
Using Balancing Methods to Improve Glycaemia-Based Data Mining
Diogo Machado
1,3
, Vítor Santos Costa
2,3
and Pedro Brandão
1,3
1
Instituto de Telecomunicações, Portugal
2
INESC-TEC, Portugal
3
Faculty of Science of the University of Porto, Portugal
Keywords:
Data Mining, Diabetes, Data-Balance, over-Sampling, Under-Sampling.
Abstract:
Imbalanced data sets pose a complex problem in data mining. Health related data sets, where the positive class
is connected to the existence of an anomaly, are prone to be imbalanced. Data related to diabetes management
follows this trend. In the case of diabetes, patients avoid situations of hypo/hyperglycaemia, which is the
anomaly we want to detect. The use of balancing methods can provide more examples of the minority class, and
assist the classifier by clearing the decision boundary. Nevertheless, each over-sampling and under-sampling
method can affect the data set uniquely, which will influence the classifier’s performance. In this work, the
authors studied the impact of the most known data-balancing methods applied to the Ohio and St. Louis
diabetes related data sets. The best and most robust approach was the use of ENN with SMOTE. This hybrid
method produced significant performance gains on all the performed tests. ENN in particular had a meaningful
impact on all the tests. Given the limited volume of glycaemia-based data available for diabetes management,
over-sampling methods would be expected to have a greater role in improving the classifier’s performance. In
our experiments, the clearing of noise values by the under-sampling methods, produced better results.
1 INTRODUCTION
Data mining can be regarded as the computational pro-
cess that analyses and extracts knowledge from a data
set (Raval, 2012). Data mining has contributed posit-
ively in many health-domains such as liver diseases,
cardiovascular diseases, coronary diseases, cancer, and
diabetes (Shukla et al., 2014).
In this work we focus on data mining applied to
diabetes. This chronic disease, characterised by high
glycaemia levels (blood glucose), affected 463 million
people around the world in 2019, and it is projected
to reach 700 million by 2045 (International Diabetes
Federation, 2019).
Diabetic patients should have a strict manage-
ment of their disease to avoid both hyperglycaemia
(very high glycaemia levels) and hypoglycaemia (very
low glycaemia levels). Both can lead to severe con-
sequences. In the long term, repeated hyperglycaemia
episodes damage the large and small blood vessels,
and may lead to blindness, heart problems, increased
risk of having a stroke, among other serious health
issues. In the short term, extreme hyperglycaemia and
hypoglycaemia events can make the person go into a
coma, and even death (Mouri and Badireddy, 2021;
Seery, 2019b,a).
Given the severity of these occurrences, most dia-
betic patients attempt, to their best ability, to stay
within a glycaemia range considered normal. There-
fore, in data mining approaches, the data sets have an
imbalance, as most observations will be normal. Im-
balanced data is common in health related data sets,
where the data target consists of values that are either
designated as "normal" or "abnormal" (Li et al., 2010).
This reality causes the use of data mining algorithms
to be more complex, since data imbalances cause bias
towards the majority class (Li et al., 2010).
Glucose values were traditionally obtained only
through finger-pricking. This invasive method, accord-
ing to medical doctors, should be performed at least
six times per day, before and after the three main meals
(breakfast, lunch, and dinner). Due to the small num-
ber of samples, using finger-based glucose values for
data mining is challenging.
Nowadays, diabetic patients have a more conveni-
ent method of glycaemia tracking, continuous-glucose
monitoring (CGM). CGM uses a painless, one-time
sensor device application under the skin of the belly
or the back of the arm, that must be replaced period-
ically (around every seven to ten days). While less
188
Machado, D., Costa, V. and Brandão, P.
Using Balancing Methods to Improve Glycaemia-Based Data Mining.
DOI: 10.5220/0011797100003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 5: HEALTHINF, pages 188-198
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

convenient, the traditional finger-prick method is more
accurate than CGM (Siegmund et al., 2017; Cengiz and
Tamborlane, 2009; Medtronic Diabetes, 2014). This is
a consequence of the physiological delay between in-
terstitial glucose and blood-glucose (Cengiz and Tam-
borlane, 2009). Even so, CGM devices are able to
sample a new glycaemia value each five minutes, with
some devices having an even higher sample rate. This
amount of data would, realistically, be impossible to
obtain using the traditional finger-pricking method. In
terms of data mining, CGM devices can give a better
perspective of the diabetic patient’s glycaemia val-
ues and trends throughout the day. While more data
is beneficial, CGM data continues to be imbalanced.
Data balancing methods are often used to improve
learning performance. In this work, we study how
different methods perform on two different data sets.
The first reports finger-prick-based glycaemia data,
the second reports CGM-based glycaemia. Although
CGM is now prominent in data mining, having both
types of glycaemia sampling methods as target will
allow a better evaluation of the impact of data balan-
cing (Machado et al., 2022).
Given the scope of this work, we decided to limit
the number of classification methods, to broaden the
number of data balancing method variations. For the
classification of glycaemia values, we chose the Ran-
dom Forest (RF) ensemble learning method. RF allows
the evaluation of the balancing method’s effects, but
also the assessment of the impact of different features.
The article starts with a brief presentation and stat-
istics of the used data sets; it follows with an explana-
tion of the data balance methods present in this work;
after we discuss the few articles of related work; we
then describe the experiments’ and their results; and
finally, we present the work’s conclusions.
2 DATA SETS
In this work, two very distinct data sets were used to
evaluate different balancing techniques.
The data set from the University of St. Louis, avail-
able on the UCI Machine Learning Repository (Dua
and Graff, 2017)
1
contains data from 70 different pa-
tients, over several weeks to months (minimum eight
days, maximum 288 days). This data set possesses
data concerning the patient’s glycaemia values (ob-
tained through finger-prick testing), insulin adminis-
tration (short, medium, and long duration), and meal
and exercise classifications (regular, more-than-usual
or less-than-usual quantities). In terms of total records,
1
https://archive.ics.uci.edu/ml/datasets/diabetes
the St. Louis data set has 11737 finger-prick-based
glycaemia records, 317 meal records, 8110 insulin
administration records, and 201 exercise records. In
terms of glycaemia record variance, the minimum num-
ber of records by a single user in the data set is 25
and the maximum is 616. Regarding the glycaemia
value classification: 11.62% of values represent hy-
poglycaemia; 14.51% represent hyperglycaemia; and
finally 73.87% of the values are considered as normal.
The Ohio data set (Marling and Bunescu, 2020)
contains eight weeks of data from 12 different pa-
tients. This data set possesses both sensor (continuous
monitoring) and finger-prick tested glycaemia values,
administrated insulin dosages (basal and regular), exer-
cise, and other physiological parameters e.g. the heart
rate, for patients that used a proper sensor. The Ohio
data set has a total of 166533 sensor-based glycaemia
records, 4566 finger-prick-based glycaemia records,
2773 meal records, 3731 insulin records, and 221 ex-
ercise records. This considerable amount of records,
compared to the St. Louis data set, occurs because
the CGM device used by the patients automatically
recorded a new glycaemia value every five minutes.
In terms of glycaemia value classification, the values
must be divided, having into account the glycaemia
sampling method (finger or sensor-based). In terms
of glycaemia values that were obtained through a
continuous monitor (sensor): 3.28% of values repres-
ent hypoglycaemia; 8.48% represent hyperglycaemia;
and, finally, 88.23% of the values are considered as
normal. Regarding the glycaemia values, obtained
through finger-pricking: 4.58% of values represent hy-
poglycaemia; 12.07% represent hyperglycaemia; and,
finally, 83.36% of the values are considered as normal.
While the St. Louis data set contains fewer fea-
tures and sparser data, the Ohio data set is richer and
contains continuous glycaemia data from a glycaemia
sensor. Figures 1 to 3 display statistics about: the pa-
tient’s ID, the number of days recorded, number of
hypoglycaemia, number of normal glycaemia, number
of hyperglycaemia, and the HbA1c correspondent to
the glycaemia values recorded. Considering the in-
formation in these figures, the patients that constitute
St. Louis data set seem to have glycaemia values less
controlled than the ones present in the Ohio data set.
The St. Louis data set contains, in percentage, more
hypoglycaemia and hyperglycaemia. Although the
level of HbA1c seems on par with the Ohio data set,
the high standard deviation refutes this idea.
The referred data sets contain raw records. The two
data sets used in this work result from preprocessing
the St. Louis and Ohio data sets and have different
targets and features. The first generated data-set targets
finger-based glycaemia. It concatenates the St. Louis
Using Balancing Methods to Improve Glycaemia-Based Data Mining
189
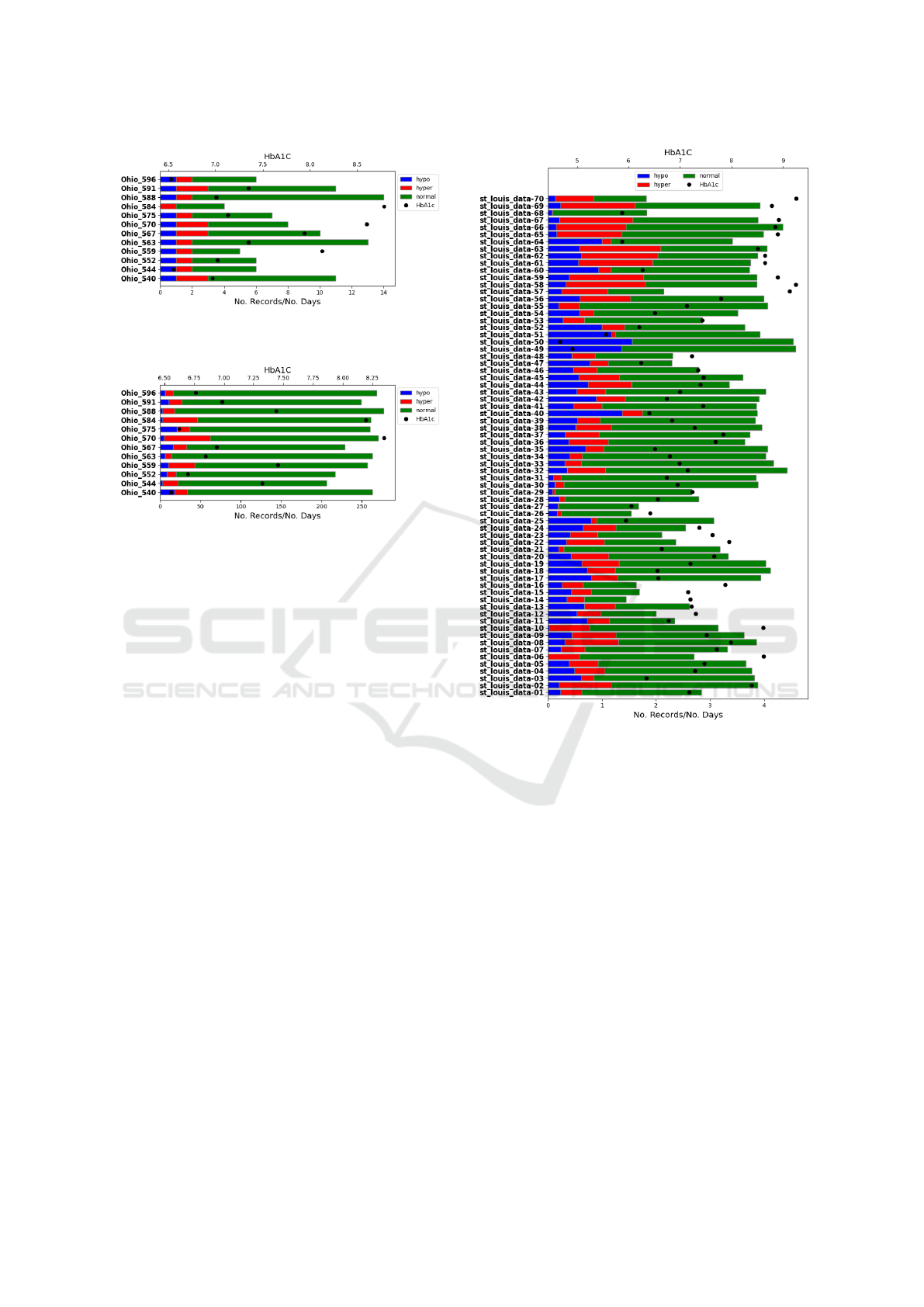
Figure 1: Ohio raw finger-prick based glycaemic data statist-
ics.
Figure 2: Ohio raw sensor-based glycaemic data statistics.
and the Ohio data sets, and it is restricted to the more
limited set of available features in the S Louis data.
The second data-set targets CGM-based glycaemia,
only available in the Ohio data set.
The Ohio-St. Louis joint data set, with finger-prick
glycaemia as target, is composed of 43 features. Of
these, three of them are parameters commonly used
by medical doctors to evaluate the patients’ diabetes
management.
The time-in-range represents the number of gly-
caemia records within the range of [70-180] divided by
the total amount of glycaemia records. Values of time-
in-range above 0.7 (70%) are considered good. The
glycaemia value tendency is a metric that represents
the general tendency of the glycaemia values. This
parameter is specially important when the glycaemia
values are reaching hypo/hyperglycaemia thresholds.
To calculate this parameter, the authors utilised the
least square polynomial regression. The variation
coefficient represents the standard value deviation di-
vided by the value average, then multiplied by 100.
This coefficient is more used than the standard devi-
ation parameter since it is less dependent of the average
value. In terms of glycaemia value management, the
variation coefficient should be below 36%.
The features present in the conjunct data set are
the glycaemia designation classified as: hypergly-
caemia (glycaemia values above 250), hypoglycaemia
(glycaemia values under 70), and normal (glycaemia
values between 70 and 250); the weekday and time
of day of the target glycaemia; past finger-based
Figure 3: St. Louis finger-prick glycaemic data statistics.
glycaemia value, time in range, tendency, variation
coefficient, average, and the existence of prior hypo-
/hyperglycaemia. These features are sampled in differ-
ent time intervals: 30 minutes to one hour, one to three
hours, three to 12 hours, 12 to 24 hours, and 24 to 48
hours. We calculate the existence of recorded meals in
the last three hours, and in the last three to six hours;
the existence of recorded insulin bolus in the last hour,
in the last hour to three hours, in the last three to six
hours, and in the last six to 12 hours; the existence of
recorded exercises in the last hour. These features are
displayed in Listing: 1.
The Ohio sensor-based data set is composed of 81
features, the 43 already present in the Ohio-St. Louis
conjunct data set, 33 features related to sensor-based
glycaemia values, and five new features. The new fea-
tures contain information not available in the St. Louis
data set: if the user paused, reduced or increased the
basal insulin; the correlation between recorded carbo-
hydrates and the insulin bolus; and the physical efforts
HEALTHINF 2023 - 16th International Conference on Health Informatics
190

glycaemia_designation , wee kday ,
time _day ,
fin g e r _ g l y c a e mia _ v a l u e _ 3 0 m _1h , [.. .] ,
fin g e r _ t i m e _ i n _ r ang e _ 3 0 m _ 1 h , [...] ,
finger_te n d e n c y _ 3 0 m _ 1 h , [.. .] ,
fin g e r _ v a r i a t i o n _co f _ 3 0 m _ 1 h , [...] ,
finger_average_30m_1h , [.. .] ,
finger _ h a d _ h y p e r _ 3 0 m _ 1 h , [...] ,
finger_ha d _ h y p o _ 3 0 m _ 1 h , [.. .] ,
had_meal_last _ 3 h , h a d _ meal_3h_6h ,
had_insulin_last_1h , [... ] ,
ex e r c i s e _ l a st _ 1 h
Listing 1: Part of the features in the Ohio-St. Louis joint data
set.
detected. Efforts are an experimental measure, calcu-
lated by the authors through the heart rate information
present on the Ohio data set. The features present on
the Ohio sensor-based data set are displayed in List-
ing: 2.
glycaemia_designation , wee kday ,
time _day ,
glycaemia _ v a l u e _ 3 0 m _ 1 h , [.. .] ,
time_in_range_30m_1h , [ ...] ,
tendency_30 m _ 1 h , [.. .] ,
average_3 0 m _ 1 h , [ ...] ,
variation_cof_30m_1h , [ ...] ,
had_hyper_30m _ 1 h , [...] ,
had_hypo_30 m _ 1 h , [.. .] ,
fin g e r _ g l y c a e mia _ v a l u e _ 3 0 m _1h , [.. .] ,
fin g e r _ t i m e _ i n _ r ang e _ 3 0 m _ 1 h , [...] ,
finger_te n d e n c y _ 3 0 m _ 1 h , [.. .] ,
fin g e r _ v a r i a t i o n _co f _ 3 0 m _ 1 h , [...] ,
finger_average_30m_1h , [.. .] ,
finger _ h a d _ h y p e r _ 3 0 m _ 1 h , [...] ,
finger_ha d _ h y p o _ 3 0 m _ 1 h , [.. .] ,
n_basal_paused_1h_3h ,
n_basal_reduced_1h_3h ,
n_basa l _ i n c r e a s e d _ 1 h _ 3 h ,
ca r b s _ i n s u l in_ c o r r e l a t i on _ 1 h _ 3 h ,
had_meal_last _ 3 h , h a d _ meal_3h_6h ,
had_insulin_last_1h , [... ] ,
exercise_last _ 1 h ,
eff o r t _ l a s t _ 1 h
Listing 2: Part of the features in the Ohio data set.
3 DATA BALANCING METHODS
Glycaemia-based data is particularly imbalanced. Tak-
ing the St. Louis data set as an example, the hy-
poglycaemia values represent only 10% of the total
values, and the hyperglycaemia values represent 13%,
leaving the denominated normal values to represent
77% of the data set’s glycaemia values. This trend,
also visible in the Ohio data set, should be expected,
given that both hypoglycaemia and hyperglycaemia
values represent health risks that should be prevented
by patients.
Imbalanced data sets are a known problem in data-
mining. A common approach is the use of over-
sampling and/or under-sampling, to achieve some sort
of balance within the data set’s classes.
3.1 Over-Sampling Methods
Over-sampling is a method that produces new values
in order to increase the representation of the minor-
ity classes. There are two base approaches to over-
sampling: by value duplication, and by creating syn-
thetic values. Value duplication can virtually balance
the weight of every class, but since each new value
does not include additional information to the model,
it can create value bias. Within the over-sampling
methods, that create synthetic values, there are some
different approaches to consider:
Synthetic Minority Oversampling TEchnique
(
SMOTE
) (Bowyer et al., 2011): This technique man-
ufactures values similar to the ones already present
in the imbalanced class using the k-Nearest Neigh-
bours (
KNN
) algorithm. To achieve this,
SMOTE
chooses a random value, from the minority class, and
k of the nearest neighbours. The method then creates
synthetic values within the chosen values’ domain.
Borderline-SMOTE (Han et al., 2005): Classifiers’
decisions are dependant of the clarity of the decision
border. The line that divides and enables classific-
ation is present where values from different classes
congregate. The values in this location implicitly as-
sume a greater importance in the data set. While the
SMOTE
method creates values at random positions,
the Borderline-SMOTE chooses, at random, values
close to the border between classes to run the
SMOTE
value creation method.
Borderline-SMOTE SVM (Nguyen et al., 2011):
Similar to Borderline-SMOTE, this method, instead of
KNN
, uses Support-Vector Machine (
SVM
) to choose
values at the decision border. Additionally, this method
selects regions in the minority class that have less value
density. The method then creates values towards the
class boundary.
Adaptive Synthetic Sampling (
ADASYN
) (He et al.,
2008): It is a more generalised form of the
SMOTE
method. The main difference between these methods
is the use of the density distribution. This method
searches low density data areas, in minority classes,
and uses these empty spaces to generate synthetic val-
ues. It re-shapes the decision boundary, using syn-
thetic values, according to their learning difficulty
level. More data will be generated for classes that
Using Balancing Methods to Improve Glycaemia-Based Data Mining
191

are harder to learn.
3.2 Under-Sampling Methods
Contrary to over-sampling, under-sampling method
achieves class equilibrium by excluding values in the
majority class. This sampling can be done at ran-
dom, by removing random values until the classes
have equal weight, or it can be done while aware of
the removed values. Some known, and used, under-
sampling methods are:
Near-Miss Under-Sampler: It is a collection of under-
sampling methods based on the work of Mani and
Zhang (2003), and available on the imblearn library
that uses
KNN
to select examples from the majority
class. Near-Miss has three versions:
•
NearMiss-1 retains values with the minimum av-
erage distance to the three closest minority class
values;
•
NearMiss-2 retains values with the minimum av-
erage distance to the three furthest minority class
values;
•
NearMiss-3 retains values with the minimum dis-
tance to each minority class value.
Condensed Nearest Neighbors (
CNN
) Rule Under-
Sampling (Hart, 1968):
CNN
algorithm uses the one
nearest neighbour rule to decide, at each sample, if a
sample should be added or removed from the data set.
The
CNN
algorithm creates a sub-set containing all the
minority class values, and a sample from the majority
class. Then, the values from the majority class are
iterated and classified, using the current sub-set and
the one nearest neighbour rule. The samples that are
not correctly classified from the majority class set, are
added to the sub-set. In the end, this method is able
to obtain a sub-set containing all the minority class
values and the inconsistent majority class values.
Tomek-links for Under-Sampling (Tomek et al.,
1976): Based on
CNN
, Tomek-links addresses two
lacking elements of the
CNN
method, according to the
author. The first point that was criticised by Tomek
et al. (1976) was the fact that the values, at the be-
ginning of the
CNN
method, are chosen at random.
Only later in the process, does the method become
less random and collects values closer to the decision
boundary. This process returns a sub-set containing
redundant samples, that could be eliminated. The pro-
posed solution is to find links between pair values of
different classes that, together, have the smallest Eu-
clidean distance. This method can be used to erase
possible ambiguous values by finding and removing
examples in the majority class, that are closest to the
minority class. This action clears the borderline area
and facilitates class division.
Edited Nearest Neighbors (
ENN
) Rule for Under-
Sampling (Wilson, 1972): This method for re-
sampling and classification can be used as an under-
sampling method. This method uses
KNN
to identify
and remove incoherent values in the data set. The pro-
cess begins with a set equal to the training set. Each
value in this set is compared to its neighbours using
a
KNN
method, by default with k equal to three. If
the selected value differs from the majority, then the
value is removed. Similarly to Tomek-links, the
ENN
rule for under-sampling removes noisy and ambigu-
ous values from the class boundary, thus supporting
classification methods.
3.3 Joint Methods
One-Sided Selection for Under-Sampling
(OSS) (Kubat et al., 1997): This method com-
bines the Tomek-links and the
CNN
method. The
Tomek-links method is used as an under-sampling
method to remove noisy and borderline values from
the majority class. The
CNN
method is used to
remove values from the majority class that are distant
from the decision border.
Neighbourhood Cleaning Rule for Under-Sampling
(NCR): This is an under-sampling method, designed
to prioritise data quality, in detriment of data bal-
ance (Laurikkala, 2001). It combines the
CNN
method
and the
ENN
rule methods to remove redundant noise
values. By removing close border points, this method
is able to smooth the decision boundary (Gu et al.,
2008).
SMOTE and Tomek-links: Batista et al. (2004) pro-
pose the application of SMOTE to balance the data set,
followed by the Tomek-links method. In this proposal,
the Tomek-links method would not be used to under-
sample the majority class. Instead, this method is used
to remove values from every class. By doing this, the
authors claim that this method is capable of producing
a balanced data set, with well-defined class clusters.
SMOTE and
ENN
: Similar to the previous proposal,
Batista et al. (2004) also proposed the use of SMOTE
and
ENN
, instead of the Tomek-links method. They
affirm that, since
ENN
tends to remove more examples
than Tomek-links, it should be expected to obtain a
clearer decision boundary.
The different balancing techniques attempt to reach
a similar goal: to clear the decision boundary and fa-
cilitate the distinction between classes. Over-sampling
techniques, while trying to fulfil the previous objective,
focus on increasing the amount of available data on
the minority classes. Unfortunately, there is neither a
HEALTHINF 2023 - 16th International Conference on Health Informatics
192

method, nor a junction of methods, that reigns above
the rest. Depending on the available data, the balan-
cing methods applied will create different impacts.
4 STATE OF THE ART
In recent times, the use of data mining and machine
learning applied to the field of diabetes seems to be
focused on glycaemia value and/or hypo or hyper-
glycaemia occurrence prediction. These approaches
mostly use temporal-based approaches that do not re-
quire balancing. Reviewing the blood glucose level
prediction challenge papers of the Knowledge Discov-
ery in Healthcare Data 2020, we found none of the 17
articles to use any type of data balancing.
However, exceptions do exist. The work by Mayo
et al. (2019) uses over-sampling to increase the per-
formance of a machine learning-based method for
short-term glycaemia prediction. In this work ran-
dom over-sampling,
SMOTE
,
ADASYN
were used.
This work only concludes that there are performance
gains from using over-sampling. It is never mentioned
which balancing approach benefited more the machine
learning method.
The work by Berikov et al. (2022) developed a
machine learning method for the prediction of noc-
turnal hypoglycaemia in hospitalised patients. This
proposal used a personalised approach to both under
and over-sampling. The over-sampling method used
Gaussian noise to introduce new synthetic CGM val-
ues with a nocturnal hypoglycaemia occurrence. The
under-sampling, was used to select records that do
not contain events of nocturnal hypoglycaemia. The
method consisted of a k-medoids algorithm with a
number of clusters equal to the number of nocturnal
hypoglycaemia occurrences. This process then returns
a group of medoids that do not contain nocturnal hy-
poglycaemia events. This methodology is rather un-
usual, as it is composed of a somewhat random over-
sampler, and, for over-sampling, selects a group of
values that do not contain the study’s target. In the
end, the study concluded that the use of sampling was
insignificant. The authors affirm that the influence of
data balancing, according to their results, depends on
the machine learning method being applied. They refer
that the use of over-sampling alone resulted on a slight
increase in performance.
Alashban and Abubacker (2020) studied the use
of over-sampling and the joint use of over-sampling
and under-sampling to glycaemia-based data. This
work’s objective is to classify patients as normal, pre-
diabetic, or diabetic. This study does not directly
correlate to diabetes management, nonetheless, it of-
fers a significant view on the results of applying dif-
ferent balancing approaches to glycaemia-based data.
The over-sampling methods used in this work were:
random over-sampling, and
SMOTE
. The hybrid ap-
proach tested
SMOTE
with Tomek-link, and
SMOTE
with
ENN
. This study concluded that, although every
sampling technique is better than the use of an im-
balanced data set, the random over-sampling method
achieved better performance gains.
Although existent, the analysis of the impact of
balancing techniques on diabetes management data
is insufficient. The work by Alashban and Abu-
backer (2020) was the more complete study, and even
this study only evaluated four possible approaches to
data balancing.
To have a clearer idea of the real impact of over-
sampling and under-sampling on glycaemia-based
data, we will test the most known methods for over-
sampling and under-sampling, as well as the hybrid
combination of said methods.
5 METHODOLOGY
Balancing data can be approached by applying a single
under-sampling, or over-sampling method, or by ap-
plying both methods. To have a complete depiction
of the impact of the use of balancing methods on dia-
betes’ data, every possible combination of single and
conjunct method was tested, including applying no
balancing methods. In the conjunct method, over-
sampling is applied in order to produce more examples
in the minority classes, and then under-sampling is
applied to filter the majority class and balance the data
set. The balancing methods were only applied to train-
ing sets. The test set remains imbalanced, to determine
the classifier’s performance with real data.
The classification method used to test the impact
of balancing was the random forest.
Random forest is an ensemble learning method
commonly used for classification and regression. This
method consists of a collection of decision trees. The
decisions made by this method are obtained through
majority vote of the created decision trees. The ran-
dom forest method was chosen for this work for its
robustness. It inherits from the decision trees method
its scale in-variance, robustness to irrelevant features,
and, contrary to decision trees, random forests tend to
not over-fit as much.
The available data sets are composed by data from
different individuals. To evaluate the impact of each
balancing technique, on individual and on community
sets, two tests were carried out: (i) an individual test,
that applies an 80-20 cut on a single data set and runs
Using Balancing Methods to Improve Glycaemia-Based Data Mining
193

the random forest classifier; (ii) and a leave-one-out
test, that singles out a user to be the test set and estab-
lishes the remaining users’ data as the training set.
To rank the different balancing methods and ap-
proaches, the F1-score was calculated. Although other
metrics are usually also applied in data mining ana-
lysis, F1 is the most commonly used metric in cases
of imbalanced data sets.
Precision is a metric that quantifies the number
of correct positive predictions over all positive pre-
dicted values. The precision metric is represented on
equation 1, where
T
p
defines true positives,
F
p
false
positives, and
F
n
false negatives. Recall, displayed on
equation 2 quantifies the number of correct positive
predictions over all possible positive predictions. The
F1-score, represented on equation 3 is the precision
and recall’s harmonic mean. Rather than focusing on
pure accuracy, the F1-score focuses on the positive
class, which, in an imbalanced data set, has a greater
impact than accuracy. Higher F1-score values are as-
sociated with a better performance of the model.
P =
T
p
(T
p
+ F
p
)
(1)
R =
T
p
(T
p
+ F
n
)
(2)
F1 =
2 ∗ P ∗ R
P + R
(3)
AP =
∑
n
(R
n
− R
n−1
)P
n
(4)
Each data set, for each test, will be divided in three
data sets for binary classification, where each class is
set as positive, against the remaining two (negative)
classes. The “normal” class is an exception. Instead
of considering the “normal” class against hypo and
hyperglycaemia, the classes that represent non-normal
glycaemia were set as the positive against the “nor-
mal” class. The F1-score and the F1-score’s standard-
deviation (SD) is then calculated for each set. As an
evaluation metric, the F1-scores of each set are av-
eraged by balancing approach and ordered from best
to worst. This way, it is possible to evaluate the best
approaches on average, and the best approach for a
particular class.
6 RESULTS
As previously mentioned, the following results are
composed of the F1-scores and their respective SD for
the hypoglycaemia focused set, the hyperglycaemia fo-
cused set, and the not normal focused set. In Tables 1,
through 4 these parameters are presented.
The approaches are ordered from best to worst,
considering their hypoglycaemia F1-score and SD.
As previously mentioned, episodes of hypoglycaemia
pose a more immediate and concerning danger than
hyperglycaemia, or other intermediate glycaemia level
states. Considering this fact, it is important to prioritise
hypoglycaemia as the main focus for classification.
While the leave-one-out approach encompasses
most of the available data, and it is capable of running
with no issues, some individual sets with finger-prick-
based glycaemia lack enough data to run balancing
techniques and the random forest classifier. Sensor
data, with a higher volume of data, was able to com-
plete all the individual and the leave-one-out tests.
The balancing methods are displayed from top to
bottom according to their evaluation rank.
The results of the individual tests using finger-
prick-based glycaemia values as target (displayed on
Table 1) have near miss as the best approach. ENN and
near-miss alone, or used together with over-sampling
methods, constitute nine of the top ten scoring ap-
proaches. In terms of over-sampling approaches, in the
top ten, the adaptive synthetic sampling method was
the most represented, appearing three times, associ-
ated with under-sampling methods. As a sole method,
over-sampling methods achieved poor results, being at
the bottom of the result’s table.
The results of the individual tests using CGM-
based glycaemia values as target (see Table 2) have
the conjunct method, using borderline SMOTE fol-
lowed by ENN, as the best approach. In this test
the ENN dominates the best approaches. As a stan-
dalone method, ENN is ranked fourth. The near miss
approach, which was first on the previous test, did
not achieve the same success. Although near miss
with SMOTE rank sixth, the remaining near miss ap-
proaches rank on the bottom half of the results table.
In this test, applying no balancing approaches achieved
the worse results.
The results of the leave-one-out tests using finger-
prick-based glycaemia values as target are displayed
on Table 3.
As the individual finger-prick-based results, this
test has the near miss method as the best balancing
approach. The following three best approaches have
ENN as a common denominator. Near miss seems to
falter when applied together with over-sampling. This
fact hints to an inability of the near miss approach
to clear the noise introduced by the over-sampling
methods. In contrast, ENN only has a positive impact
when applied in combination with an over-sampling
method. ENN in this test was inconsistent. While
used with adaptive synthetic sampling, SMOTE, and
borderline SMOTE, ENN achieved very good results,
close to the ones obtained by the top ranking method.
HEALTHINF 2023 - 16th International Conference on Health Informatics
194

Table 1: Data balancing methods results for the individual tests, using data based on the Ohio and St. Louis data sets, sampled
considering finger-prick-based glycaemia-based values as target.
Under-sample Over-sample
hypo hyper not normal
F1(SD)
near miss 0.20 (0.13) 0.29 (0.16) 0.38 (0.14)
enn smote 0.20 (0.17) 0.35 (0.16) 0.40 (0.18)
enn
borderline svm
smote
0.20 (0.18) 0.22 (0.16) 0.18 (0.17)
enn
adaptive syn-
thetic sampling
0.19 (0.16) 0.28 (0.15) 0.36 (0.18)
enn
borderline smote
0.17 (0.16) 0.28 (0.20) 0.38 (0.20)
. . .
tomek links
borderline svm
smote
0.09 (0.15) 0.16 (0.17) 0.25 (0.16)
smote 0.09 (0.13) 0.16 (0.18) 0.28 (0.20)
no balancing 0.06 (0.13) 0.12 (0.18) 0.22 (0.21)
tomek links 0.06 (0.13) 0.14 (0.19) 0.25 (0.21)
Table 2: Data balancing methods results for the individual tests, using data based on the Ohio data set, sampled considering
CGM-based glycaemia-based values as target.
Under-sample Over-sample
hypo hyper not normal
F1(SD)
enn
borderline smote
0.25 (0.16) 0.55 (0.09) 0.47 (0.08)
enn smote 0.22 (0.14) 0.51 (0.14) 0.42 (0.14)
enn
adaptive syn-
thetic sampling
0.20 (0.15) 0.52 (0.11) 0.41 (0.13)
enn 0.19 (0.15) 0.54 (0.09) 0.46 (0.10)
enn
borderline svm
smote
0.19 (0.15) 0.48 (0.13) 0.38 (0.16)
. . .
near miss
adaptive syn-
thetic sampling
0.10 (0.13) 0.43 (0.16) 0.36 (0.16)
tomek links
borderline svm
smote
0.09 (0.14) 0.46 (0.12) 0.39 (0.13)
no balancing 0.07 (0.10) 0.41 (0.15) 0.34 (0.16)
As a sole approach, ENN is on the bottom half of
the table, being one of the top five worse approaches.
With borderline SVM SMOTE, ENN even reaches the
bottom of the table, under the no balancing technique.
The results of the leave-one-out tests using CGM-
based glycaemia values as target (displayed on Table 4)
have ENN as the best balancing approach. Apart from
when applied together with borderline SVM SMOTE,
ENN is present in the top four best approaches. Con-
trasting with previous tests, the leave-one-out CGM-
based test had near miss as the worse approach, under
the use of no balancing techniques.
Considering all the obtained results, the method
with greater impact was the ENN under-sampling tech-
nique. This under-sampling method, while used to-
gether with over-sampling, is present on the best ap-
proaches of every test.
In terms of over-sampling as a sole approach, there
Using Balancing Methods to Improve Glycaemia-Based Data Mining
195

Table 3: Data balancing methods’ results for the leave-one-out test, using data based on the Ohio and St. Louis data sets,
sampled considering finger-prick-based glycaemia-based values as target.
Under-sample Over-sample
hypo hyper not normal
F1(SD)
near miss 0.22 (0.13) 0.28 (0.15) 0.43 (0.16)
enn
adaptive syn-
thetic sampling
0.20 (0.16) 0.29 (0.14) 0.43 (0.16)
enn smote 0.20 (0.16) 0.29 (0.15) 0.43 (0.16)
enn
borderline smote
0.20 (0.16) 0.28 (0.15) 0.43 (0.16)
near miss smote 0.19 (0.20) 0.27 (0.19) 0.38 (0.20)
. . .
tomek links 0.13 (0.22) 0.13 (0.20) 0.22 (0.21)
no balancing 0.13 (0.22) 0.13 (0.21) 0.19 (0.21)
enn
borderline svm
smote
0.13 (0.19) 0.16 (0.17) 0.16 (0.16)
Table 4: Data balancing methods’ results for the leave-one-out test, using data based on the Ohio set, sampled considering
CGM-based glycaemic values as target.
Under-sample Over-sample
hypo hyper not normal
F1(SD)
enn 0.32 (0.13) 0.58 (0.09) 0.52 (0.11)
enn
adaptive syn-
thetic sampling
0.31 (0.13) 0.57 (0.07) 0.50 (0.09)
enn smote 0.31 (0.14) 0.58 (0.08) 0.51 (0.09)
enn
borderline smote
0.30 (0.14) 0.58 (0.08) 0.51 (0.09)
borderline svm
smote
0.28 (0.16) 0.59 (0.08) 0.53 (0.10)
. . .
tomek links 0.26 (0.14) 0.59 (0.08) 0.53 (0.11)
no balancing 0.11 (0.09) 0.57 (0.08) 0.48 (0.11)
near miss 0.11 (0.06) 0.34 (0.13) 0.35 (0.09)
was no clear best method. The creation of synthetic
values, if not filtered, introduces noise and thus, under-
mines the classifier’s results.
Overall the best approach was the use of ENN with
SMOTE. While SMOTE produces synthetic values
that create more training opportunities, the ENN under
sampler clears the decision border for a better clas-
sification. This combination in theory produces an
balanced and favourable environment to train a clas-
sifier. In reality, as these tests prove, it is a good,
balanced, and robust method.
In terms of performance gains, the difference
between the top scoring features is not significant.
The exception to this occurred in the first test, where
the approach using ENN with SMOTE achieved a su-
perior overall result, compared to the remaining top
three approaches. Considering hyperglycaemia classi-
fication, this approach is six percentage points better
than the second approach and, considering not-normal
glycaemia, the first and second approach are respect-
ively 20 and 22 percentage points superior to the third
best approach.
Considering the success of sole under sampling ap-
proaches, it is possible to assume that diabetes manage-
ment data by default contains noise values. Nonethe-
less, additional noise produced by an over-sampling
HEALTHINF 2023 - 16th International Conference on Health Informatics
196

method, can still be compensated by the ENN method.
As expected, applying balancing is preferable,
except for some particular cases. Table 5 shows,
for hypoglycaemia, hyperglycaemia and, not-normal
glycaemia, the performance gains obtained, compar-
ing the best balancing technique and the use of no
balancing. The gain obtained using the best perform-
ing balancing method, compared to the no balance
approach, in the individual finger-prick-based tests,
in terms of hypoglycaemia classification, was 14%,
and in CGM-based tests 18%. The highest perform-
ance gain occurred in the leave-one-out test for hy-
poglycaemia classification. The difference between
the use of no balancing and the use of ENN is 21%.
The lowest benefit was 9%, in the context of the finger-
prick-based leave-one-out test.
Overall, the advantages of using over-sample and
under-sample are substantial.
Table 5: Performance gains obtained comparing the best
balancing approach and applying no balancing techniques.
test type
glycaemia
sampling
type
class
perf.
gain
individual
finger
hypoglycaemia 14%
hyperglycaemia 17%
not-normal 16%
CGM
hypoglycaemia 18%
hyperglycaemia 14%
not-normal 13%
leave-
one-out
finger
hypoglycaemia 9%
hyperglycaemia 15%
not-normal 24%
CGM
hypoglycaemia 21%
hyperglycaemia 1%
not-normal 4%
7 CONCLUSION
Diabetes data sets tend to be scarce and skewed. Data
balancing is an extremely useful tool, as it can both
reproduce minority class examples, and clean detri-
mental examples in the decision boundary.
In the literature, a definitive approach to glycae-
mia-based data balancing does not exist. In this work,
we tested several balancing methods on finger-prick
and CGM-based glycaemia data to conclude which
approach is the most suited for glycaemia-based data
sets. The
ENN
under-sampling method was overall
the method with the greatest impact on classification
success. Contrary to what would be expected, over-
sampling is not always a good solution. If applied,
over-sampling should be complemented with under
sampling to achieve satisfactory results. Overall, the
use of joint methods is the better approach. The best
approach to glycaemia-based data is ENN and SMOTE.
This hybrid approach is the most consistent as it is
present as one of the top three best approaches in all
tests. It should be noted that, although this method
does not appear as the best approach in any test, the
difference in F1-score to the best approach is not sig-
nificant.
The individual, smaller data sets have more consist-
ent performance gains throughout all classes. Larger
data sets have significant, but uneven performance
benefits, depending on the class. This fact could be
justified by the lesser interference of data from other
sets that could introduce further noise. Usually, each
patient’s diabetes management is a unique case with
particular characteristics. This could also justify the
greater impact of under-sampling methods, as they are
responsible for clearing noise values and improve the
decision boundary.
In this study, CGM-based glycaemia data was only
available in the Ohio data set. As these sensor-based
data becomes ever so preponderant, it would be import-
ant to study the use of balancing methods in a larger
data set to verify if the conclusions obtained by the
CGM-based tests persist. The lack of available data
is a tremendous obstacle to the study and use of data
mining in diabetes.
ACKNOWLEDGEMENTS
This work is funded by FCT/MCTES through national
funds and when applicable co-funded EU funds under
the project UIDB/50008/2020. Diogo Machado is
funded by PhD Grant DFA/BD/6666/2020.
REFERENCES
Alashban, M. and Abubacker, N. F. (2020). Blood glucose
classification to identify a dietary plan for high-risk pa-
tients of coronary heart disease using imbalanced data
techniques. In Lecture Notes in Electrical Engineering,
pages 445–455. Springer Singapore.
Batista, G. E. A. P. A., Prati, R. C., and Monard, M. C.
(2004). A study of the behavior of several methods for
balancing machine learning training data. SIGKDD Ex-
plor. Newsl., 6(1):20–29.
Berikov, V. B., Kutnenko, O. A., Semenova, J. F., and Kli-
montov, V. V. (2022). Machine learning models for noc-
turnal hypoglycemia prediction in hospitalized patients
Using Balancing Methods to Improve Glycaemia-Based Data Mining
197

with type 1 diabetes. Journal of Personalized Medicine,
12(8):1262.
Bowyer, K. W., Chawla, N. V., Hall, L. O., and Kegelmeyer,
W. P. (2011). SMOTE: synthetic minority over-sampling
technique. CoRR, abs/1106.1813.
Cengiz, E. and Tamborlane, W. V. (2009). A tale of two com-
partments: Interstitial versus blood glucose monitoring.
Diabetes Technology & Therapeutics, 11(S1):11–16.
Dua, D. and Graff, C. (2017). UCI machine learning reposit-
ory.
Gu, Q., Cai, Z., Zhu, L., and Huang, B. (2008). Data mining
on imbalanced data sets. In 2008 International Confer-
ence on Advanced Computer Theory and Engineering,
pages 1020–1024. IEEE.
Han, H., Wang, W.-Y., and Mao, B.-H. (2005). Borderline-
smote: A new over-sampling method in imbalanced data
sets learning. In Huang, D.-S., Zhang, X.-P., and Huang,
G.-B., editors, Advances in Intelligent Computing, pages
878–887, Berlin, Heidelberg. Springer Berlin Heidelberg.
Hart, P. (1968). The condensed nearest neighbor rule
(corresp.). IEEE Transactions on Information Theory,
14(3):515–516.
He, H., Bai, Y., Garcia, E. A., and Li, S. (2008). ADASYN:
Adaptive synthetic sampling approach for imbalanced
learning. In 2008 IEEE International Joint Conference
on Neural Networks (IEEE World Congress on Computa-
tional Intelligence), pages 1322–1328.
International Diabetes Federation (2019). IDF Diabetes
Atlas, 9th edn. Accessed on: 19/07/2021.
Kubat, M., Matwin, S., et al. (1997). Addressing the curse of
imbalanced training sets: one-sided selection. In ICML,
volume 97, pages 179–186. Nashville, USA.
Laurikkala, J. (2001). Improving identification of difficult
small classes by balancing class distribution. In Quaglini,
S., Barahona, P., and Andreassen, S., editors, Artificial
Intelligence in Medicine, pages 63–66, Berlin, Heidelberg.
Springer Berlin Heidelberg.
Li, D., Liu, C., and Hu, S. C. (2010). A learning method
for the class imbalance problem with medical data sets.
Computers in Biology and Medicine, 40(5):509–518.
Machado, D., Costa, V. S., and Brandão, P. (2022). Im-
pact of the glycaemic sampling method in diabetes data
mining. In 2022 IEEE Symposium on Computers and
Communications (ISCC), pages 1–6.
Mani, I. and Zhang, I. (2003). kNN approach to unbalanced
data distributions: a case study involving information
extraction. In Proceedings of workshop on learning from
imbalanced datasets, volume 126. ICML United States.
Marling, C. and Bunescu, R. (2020). The OhioT1DM data-
set for blood glucose level prediction: Update 2020. In
CEUR workshop proceedings, volume 2675, page 71.
NIH Public Access.
Mayo, M., Chepulis, L., and Paul, R. G. (2019). Glycemic-
aware metrics and oversampling techniques for predicting
blood glucose levels using machine learning. PLOS ONE,
14(12):e0225613.
Medtronic Diabetes (2014). Why sensor glucose does not
equal blood glucose. Accessed on: 29/03/2022.
Mouri, M. and Badireddy, M. (2021). Hyperglycemia. Stat-
Pearls [Internet]. [Updated 2021 May 10].
Nguyen, H. M., Cooper, E. W., and Kamei, K. (2011). Bor-
derline over-sampling for imbalanced data classification.
International Journal of Knowledge Engineering and Soft
Data Paradigms, 3(1):4–21.
Raval, K. M. (2012). Data mining techniques. International
Journal of Advanced Research in Computer Science and
Software Engineering, 2(10).
Seery, C. (2019a). Diabetes complications. guide on dia-
betes.co.uk. Accessed on: 20/07/2021.
Seery, C. (2019b). Short term complications. guide on
diabetes.co.uk. Accessed on: 20/07/2021.
Shukla, D., Patel, S. B., and Sen, A. K. (2014). A literature
review in health informatics using data mining techniques.
International Journal of Software and Hardware Research
in Engineering, 2(2):123–129.
Siegmund, T., Heinemann, L., Kolassa, R., and Thomas,
A. (2017). Discrepancies between blood glucose and in-
terstitial glucose—technological artifacts or physiology:
Implications for selection of the appropriate therapeutic
target. Journal of Diabetes Science and Technology,
11(4):766–772.
Tomek, I. et al. (1976). Two modifications of cnn. IEEE
Transactions on Systems, Man, and Cybernetics, SMC-
6(11):769–772.
Wilson, D. L. (1972). Asymptotic properties of nearest
neighbor rules using edited data. IEEE Transactions on
Systems, Man, and Cybernetics, SMC-2(3):408–421.
HEALTHINF 2023 - 16th International Conference on Health Informatics
198
