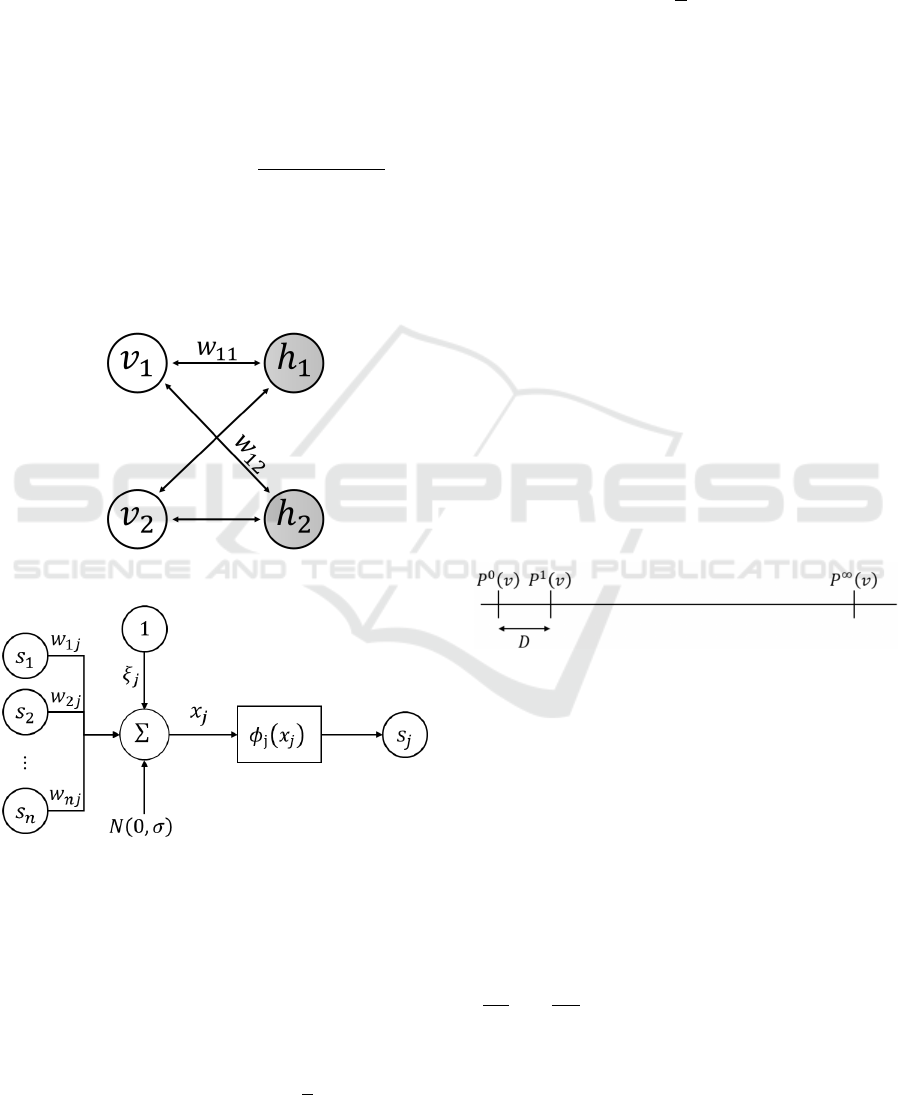
3 METHODOLOGY
3.1 Continuous Restricted Boltzmann
Machine
Chen and Murray proposed another Boltzmann ma-
chine (BM) approach with continuous neuron in
(Chen and Murray, 2003): the CRBM, a restricted
Boltzmann machine using the neuron structure de-
picted in figure 2. In the CRBM, the activation func-
tion is unique for each neuron and given by:
s
j
= φ
j
(X
j
) = θ
L
+ (θ
H
−θ
L
)
1
1 + exp(−a
j
X
j
)
(8)
where θ
L
and θ
H
are, respectively, the function’s
lower and upper bounds. a
j
is a slope parameter of
φ
j
(.). The continuous behavior for the hidden units
allows us to capture more information than binary
units.
(a) Continuous restricted Boltzmann machine.
(b) Structure of the neuron j of a CRBM.
Figure 2: White neurons are visible neurons and gray neu-
rons are hidden neurons in the CRBM. the coefficient w
i j
refers to the weight of the symmetric link between the i−th
visible unit v
i
and the j−th hidden unit h
j
..
We note W ∈ IR
(m×ℓ)
the transfer matrix between
the two layers and ξ
v
and ξ
h
the bias vectors of, re-
spectively, the visible layer and the hidden layer. The
energy function of the CRBM is:
E(v, h) = −v
T
W h −v
T
ξ
v
−h
T
ξ
h
+
∑
i
1
a
i
R
s
i
0
φ
−1
(s
′
)ds
′
,
(9)
with φ
−1
(.) the inverse of the activation for a coeffi-
cient slope a
i
= 1. The energy E(s) of a CRBM is
associated with the joint probability of the state of the
neurons P
cRBM
(s) defined as
P
CRBM
(s) =
1
Z
exp(−E(s)), (10)
where Z is a marginalization constant. Training a
CRBM is performed in minimizing the energy func-
tion Eq. (9), which itself requires sampling the hidden
units.
The CRBM training uses the contrastive diver-
gence algorithm (see (Hinton, 2012)). The training set
D = {v
k
}
1≤k≤n
is composed of n observations used to
find the best set of parameters P = {W, ξ}, ξ regroup-
ing visible and hidden bias vectors.
Minimizing directly the joint log-likelihood
∑
n
k=1
logP
CRBM
(v
k
) to update the parameters is diffi-
cult due to the presence of the constant Z. Then it
is replaced by the minimization of the contrastive di-
vergence (MCD) (Hinton, 2002) that minimizes the
contrast D between two successive Kullback-Leibler
(KL)-divergences:
D = KL(P
0
(v),P
∞
(v)) −KL(P
q
(v),P
∞
(v)), (11)
where P
0
(v),P
∞
(v),P
q
(v) are the distribution func-
tion of the visible units over respectively the training
set, the equilibrium state and after q steps of Gibbs
sampling (Hinton, 2012) (Fig. 3).
Figure 3: An intuitive idea is to minimize the KL divergence
between P
0
(v)P
∞
(v). But P∞(v) is intractable. We prefer
to minimize D. If D = 0, then P
0
(v) = P
1
(v) and then :
P
0
(v) = P
∞
(v).
An important observation is that any linear com-
bination of measures of discrepancy with positive co-
efficients is also a measure of discrepancy.
KL(P
0
(v),P
∞
(v)) −KL(P
q
(v),P
∞
(v))
+λ(BD
0
(v) −BD
q
(v)),
(12)
with λ a regularization parameter. And thus, Eq. (12)
can be used as a measure of discrepancy.
In particular, the observations are normalized: v =
(
t
2
i1
t
i·
t
2
·1
,. . .,
t
2
im
t
i·
t
2
·m
)
T
(see section 2.3).
In the next section, a CRBM driven by the loss
function (12) ranks rugby players according to their
performances measured by metabolomics.
A Rank Aggregation Algorithm for Performance Evaluation in Modern Sports Medicine with NMR-based Metabolomics
335
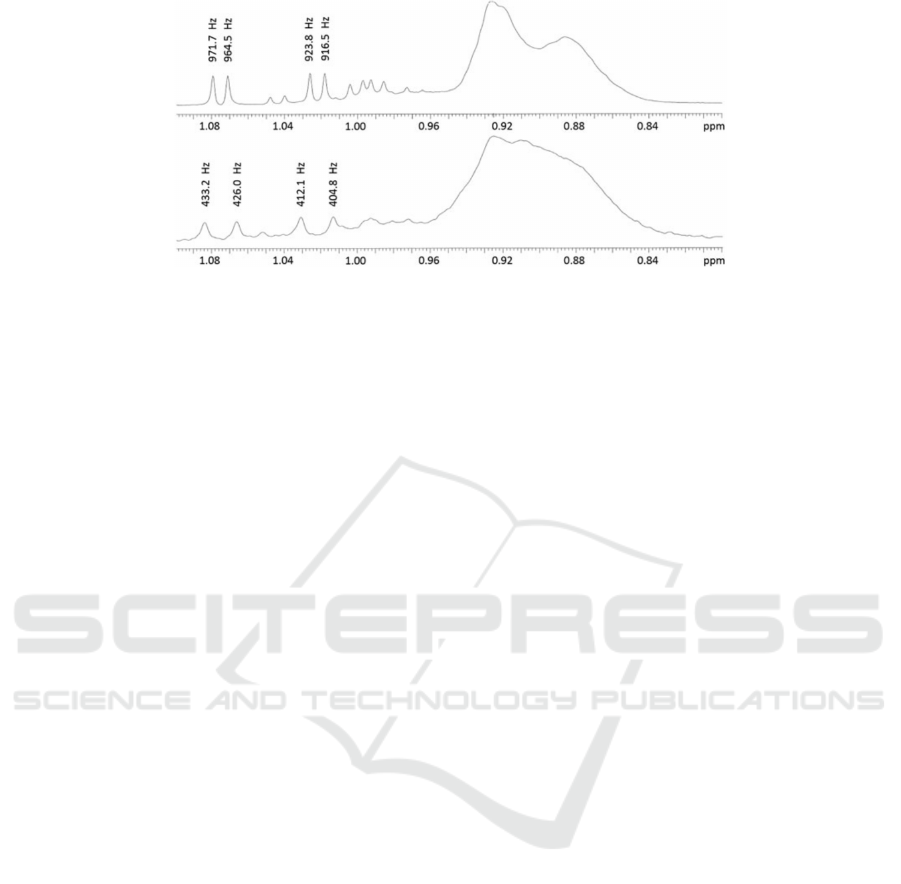
Figure 4:
1
H NMR spectrum of human plasma acquired at 900 (top) and 400 MHz (bottom), from (Louis et al., 2017).
4 EXPERIMENTS WITH
METABOLOMICS DATA
Endurance is a widely practiced sporting activity,
from novice to champion. It is defined as maintaining
an effort for a prolonged period. This effort originates
from significant physiological and metabolic stress
leading to organism adaptations. If this effort is too
great, it can cause metabolic and locomotor disorders.
The objective is to optimize training methods that will
protect the health of athletes, young or old, efficient or
less efficient. We adopt an integrative approach that
simultaneously studies the physiological responses to
exercise and the molecular and metabolic signals.
From multivariate statistical analysis of biofluids such
as urine, serum, plasma, saliva, sweat, etc., it is pos-
sible to generate metabolomic profiles or biomark-
ers. See (Khoramipour et al., 2022) for a review of
metabolomics practice in sports medicine.
Since the 00’s metabolomics investigates quanti-
tatively the metabolome of living systems in response
to pathophysiological stimuli or genetic modification
(Amara et al., 2022).
Nuclear magnetic resonance (NMR) is tradition-
ally used to elucidate molecular structures. It takes
advantage of the energy transition of nuclear spins in
a strong magnetic field to identify and elucidate the
structure of organic molecules and specific metabo-
lites. Metabolites are intermediate organic com-
pounds resulting from metabolism. To understand the
metabolomic changes induced by endurance exercise
and training of rugby players according to the inten-
sity and duration of the activity, we study the physio-
logical modulation of rugby players according to their
positions.
The study focuses on the activity variability be-
tween the forwards - more intense and intermittent ef-
forts - and the rears - greater distance covered, more
running, more rest time, etc. (Paul et al., 2022). It
aims to answer whether, during matches, (a) the uri-
nary metabolites are identical before and after 80 min-
utes of a match? (b) this metabolomic modulation is
of the same order depending on the player’s position?
No study to date has investigated how to predict
physiological exertion in rugby or how to classify a
player according to its physiological parameters.
The experimental protocol is as follows: the urine
of 80 players (40 forwards and 40 rears) is analyzed
by NMR to identify the metabolites present in the
two situations described above. The variations in
metabolism explain the variations in physiological pa-
rameters as a function of the time and position factors.
NMR spectra contain more than 10,000 values. See
Fig 4 for an example of NMR spectrum.
19 variables represent the physiological vari-
ables, among which: forward/backward position
of the player during the match, body mass in-
dex, experience, playing time, distance covered on
the playground but also plasmatic metabolite rates
in phenylalanine, tyrosine, glucose, creatinine, β-
hydroxybutyrate, lactate, pyruvate, N-acetyl glyco-
protein, lipids (Table 1). This set of variables consti-
tutes the criteria that are used to rank the rugby play-
ers.
There are 70 samples in the training set and 10 in
the test set. The structure of the CRBM is: 19 visi-
ble and 3 hidden units. Due to the small training and
test data, we did not divide the data into mini-batches
during the experiment. All the data were divided
into eight groups for the seven-fold cross-validation
method. Seven groups were selected as the training
set each time, and the remaining group was the test
set. This process was repeated until each group be-
came a test set. The number of iterations was 200 for
each CRBM. For training, the CRBMs were initial-
ized with small random weights and zero bias param-
eters. The learning rate was η = 0.1 when training
with CD in Eq. (12) and λ = 0.005. CRBMs models
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
336

Table 1: Data description. Notice that variable 17 precises the position of the players. Forwards: pillars, hooker, 2nd lines.
Backward: 3rd lines, 9 and 10, backs.
Variables type (if real: µ± std.err.) physiological signification
1 term binary 1st term=1, 2nd term=0
2 P binary position forward=1, backward=0
3 A 28,4 ±0.97 Age
4 H 179.95 ±1.0 height
5 W 89.75 ±2.81 weight
6 BMI 27.65 ±0.756 body-mass index
7 X 41,3 ±1.01 game experience
8 Ty 143,9 ±1.89 tyrosine
9 Glu 269,5 ±2.59 glucose
10 β 414, 1 ±3.22 β-hydroxybutyrate
11 Creat 6.57 ±0.22 creatinine
12 Gly 5069 ±11.25 glycoprotein
13 D 2.238 ±7.48 distance covered on the playground
14 L 51,6 ±1.13 lipids
16 R {1,2, 3} rolling position
17 PHC0 −86.98 ±15.73 phenylalanine 0
18 PHC1 23.36 ±9.19 phenylalanine 1
19 SNR 41.03 ±3.56 signal to noise ratio
Table 2: Test results comparing CRBM with classic MCDMs algorithms VIKOR and ELECTRE IV. Float numbers are issued
from the models and the ranks in sorting these numbers.
Players VIKOR ELECTRE IV CRBM+Bhattacharyya
1 3.608 7 3.127 1 3.291 1
2 5.123 1 6.324 6 6.074 7
3 4.639 6 5.094 8 4.925 8
4 5.378 8 4.923 7 5.088 6
5 5.811 3 6.147 4 5.886 3
6 4.033 2 3.401 3 3.693 4
7 3.468 4 3.567 10 3.467 5
8 4.254 10 3.411 5 3.663 10
9 5.6220 9 6.2400 9 5.9600 9
10 5.8110 5 6.3240 2 6.0740 2
are updated while the new training example is com-
pleted.
The results obtained with VIKOR, ELECTRE IV,
and the CRBM are gathered in Table 2 for compar-
ison. For each MCDM algorithm, the first column
measures the discrepancy of the model Eq. (12) and
the second column the rank of the rugby player (in
bold). They provide an interpretation that the CRBM
method is closer to the actual results and far from
the ELECTRE IV method (Roy, 1985). VIKOR is
a MCDMs, ranking preferences among a set of al-
ternatives in the presence of conflicting criteria un-
der the concept of group regrets (Guiwu et al., 2020).
ELECTRE IV assumes that all requirements (actually
pseudo-criteria) have the same importance.
At first glance, the rankings are not similar but not
so different either. The rankings order the players ac-
cording to the sportive qualities recorded in Tab. 1.
For example, ELECTRE IV and CRBM would select
player one first, then player 10. But ELECTRE IV
would prefer player 6 in the third place while CRBM
would choose player 5.
The discriminant representation provided by the
hidden layer is the most determining factor in favor
of CRBM against VIKOR or ELECTRE. The players
were ranked by the first hidden neuron of the CRBM.
The scatter-plot Figure 5 uses the values of the first
two hidden neurons. This means the hidden neurons
capture a latent representation capable of discriminat-
ing between the two classes, "rears" and "forwards."
A Rank Aggregation Algorithm for Performance Evaluation in Modern Sports Medicine with NMR-based Metabolomics
337
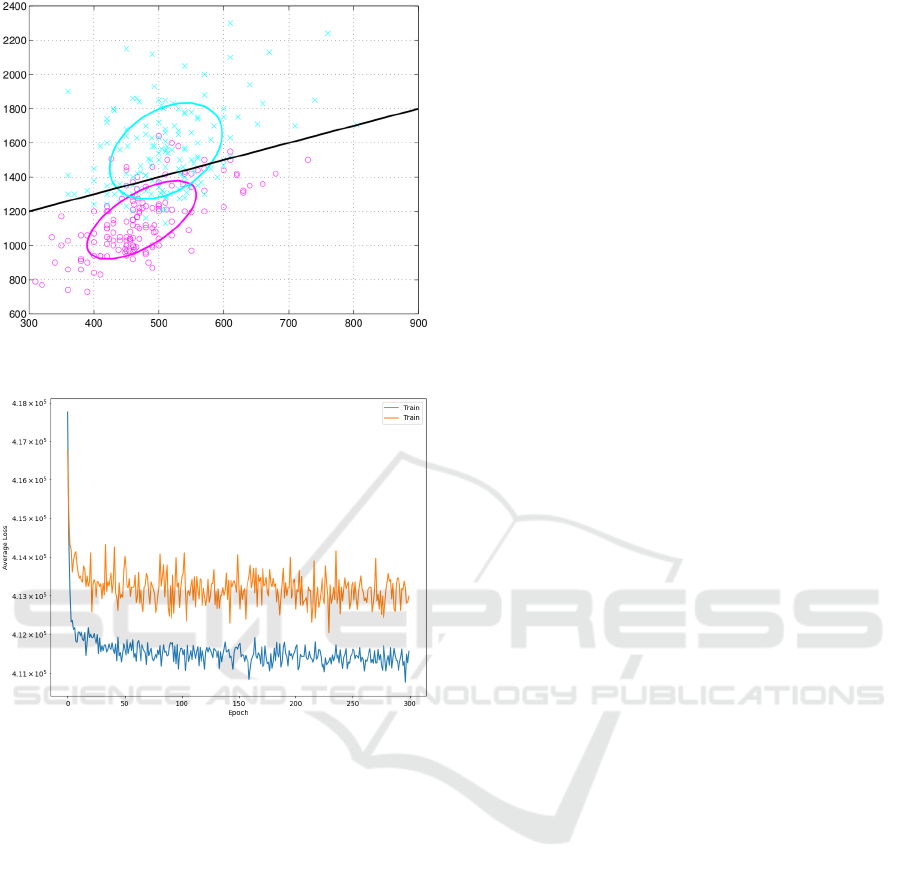
(a) Separated classes (before and after the match) obtained
from the values of the hidden neurons.
(b) Reconstruction error for the training (blues) and the test
(red) sets.
Figure 5: Score plot obtained with the two hidden neurons
of the restricted Boltzmann machine (RBM): the samples
separate each other before and after the match.
5 CONCLUSION AND
DISCUSSIONS
CRBMs are domain-independent feature extractor
that transforms raw data into latent variables. The
most relevant questions are: how to dimension the
hidden layer h optimally? And how do the neurons
interact?
Our generative network is relatively small. Hence
to compute p(h|v) is an affordable problem for small
RBM, but once we have a large number of hidden
neurons, it becomes impossible to compute all pos-
sible p(h|v). The more neurons, the more computa-
tional efforts are needed: massive networks should
not be the only way to reduce the modeling error. The
choice of dimension remains today an unsolved issue.
In addition, for each configuration v, some hidden
neurons have a probability close to 0 or 1, meaning
that for each v, some states of h are irrelevant.
Besides being energy-consuming, a significant di-
mension network requires much time to learn. In
many papers, authors focus on comparing the per-
formance between models but barely reach compu-
tational efforts between models. The issue of compu-
tational efforts can have a significant impact, partic-
ularly in the real-time system. Still, it is enormously
dependent on the data, the application, and the used
hardware.
To reduce the bias between the data distribution
P
data
(x) and the estimated data distribution P
model
(x)
the cost function was modified (Eq. 12).
The next step is constructing a deeper network,
such as deep belief network (DBN), that may provide
more explainability hints.
REFERENCES
Amara, A., Frainay, C., Jourdan, F., Naake, T., Neumann,
S., Novoa-Del-Toro, E. M., Salek, R., Salzer, L.,
Scharfenberg, S., and Witting, M. (2022). Networks
and Graphs Discovery in Metabolomics Data Anal-
ysis and Interpretation. Frontiers in Molecular Bio-
sciences, 9.
Benson, D. (2016). Representations of Elementary Abelian
p-Groups and Vector Bundles. Cambridge tracts in
mathematics. Cambridge University Press, 1 edition.
Blackburn, D. and Ukhov, A. (2013). Individual vs. ag-
gregate preferences: The case of a small fish in a big
pond. Management Science, 59:470–484.
Brüggemann, R. and Patil, G. (2011). Ranking and Prior-
itization for Multi-indicator Systems: Introduction to
Partial Order Applications. Environmental and Eco-
logical Statistics. Springer-Verlag New York.
Burges, C., Ragno, R., and Le, Q. (2007). Learning to
rank with nonsmooth cost functions. In Schölkopf, B.,
Platt, J., and Hoffman, T., editors, Advances in Neu-
ral Information Processing Systems, volume 19. MIT
Press.
Chaudhuri, S. and Tewari, A. (2015). Perceptron like
algorithms for online learning to rank. ArXiv,
abs/1508.00842.
Chavhan, S., Raghuwanshi, M. M., and Dharmik, R. C.
(2021). Information retrieval using machine learning
for ranking: A review. Journal of Physics: Conference
Series, 1913(1):012150.
Chen, H. and Murray, A. (2003). Continuous restricted
boltzmann machine with an implementable training
algorithm. IEE Proceedings-Vision, Image and Sig-
nal Processing, 150(3):153–158.
Freund, D. and Williamson, D. (2015). Rank aggregation:
New bounds for MCx. CoRR, abs/1510.00738.
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
338

Gehrlein, W. and Lepelley, D. (2011). Voting Paradoxes and
Group Coherence: The Condorcet Efficiency of Voting
Rules. Studies in Choice and Welfare. Springer-Verlag
Berlin Heidelberg, 1 edition.
Guiwu, W., Jie, W., Jianping, L., Jiang, W., Cun, W., Fuad,
E., and Tasawar, H. (2020). Vikor method for multiple
criteria group decision making under 2-tuple linguis-
tic neutrosophic environment. Economic Research-
Ekonomska Istraživanja, 33(1):3185–3208.
Hinton, G. (2002). Training products of experts by mini-
mizing contrastive divergence. Neural computation,
14(8):1771–1800.
Hinton, G. (2012). A practical guide to training restricted
boltzmann machines. In Montavon, G., Orr, G. B.,
and Müller, K.-R., editors, Neural Networks: Tricks
of the Trade (2nd ed.), volume 7700, pages 599–619.
Springer.
Khoramipour, K., Sandbakk, O., Keshteli, A., Gaeini, A.,
Wishart, D., and Chamari, K. (2022). Metabolomics
in exercise and sports: A systematic review. Sports
Med., 52(3):547–583.
Li, X., Wang, X., and Xiao, G. (2017). A comparative study
of rank aggregation methods for partial and top ranked
lists in genomics applications. Briefings in Bioinfor-
matics, 20(1):178–189.
Louis, E., Cantrelle, F., Mesotten, L., Reekmans, G., and
Adriaensens, P. (2017). Metabolic phenotyping of hu-
man plasma by (1) h-nmr at high and medium mag-
netic field strengths: a case study for lung cancer.
Magnetic resonance in chemistry : MRC, 55.
Nielsen, F. (2022). Statistical divergences between densi-
ties of truncated exponential families with nested sup-
ports: Duo bregman and duo jensen divergences. En-
tropy, 24(3).
Paul, L., Naughton, M., Jones, B., Davidow, D., Patel, A.,
Lambert, M., and Hendricks, S. (2022). Quantifying
collision frequency and intensity in rugby union and
rugby sevens: A systematic review. Sports Medicine -
Open, 8.
Rahman, A., Sungyoung, L., and Tae, C. (2017). Accurate
multi-criteria decision making methodology for rec-
ommending machine learning algorithm. Expert Sys-
tems with Applications, 71:257–278.
Roy, B. (1985). Methodologie multicritere d’aide a la deci-
sion. Economica, Paris,.
Savage, R. (1956). Contributions to the theory of rank-order
statistics – the trend case. The Annals of Mathematical
Statistics, 27(3):590–615.
Sharma, P., Yadav, D., Thakur, R. N., Reddy, K., and
Praveen, M. (2022). Web page ranking using web
mining techniques: A comprehensive survey. Mob.
Inf. Syst., 2022.
Thakkar, J. (2021). Multi-Criteria Decision Making.
Springer, first edition.
Verma, S., Patel, P., and Majumdar, A. (2019). Col-
laborative Filtering with Label Consistent Re-
stricted Boltzmann Machine. arXiv e-prints, page
arXiv:1910.07724.
Vigneron, V. and Tomazeli Duarte, L. (2018). Rank-order
principal components. A separation algorithm for or-
dinal data exploration. In 2018 International Joint
Conference on Neural Networks, IJCNN 2018, Rio de
Janeiro, Brazil, July 8-13, 2018, pages 1–6.
Vrábel, J., Po
ˇ
rízka, P., and Kaiser, J. (2020). Restricted
boltzmann machine method for dimensionality reduc-
tion of large spectroscopic data. Spectrochimica Acta
Part B: Atomic Spectroscopy, 167:105849.
Yadav, N.. author. abd Yadav, A. and Kumar, M. (2015). An
Introduction to Neural Network Methods for Differen-
tial Equations. Springer, Gurgaon, Haryana, India.
Yazdani, M., Fomba, S., and Zaraté, P. (2017). A Decision
Support System for Multiple Criteria Decision Mak-
ing Problems. In 17th International Conference on
Group Decision and Negotiation (GDN 217), pages
67–75, Stuttgart, Germany.
Yin, J., Lv, J., Sang, Y., and Guo, J. (2018). Classifica-
tion model of restricted boltzmann machine based on
reconstruction error. Neural Computing and Applica-
tions, 29:1–16.
A Rank Aggregation Algorithm for Performance Evaluation in Modern Sports Medicine with NMR-based Metabolomics
339
