
Simulating Ultrasound Images from CT Scans
Sahar Almahfouz Nasser
a
and Amit Sethi
b
Electrical Engineering Department, Indian Institute of Technology Bombay, Mumbai, Maharashtra, India
Keywords:
Ultrasound, Simulation, Speckle Noise, CT, Stride, Reconstruction, Wave-equation, Devito.
Abstract:
Anatomical information in ultrasound (US) imaging has not been exploited fully because its wave interference
pattern (WIP) has been viewed as speckle noise. We tested the idea that more information can be retrieved
by disentangling the WIP rather than discarding it as noise. We numerically solved the forward model of
generating US images from computed tomography (CT) images by solving wave-equations using the Stride
library. By doing so, we have paved the way for using deep neural networks to be trained on the data generated
by the forward model to simulate the solution of the inverse problem, which is generating the CT-style and
CT-quality images from a real US image. We demonstrate qualitative features of the generated images that are
rich in anatomical details and realism.
1 INTRODUCTION
1.1 Background
Ultrasound is a non-ionizing imaging modality that
makes it a vital tool for medical imaging and image-
guided interventions. It is also portable and realtime,
unlike other imaging modalities, such as magnetic
resonance imaging (MRI) and computed tomography
(CT), which are rich in detail but are bulky, unweidly
and not real time. However, the presence of speckle
noise-like artifacts, blurring, and shading issues re-
duce the diagnostic value of US as an imaging modal-
ity.
Developing methods for US denoising is essential
to conduct a better diagnosis, assessment, and image-
guided interventions in real time (Duarte-Salazar
et al., 2020). The main artifact in US is often said
to be speckle noise. Speckle noise is a granular noise
with a multiplicative nature (Wagner, 1983), and (Ka-
plan and Ma, 1994). For instance, In synthetic-
aperture radar (SAR) images the observed signal, can
be described according to (Mather and Tso, 2016) as
follows:
f (x, y) = g(x, y)× n(x, y) + w(x, y) (1)
where f (x, y) is the observed signal, g(x, y) is the
original signal, n(x, y) is a multiplicative noise, and
w(x, y) is an additive noise.
a
https://orcid.org/0000-0002-5063-9211
b
https://orcid.org/0000-0002-8634-1804
However, while the noise in US appears to be
speckled in nature, it is actually wave interference
pattern (WIP), which is produced by additive and de-
structive interference of the ultrasound waves with the
tissue – a phenomenon which is known as scattering.
There are two types of scattering: diffuse scattering
and coherent scattering. Diffuse scattering generates
speckles in the image, while the coherent one yields
clear, dark, and bright features. Speckle noise in US
is, therefore, a signal-dependent noise, which relies
on the structure and the imaging factors of the imag-
ing system (Singh et al., 2017).
In this work, we describe a simulation method for
US images starting from 2D CT images. The main
purpose of this simulation is to generate paired im-
ages to learn the inverse model from US to CT, so
that deep neural networks can be trained for real time
and portable simulation of CT-like images with rich
anatomical details from regular real time US images
and videos. Such a simulation will bring the best of
the two modalities – portability and real time nature
of US with the clarity and details of CT – to diagnos-
tics and surgical intervention. Surprisingly, even the
forward model to simulate US from CT had not been
fully described in one place, and nor made available
as a software before our work.
In the rest of the paper, we describe US physics,
related work, and our proposed method. Then, we
show qualitative results of our method and conclude
with possible directions for future work.
138
Nasser, S. and Sethi, A.
Simulating Ultrasound Images from CT Scans.
DOI: 10.5220/0011780700003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 2: BIOIMAGING, pages 138-145
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
1.2 Ultrasound Physics
The US is a non-ionizing type of energy, which makes
it suitable for real-time and interactive medical imag-
ing.
US generation is based on the reverse piezoelec-
tric effect while detecting it is based on the piezoelec-
tric effect. The US waves propagate in the tissue in
two ways longitudinal and transverse. In longitudinal
propagation, the wave propagates in the same direc-
tion as the perturbation causing it. However, if the
wave propagation is perpendicular to the disturbance
generating it, it is called transverse propagation.
US interacts with tissues in four ways: reflec-
tion, refraction, absorption, and scattering (Tole et al.,
2005).
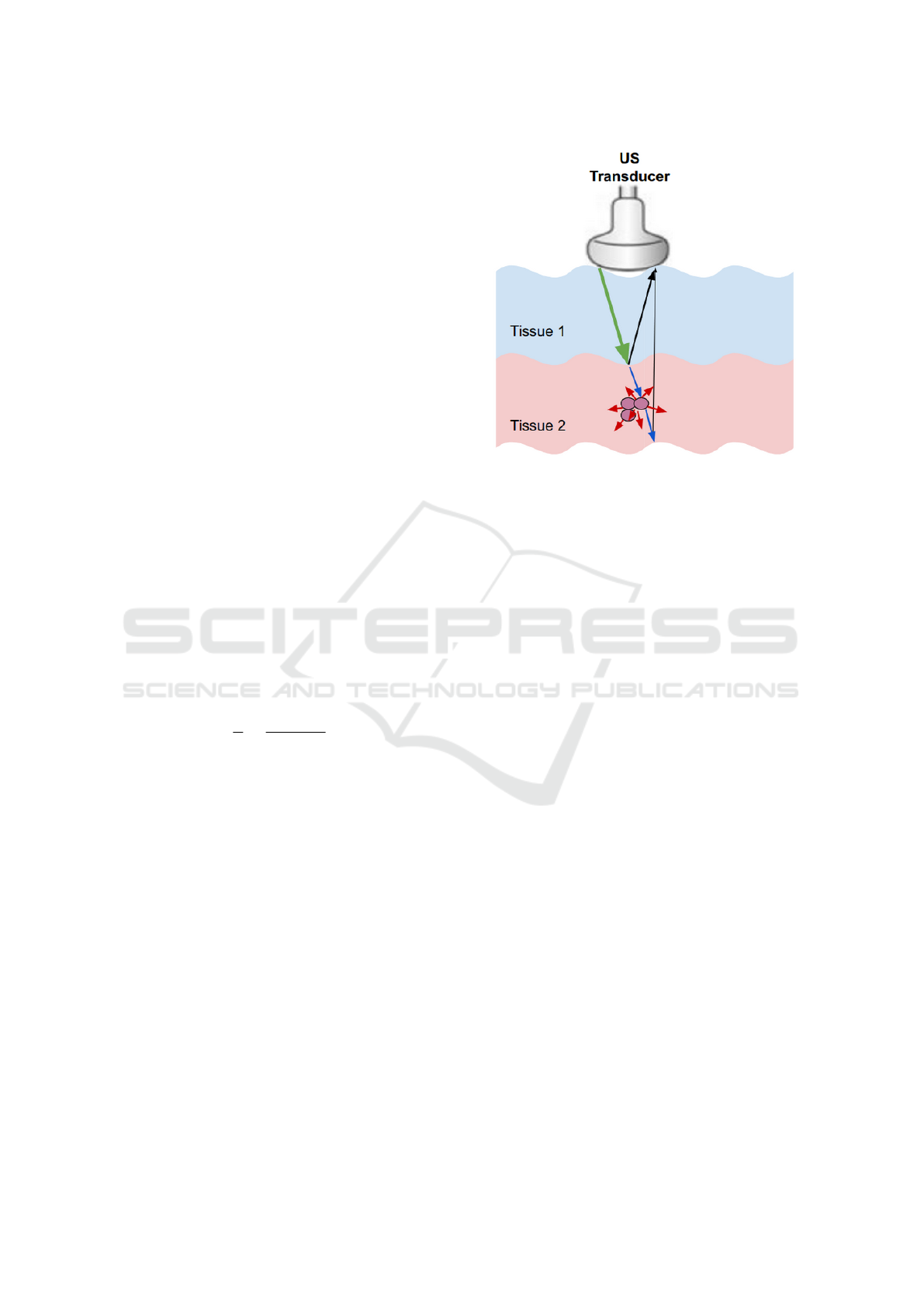
1.2.1 Reflection
Reflection happens at the boundaries between adja-
cent tissues – the acoustic boundaries. Based on the
size of the boundary relative to the US beam wave-
length, or the irregularities of the surface of the reflec-
tor, we can divide the reflection into two categories:
the specular reflection and the non-specular reflec-
tion. Specular reflection happens when the bound-
ary is smooth and longer than the beam dimensions,
while non-specular reflection occurs when the size of
the reflector is smaller than the wavelength of the ul-
trasound beam.
The reflection coefficient on the acoustic surface
is given by
I
r
I
i
=
(z
1
− z
2
)
2
(z
1
+ z
2
)
2
(2)
where I
i
is the intensity of the incident beam, I
r
is
the intensity of the reflected beam, z
1
is the acoustic
impedance of the first medium, and z
2
is the acoustic
impedance of the second medium.
The more the difference between the impedance
values (acoustic mismatch), the larger the echo.
The irregularity in the shape of the reflecting sur-
face or its small dimensions reflects the incident beam
in many directions is known as US scattering.
The scattering strongly depends on the US fre-
quency, so it increases as the frequency increases.
ν = f × λ (3)
where ν is the sound velocity, f is the frequency, and
λ is the wavelength. See Figure 1.
1.2.2 Absorption
During absorption, the US energy gets converted into
heat. Three factors affect absorption – the viscosity of
the medium, the relaxation time of the medium, and
Figure 1: Ultrasound interaction with tissues. The green,
black, red, blue arrows represent the incident wave, the
specular reflection, the non-specular reflection, and the re-
fracted wave, respectively.
the frequency of the beam. Absorption increases in
direct proportion to all three factors. The viscosity is
generated from the frictional forces between the par-
ticles. The relaxation time is the duration required for
the particles of the medium to get back to their mean
position after getting displaced by the US waves. Ab-
sorption also increases with the beam frequency, al-
though increased frequency can enhance details in the
US image.
1.2.3 Attenuation
Attenuation is the reduction of the beam intensity
caused by the total losses throughout the propagation.
Based on the power law (Cong et al., 2013), the en-
ergy attenuation of the ultrasound wave after its prop-
agation in the medium for a distance d is given by:
U(α
re f
, d) = U
in
× e
−2α
re f
d
(4)
where α is an attenuation parameter related to the
properties of the propagation medium.
2 RELATED WORK
As we mentioned in the abstract, many researchers
tried to simulate ultrasound images simply by model-
ing US with speckle noise and ignoring other interac-
tions of ultrasound with the tissue.
Goodman modeled speckle noise in laser images
by a Rayleigh distribution (Goodman, 1975). Wagner
et al (Wagner, 1983) represented the speckle noise by
Simulating Ultrasound Images from CT Scans
139

a Rician model. While Shankar (Shankar, 2000) came
up with Nakagami distribution to describe speckle
noise. Usually, Gamma distribution is the best ap-
proximation of speckle noise in SAR images (Ayed
et al., 2005). Zimmer (Zimmer et al., 2000) mod-
eled speckle noise in ultrasound liver images by a log-
normal distribution. Tao et al in (Tao et al., 2006)
proved that Gamma and Weibull distributions are bet-
ter approximations of speckle noise in clinical cardiac
ultrasound images than normal or log-normal distri-
butions.
In (Achim et al., 2001) and (Rabbani et al., 2008),
the authors proposed a method to convert the multi-
plicative noise into an additive noise by logarithmi-
cally transforming the image as follows:
I(x, y) = S(x, y)η(x, y), (5)
where I is the noisy observation (the US image), S is
the noise-free image, and η represents the multiplica-
tive speckle noise.
logI(x, y) = log(S(x, y)) + log(η
m
(x, y)) (6)
f (x, y) = g(x, y) + ε(x, y) (7)
Shams et al (Shams et al., 2008) proposed a novel
method for simulating ultrasound images from 3D CT
scans. In the proposed method, the authors started
with edge detection of the CT image to calculate the
reflection coefficients. Then they generated the scat-
tering image using FieldII (Jensen, 1996) by placing
scatterers with strength randomly chosen by Field II
from a normal distribution. The authors indicated that
using this method to create a realistic speckle pattern
is very computationally expensive. For instance, sim-
ulating a B-mode image with 128 RF scan lines takes
nearly two days.
Kutter et al (Kutter et al., 2009) proposed a
simulation-based registration pipeline of US to CT
images in real-time. They developed a simple ray-
based modeling of ultrasound images using OpenGL
software (Woo et al., 1999). They used a Lambertian
scattering model to simulate the scattered signal. And
they generated a scattering image using Field II soft-
ware. In (Reichl et al., 2009), the US intensity at the
location of the probe was adjusted at first. Then, the
amount of intensity transmitted, reflected, or absorbed
along each column (each scanline) of the image was
computed for every pixel according to the propagation
characteristics. After that, the reflection and the ab-
sorption were subtracted from the incident intensity at
every pixel. Finally, in the post-processing stage, arti-
facts such as speckle noise and blurring were added
to the ultrasound images. In their proposed work,
speckle noise was characterized by a Rayleigh distri-
bution.
Feng Gu et al. proposed a genrative adversarial
network (GAN) to model the speckle noise in syn-
thetic aperture radar (SAR) images (Gu et al., 2019).
In this work, we present a novel method for sim-
ulating the interaction pattern between the ultrasound
waves and the tissue, that is inspired by the underly-
ing physics of ultrasound image generation, starting
from CT images of different body parts.
3 PROPOSED METHOD
Our proposed method for generating ultrasound im-
ages from CT images consists of two stages – generat-
ing speed of sound images from CT images, and gen-
erating US images from speed of sound images. The
code of our proposed method is available at (Nasser
and Sethi, ).
3.1 Generating Speed of Sound Images
from CT Images
To generate the speed of sound images from CT im-
ages, we use the fact that the intensity value of a spe-
cific pixel of a 2D CT image represents the Hounsfield
unit (HU) of the underlying tissue that corresponds to
that pixel. HU is a measure of X-ray attenuation in
the tissue. Given a tissue x, the HU is given by:
HU
x
= 1000 ×
µ
x
− µ
water
µ
water
(8)
where µ
x
is the total linear attenuation coefficient of
the tissue x at a given x-ray energy. µ
x
of a tissue x can
be computed from multiplying the mass density of a
tissue x (ρ
x
) by the weighted sum of the mass attenu-
ation coefficients of all the elements which compose
the tissue x, as shown in the following equation:
µ
x
= ρ
x
∑
i
w
i
×
µ
i
ρ
i
(9)
where
µ
i
ρ
i
is the mass attenuation coefficient of the el-
ement (i) in cm
2
/g. Table 1 shows the elemental com-
position (w
i
values) of a few of the body tissues taken
from the ITIS database (ITI, ).
Given the kilovoltage peak applied to the X-ray
tube for capturing CT images is 120 kPv, and w
i
val-
ues from (ITI, ), we can compute the mass attenuation
coefficient of each element in the elemental composi-
tion of a certain tissue by using NIST software (NIS,
). NIST allows us to compute the mass attenuation
coefficients of the tissues, which, in turn, allows us to
compute their corresponding HUs by substituting the
values in equation 8.
BIOIMAGING 2023 - 10th International Conference on Bioimaging
140
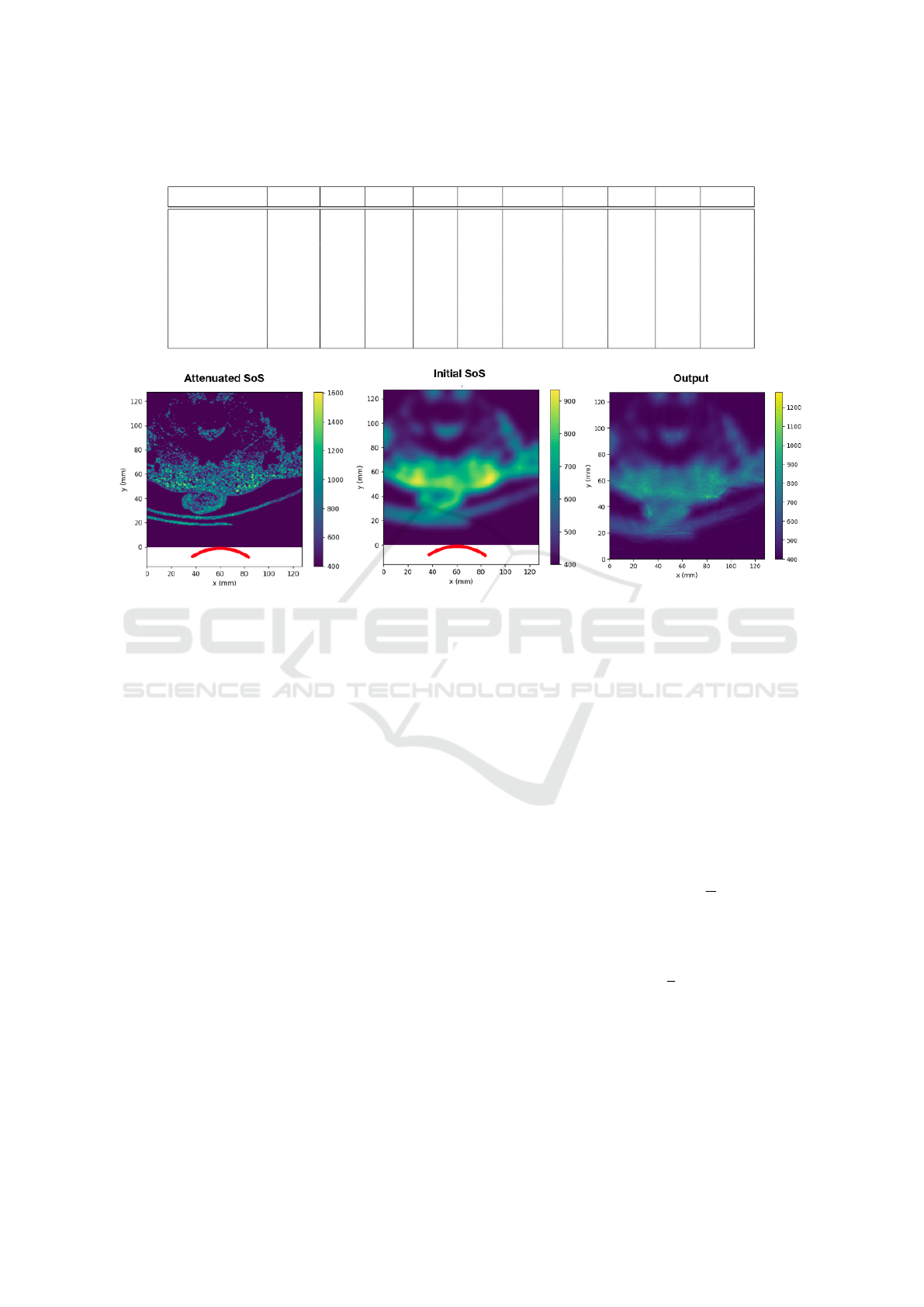
Table 1: Examples of the composition of a few of the body tissues. This table does not include the values of all the elements
of the tissue composition, other elements, such as silicon and phosphorus, can be found in (Ele, ).
Tissue Hydrogen Carbon Nitrogen Oxygen Sodium Magnesium Sulfur Chlorine Argon Potassium
Air 0 0.00015 0.78 0.21 0 0 0 0 0.0047 0
Liver 0.63 0.073 0.013 0.28 0.00054 0 0 0.0006 0.00058 0.00035
Prostate 0.64 0.046 0.011 0.3 0.00054 0.0002 0.00039 0 0 0.00032
Fat 0.5 0.37 0.003 0.13 0.00018 0.0005 0.00013 0.00012 0 0
Kidney 0.63 0.069 0.013 0.28 0.00054 0.0004 0.00039 0.00035 0 0.00032
Urinary Bladder Wall 0.64 0.049 0.011 0.29 0.00054 0.0004 0.00039 0.00052 0 0.00047
Urine 0.66 0.0025 0.0043 0.33 0.0011 0.0002 0 0.001 0 0.00031
Water 0.67 0 0 0.33 0 0 0 0 0 0
Figure 2: The images from left to right are the attenuated speed of sound (SoS) image (the input of the forward pass), the
initial speed of sound image (the input of the backward pass), and the output of Stride software.
Now having the speed of sound values and the cor-
responding HU values of the tissues at 37 Celsius, we
can generate the speed of sound images from the cor-
responding CT images.
We simulate the attenuation of US waves in tissues
using equation (4),
3.2 Generating US Images from Speed
of Sound Images
In the second stage of our proposed method, we sim-
ulate the US images from the speed of the sound
images. For simulating ultrasound images from the
speed of sound images which we generate, we use
Stride software (Cueto et al., 2022). Stride is an
open-source library for ultrasound computed tomog-
raphy. Unlike the methods based on full-waveform
inversion, Stride is not computationally expensive.
Stride is user-friendly software, and the code can be
run on CPUs and GPUs. This tool is based on a
domain-specific language called Devito which gener-
ates solvers of the wave-equation.
We can summarize the overall workflow of this
software to reconstruct the image of the tissue from
the measurements as follows:
1. The sensors produce acoustic waves, which prop-
agate throughout the medium. These propagated
waves get reflected on the acoustic boundaries of
the medium.
2. The reflected waves get captured by the receivers.
3. The acquired data is used to reconstruct the phys-
ical properties of the medium, for instance, the
speed of the wave through it and its density.
4. The reconstruction procedure minimizes the mis-
fit between the recorded measurements and the
numerically modeled ultrasound data.
Thus from a set of measurements of the pressure
wave field u, we can build an accurate model of the
discrete wave velocity C (or m =
1
c
2
) by consider-
ing it as a partial differential equation-constrained op-
timization problem, where the objective function is
given by:
minimize
m
Φ
s
(m) =
1
2
||p
r
u − d||
2
2
(10)
with u = A(m)
−1
P
t
s
q
s
, where p
r
is the sampling op-
erator of receiver locations, P
t
s
represents the injec-
tion operator at source locations, A(m) is the discrete
isotropic wave equation matrix, u is the discrete pres-
sure wave field, q
s
is the pressure source, and d is the
measured date.
Simulating Ultrasound Images from CT Scans
141
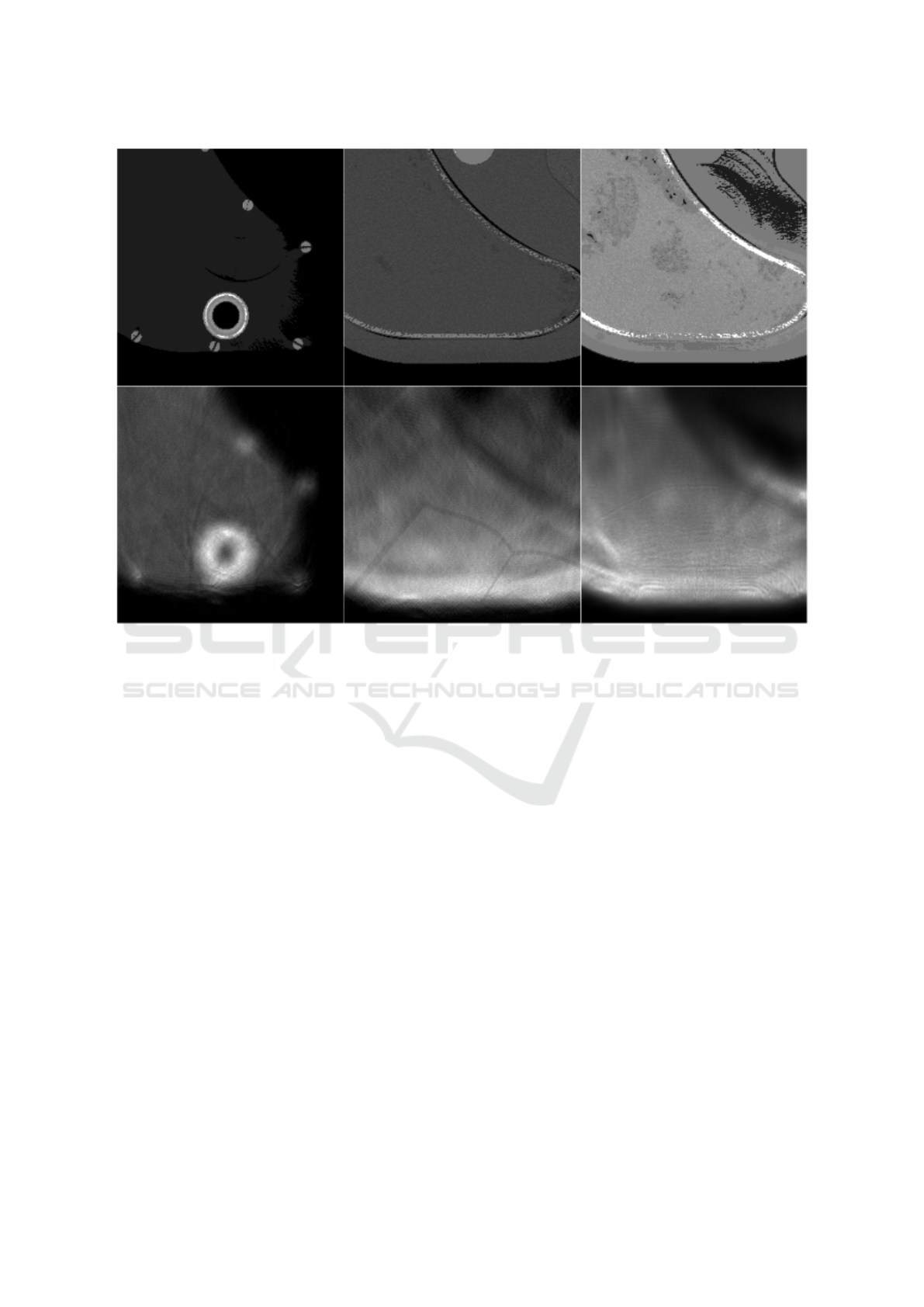
Figure 3: A few example output images of our proposed method for ultrasound simulation. The images are three different
slices of a phantom of the liver. The first row contains the speed of sound images generated from the corresponding CT images
(Ima, ). The second row contains the simulated ultrasound images. The experiments from left to right are a simulation using
a linear probe without attenuation, a simulation using a linear probe with attenuation, and a simulation using a curvilinear
probe with attenuation correspondingly.
By solving the optimization problem based on the
gradient method (Plessix, 2006) (Haber et al., 2012)
we get:
∆Φ
s
(m) = sum
n
t
t=1
u[t]ν
tt
[t] = J
T
δd
s
(11)
where n
t
is the number of steps, δd
s
= p
r
u − d is the
data residual between the measured and the modeled
data, J is the Jacobian operator, ν
tt
is the second-order
time derivative of the adjoint wave field A
T
(m)ν =
P
r
T
δd
s
.
4 RESULTS AND DISCUSSION
We designed a curvilinear transducer similar to the
one used by clinicians for abdominal imaging. The
transducer has a central frequency of 2.5 MHZ, a
number of elements equals 64, a radius of the cur-
vature equals 5R, and a curvature equals 50 mm.
The simulation consists of a forward pass and a
backward pass. For the backward pass, rather than
starting from a fixed speed of sound, we started from
an initial estimation of the speed of the sound image
to improve convergence. This initial estimation is a
blurred version of the ground truth speed of the sound
image. See Figure 2.
Figure 3 shows a few results of our proposed
method for ultrasound simulation. One can see in the
simulated US image the arc artifacts that are found in
real ultrasound images when a curvilinear transducer
is used.
In figure 4, we try to qualitatively assess our re-
sults by visually comparing the simulated images with
real US images. In figure 4, the CT images and real
US images were taken from (Ima, ), (AbU, ).
We refer to the work presented in (Kutter et al.,
2009) and (Reichl et al., 2009) for visual comparison
with the results of our proposed method. In our sim-
ulation, we use Stride software built upon the open-
source programming language (Python). While meth-
ods (Kutter et al., 2009) and (Reichl et al., 2009) use a
Matlab-based software called Field II. For a fair com-
parison between our methods and other methods, our
extended paper should incorporate a comparison be-
BIOIMAGING 2023 - 10th International Conference on Bioimaging
142
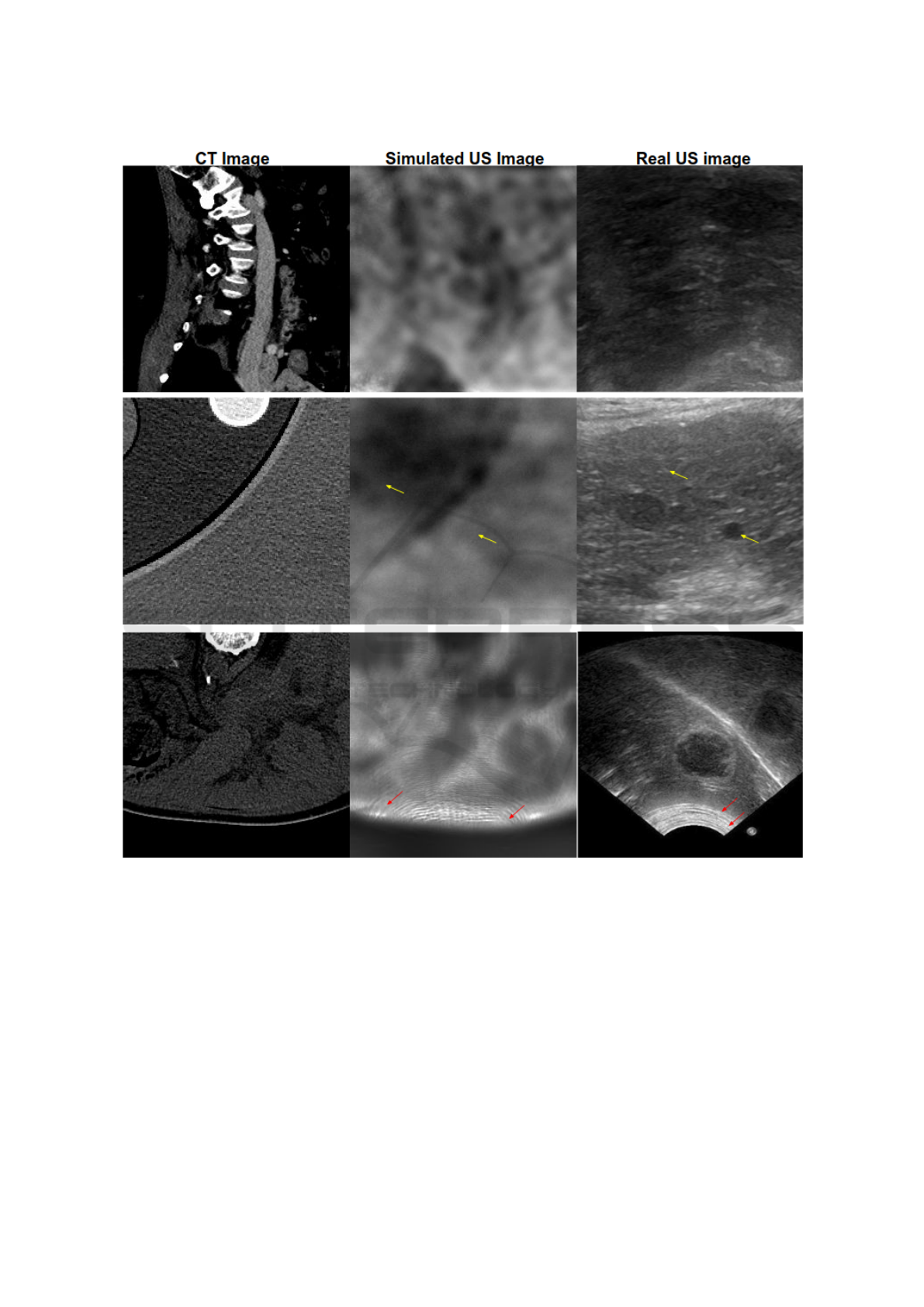
Figure 4: The first two columns contain CT images and the corresponding simulated ultrasound images, while the third
column contains real ultrasound images which do not correspond to CT images. The red arrows indicate the arc artifacts in
both real and simulated US images. The yellow arrows indicate the granular texture in simulated and real US images. For a
precise comparison between our simulated images and real US ones, we are planning further experiments in which we acquire
CT scans of a phantom to simulate ultrasound images using our proposed method. After that, we can compare the simulated
US images with the real US images of the same phantom.
tween these methods for the same ground truth CT
images, and the same US imaging probe.
5 CONCLUSIONS
In this work, we showed promising results in sim-
ulating US images from CT images. The wave in-
terference pattern and other artifacts were similar to
real US images. Our proposed method for obtain-
ing the simulated ultrasound images from the CT im-
Simulating Ultrasound Images from CT Scans
143

ages needs further improvement. After optimizing the
hyper-parameters of our simulation we will be able
to form a paired dataset (CT-Ultrasound pairs) which
can be used for training a generative adversarial net-
work GAN in a supervised manner and finally testing
it on reconstructing the CT images from the real US
images. The potential utility of this work is to train
deep neural networks for the inverse problem of sim-
ulating CT images from the given US images, which
can aid clinicians in diagnosis and surgical interven-
tion.
ACKNOWLEDGEMENT
This work would not have been possible without the
financial support of the Qualcomm Innovation Fel-
lowship Award, India. We are indebted to the devel-
oper of Stride, Mr. Carlos Cueto from Imperial Col-
lege London, for his feedback and support.
REFERENCES
Abdominalultrasonography. https://
todaysveterinarypractice.com/radiology-imaging/
imaging-essentialssmall-animal-abdominal-ultrasono
graphy-part-2-physical-principles-artifacts-false-
assumptions/. Accessed: 2021-10-24.
Cancer imaging archive. https://www.
cancerimagingarchive.net/. Accessed: 2021-10-
24.
Computing the mass attenuation coefficients of the ele-
mental composition of the tissues. https://www.nist.
gov/pml/xcom-photon-cross-sections-database. Ac-
cessed: 2021-10-24.
Element composition of the body tissues. https:
//itis.swiss/virtual-population/tissue-properties/
database/elements/. Accessed: 2021-10-24.
ITIS Foundation. https://itis.swiss/virtual-population/
tissue-properties/database/elements/. Accessed:
2021-10-24.
Achim, A., Bezerianos, A., and Tsakalides, P. (2001).
Novel bayesian multiscale method for speckle re-
moval in medical ultrasound images. IEEE transac-
tions on medical imaging, 20(8):772–783.
Ayed, I. B., Mitiche, A., and Belhadj, Z. (2005). Multire-
gion level-set partitioning of synthetic aperture radar
images. IEEE Transactions on Pattern Analysis and
Machine Intelligence, 27(5):793–800.
Cong, W., Yang, J., Liu, Y., and Wang, Y. (2013). Fast
and automatic ultrasound simulation from ct im-
ages. Computational and mathematical methods in
medicine, 2013.
Cueto, C., Bates, O., Strong, G., Cudeiro, J., Luporini, F.,
Agudo,
`
O. C., Gorman, G., Guasch, L., and Tang, M.-
X. (2022). Stride: A flexible software platform for
high-performance ultrasound computed tomography.
Computer Methods and Programs in Biomedicine,
221:106855.
Duarte-Salazar, C. A., Castro-Ospina, A. E., Becerra,
M. A., and Delgado-Trejos, E. (2020). Speckle noise
reduction in ultrasound images for improving the
metrological evaluation of biomedical applications:
an overview. IEEE Access, 8:15983–15999.
Goodman, J. W. (1975). Statistical properties of laser
speckle patterns. In Laser speckle and related phe-
nomena, pages 9–75. Springer.
Gu, F., Zhang, H., and Wang, C. (2019). A gan-based
method for sar image despeckling. In 2019 SAR in
Big Data Era (BIGSARDATA), pages 1–5. IEEE.
Haber, E., Chung, M., and Herrmann, F. (2012). An effec-
tive method for parameter estimation with pde con-
straints with multiple right-hand sides. SIAM Journal
on Optimization, 22(3):739–757.
Jensen, J. A. (1996). Field: A program for simulating ultra-
sound systems. In 10TH NORDICBALTIC CONFER-
ENCE ON BIOMEDICAL IMAGING, VOL. 4, SUP-
PLEMENT 1, PART 1: 351–353. Citeseer.
Kaplan, D. and Ma, Q. (1994). On the statistical character-
istics of log-compressed rayleigh signals: Theoretical
formulation and experimental results. The Journal of
the Acoustical Society of America, 95(3):1396–1400.
Kutter, O., Shams, R., and Navab, N. (2009). Visualization
and gpu-accelerated simulation of medical ultrasound
from ct images. Computer methods and programs in
biomedicine, 94(3):250–266.
Mather, P. and Tso, B. (2016). Classification methods for
remotely sensed data. CRC press.
Nasser, S. A. and Sethi, A. Simulat-
ing Ultrasound Images From Ct Scans.
https://github.com/SaharAlmahfouzNasser/Simulating-
Ultrasound-images-from-CT-Scans.git.
Plessix, R.-E. (2006). A review of the adjoint-state method
for computing the gradient of a functional with geo-
physical applications. Geophysical Journal Interna-
tional, 167(2):495–503.
Rabbani, H., Vafadust, M., Abolmaesumi, P., and Gazor, S.
(2008). Speckle noise reduction of medical ultrasound
images in complex wavelet domain using mixture pri-
ors. IEEE transactions on biomedical engineering,
55(9):2152–2160.
Reichl, T., Passenger, J., Acosta, O., and Salvado, O.
(2009). Ultrasound goes gpu: real-time simulation
using cuda. In Medical Imaging 2009: Visualiza-
tion, Image-Guided Procedures, and Modeling, vol-
ume 7261, pages 386–395. SPIE.
Shams, R., Hartley, R., and Navab, N. (2008). Real-time
simulation of medical ultrasound from ct images. In
International Conference on Medical Image Comput-
ing and Computer-Assisted Intervention, pages 734–
741. Springer.
Shankar, P. M. (2000). A general statistical model for ultra-
sonic backscattering from tissues. IEEE transactions
on ultrasonics, ferroelectrics, and frequency control,
47(3):727–736.
BIOIMAGING 2023 - 10th International Conference on Bioimaging
144

Singh, P., Mukundan, R., and de Ryke, R. (2017). Synthetic
models of ultrasound image formation for speckle
noise simulation and analysis. In 2017 International
Conference on Signals and Systems (ICSigSys), pages
278–284. IEEE.
Tao, Z., Tagare, H. D., and Beaty, J. D. (2006). Evaluation
of four probability distribution models for speckle in
clinical cardiac ultrasound images. IEEE transactions
on medical imaging, 25(11):1483–1491.
Tole, N. M. et al. (2005). Basic physics of ultrasonographic
imaging. World Health Organization.
Wagner, R. F. (1983). Statistics of speckle in ultrasound
b-scans. IEEE Trans. Sonics & Ultrason., 30(3):156–
163.
Woo, M., Neider, J., Davis, T., and Shreiner,
D. (1999). OpenGLprogramming-
guide:theofficialguidetolearningOpenGL,version1.2.
Addison-WesleyLongmanPublishingCo.,Inc.
Zimmer, Y., Tepper, R., and Akselrod, S. (2000). A lognor-
mal approximation for the gray level statistics in ul-
trasound images. In Proceedings of the 22nd Annual
International Conference of the IEEE Engineering in
Medicine and Biology Society (Cat. No. 00CH37143),
volume 4, pages 2656–2661. IEEE.
Simulating Ultrasound Images from CT Scans
145
