Separation of Concerns in an Edge-Based Compartmental Modeling
Framework
A. Yvan Guifo Fodjo
1,2 a
, Jerry Lacmou Zeutouo
2,3 b
and Samuel Bowong
4,5
1
CNRS, UMR 7606, LIP6, Sorbonne Universit
´
e, Paris, France
2
URIFIA, Universit
´
e de Dschang, Dschang, Cameroon
3
Inria Avalon, LIP,
´
Ecole Normale Sup
´
erieure de Lyon, University of Lyon, France
4
IRD, UMI 209, UMMISCO, Bondy, France
5
D
´
epartement de Math
´
ematiques, Universit
´
e de Douala, Douala, Cameroon
Keywords:
Separation of Concerns, Compartmental Models, Contact Network, Epidemiology Modeling Tool, Edge-
Based Compartmental Network.
Abstract:
A well-known framework with strong potential for epidemic prediction and the ability to incorporate realistic
contact structures is edge-based compartmental modeling (EBCM). However, models built from this frame-
work lead to a multiplication of ordinary differential equations and many parameters to be estimated, which
make the models complex and difficult to extend or to reuse. The Kendrick approach has shown promising re-
sults in generalizing compartmental models to take into account aspects of contact networks while preserving
the separation of concerns, thus allowing to define modular, extensible and reusable models. But this general-
ization of compartmental models to contact network aspects is still limited to a few contact networks. In this
paper, we present an attempt to extend Kendrick’s approach from an approximation of EBCM models to fur-
ther support aspects of contact networks, thereby improving the predictive quality of models with significant
heterogeneity in contact structure, while maintaining the simplicity of compartmental models. This extension
consists of an integration of the basic reproductive number R
0
into the compartmental SIR framework. This
attempted is validated using Miller’s mass action and the approximation of EBCM configuration model.
1 INTRODUCTION
Mathematical modeling and computer simulation
have been widely used in epidemiology to find appro-
priate control strategies and means of control (Levin
and Durrett, 1996). One of the most common mod-
els in mathematical modeling using the compartmen-
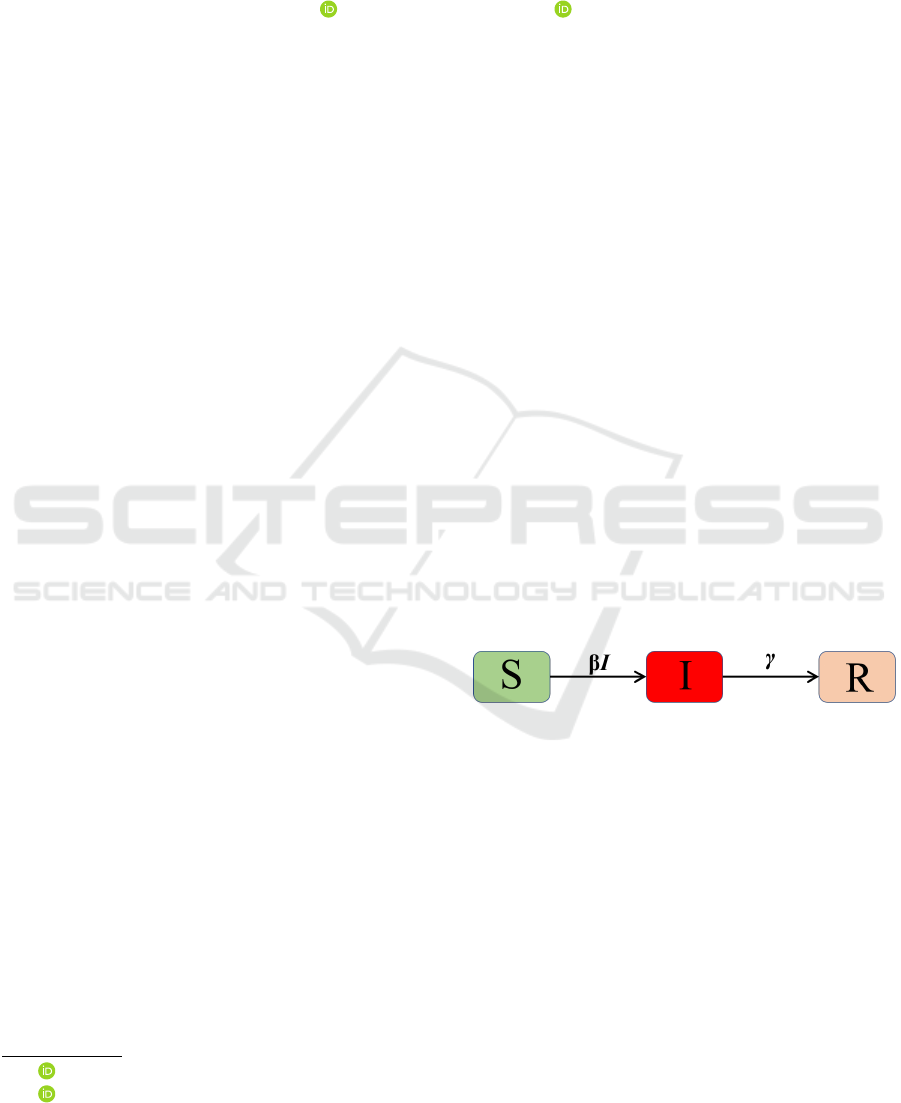
tal framework is the mass action model (see Figure
1). The basic assumption in this model is that sus-
ceptible and infectious individuals meet at random
and can spread the disease (Cuddington and Beisner,
2005; Keeling and Rohani, 2011). Susceptible indi-
viduals (S) can become infected from the force of in-
fection
1
λ(S,I,N) = βI where β is the contact trans-
mission rate and N is the population size. Infectious
individuals (I) can be recovered at the recovery rate γ.
a
https://orcid.org/0000-0002-0714-6737
b
https://orcid.org/0000-0003-4414-7453
1
This is the rate at which susceptible individuals be-
come infected.
Figure 1: Flow diagram of the mass action mathematical
model.
S
′
= −βIS
I
′
= βIS − γI
R
′
= γI
(1)
Compartmental models are typically first defined
as ordinary differential equations (ODEs) such as
Equation 1. These models can be studied ana-
lytically and/or simulated using algorithms such as
RungeKutta. However, it is considered more real-
istic to adopt a stochastic viewpoint on these mod-
els considering them as Continuous-Time Markov
Chains (CTMCs). The latter can be derived from the
ODEs modulo some widely accepted, albeit simplify-
ing, probabilistic assumptions.
Although the mass action model is known largely
for its conceptual and mathematical simplicity (Ker-
mark and Mckendrick, 1927), it has been found to
262
Guifo Fodjo, A., Zeutouo, J. and Bowong, S.
Separation of Concerns in an Edge-Based Compartmental Modeling Framework.
DOI: 10.5220/0011780200003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 3: BIOINFORMATICS, pages 262-269
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

have some shortcomings, including the fact that it in-
correctly assumes that all individuals have the same
contact rate and implicitly assumes that all partner-
ships are infinitely short (Miller et al., 2012; Wang
et al., 2018).
(Miller et al., 2012; Kiss et al., 2017; Wang et al.,
2018) built on these limitations of the mass action
model to introduce the edge-based compartmental
modelling (EBCM) framework. It is a compartment-
based model extension approach where the contacts
between individuals are materialized by the edges
(Miller et al., 2012; Wang et al., 2018). The main idea
is to integrate the heterogeneous mixture by consider-
ing the population as a network of individuals where
the contacts follow a given probability distribution.
Thus, it consists in incorporating into the ordinary dif-
ferential equations of the mass action model, a kind
of social heterogeneity (heterogeneous contact rate),
while taking into account the impact of the partner-
ship duration
2
.
The results obtained from this incorporation have
enabled realistic and more predictable contact struc-
tures to be considered. However, the modification of
the compartmental framework of mass action results
in more and more multiple and complex differential
equations, especially when new concerns (age, sex,
control strategy, etc.) and parameters are taken into
account (Balde et al., 2019). Moreover, another dif-
ficulty of the EBCM framework is that the models it
contains are not easily scalable, extensible and there-
fore not reusable.
Kendrick’s approach (Bui et al., 2016; Bui et al.,
2019) has shown its ability to define concerns (age,
sex, spatial heterogeneity, etc.) as independent (pos-
sibly incomplete) models that are then combined
into stochastic automata networks (SANs) (Plateau
and Stewart, 2000) using a tensorial sum operator.
Stochastic dependencies between concerns are then
introduced in a second phase so that independent con-
cerns can be reused and combined in other models
much more easily. In (Fodjo et al., 2022), the authors
showed that this approach, based on compartmental
models, could be extended to take into account cer-
tain aspects of contact networks while enabling the
building of reusable models. But this generalization
of compartmental models to contact network aspects
is still limited to a few contact networks (Poisson, Ex-
ponential and Scale free).
While it is well known that taking into account as-
pects of contact networks in compartmental models
leads to more realistic and predictive models, there
2
The notion of partnership, materializes here the exis-
tence of a contact (edge), between two individuals. In this
case, we say that these individuals are partners.
is also a large body of work that recognizes the cru-
cial importance of the basic reproductive number R
0
in predicting epidemics (Meyers et al., 2005; Danon
et al., 2011; Molina and Stone, 2012; Heesterbeek
et al., 2015; Zhang et al., 2015; Trapman et al., 2016;
Yang and Xu, 2019). This parameter refers to the
number of new cases caused by a single randomly in-
fected individual in a completely susceptible popula-
tion. When R
0
< 1, epidemics are impossible, while
when R
0
> 1, they are possible. (Aparicio and Pas-
cual, 2007) suggest modifying the SIR compartmen-
tal framework to incorporate the parameter R
0
into the
ordinary differential equations. In this study, in order
to take the aspects of contact networks, R
0
is approx-
imated to Poisson, Exponetial and Scale free contact
networks.
In this work, we propose an attempt to extend
Kendrick’s approach from an approximation of the
EBCM approach while maintaining the separation of
concerns and preserving the simplicity of compart-
mental models. This approximation consists in con-
structing a concern in the sense of Kendrick’s ap-
proach (i.e. as a stochastic automaton that can then
be combined) from the R
0
of each EBCM model. The
extension of the Kendrick approach is done by incor-
porating the base reproduction rate R
0
into the com-
partmental SIR framework. The simulation results
obtained are similar to those of the EBCM configu-
ration model presented in (Miller et al., 2012).
2 MILLER ET AL.’S MASS
ACTION MODEL AND
EDGE-BASED
COMPARTMENTAL
CONFIGURATION MODELS
In this section, and for the purposes of this work,
we will restrict ourselves to the standard mass ac-
tion model of (Miller et al., 2012) and the edge-based
compartmental configuration model.
2.1 Miller et Al.’s Mass Action Model
This model is constructed like the system of Equa-
tions 1) with some modifications. Such as the fact
that the authors of (Miller et al., 2012) assume that an
infected individual causes new infections at the rate
ˆ
βS(t), where
ˆ
β is the transmission rate per infected
Separation of Concerns in an Edge-Based Compartmental Modeling Framework
263
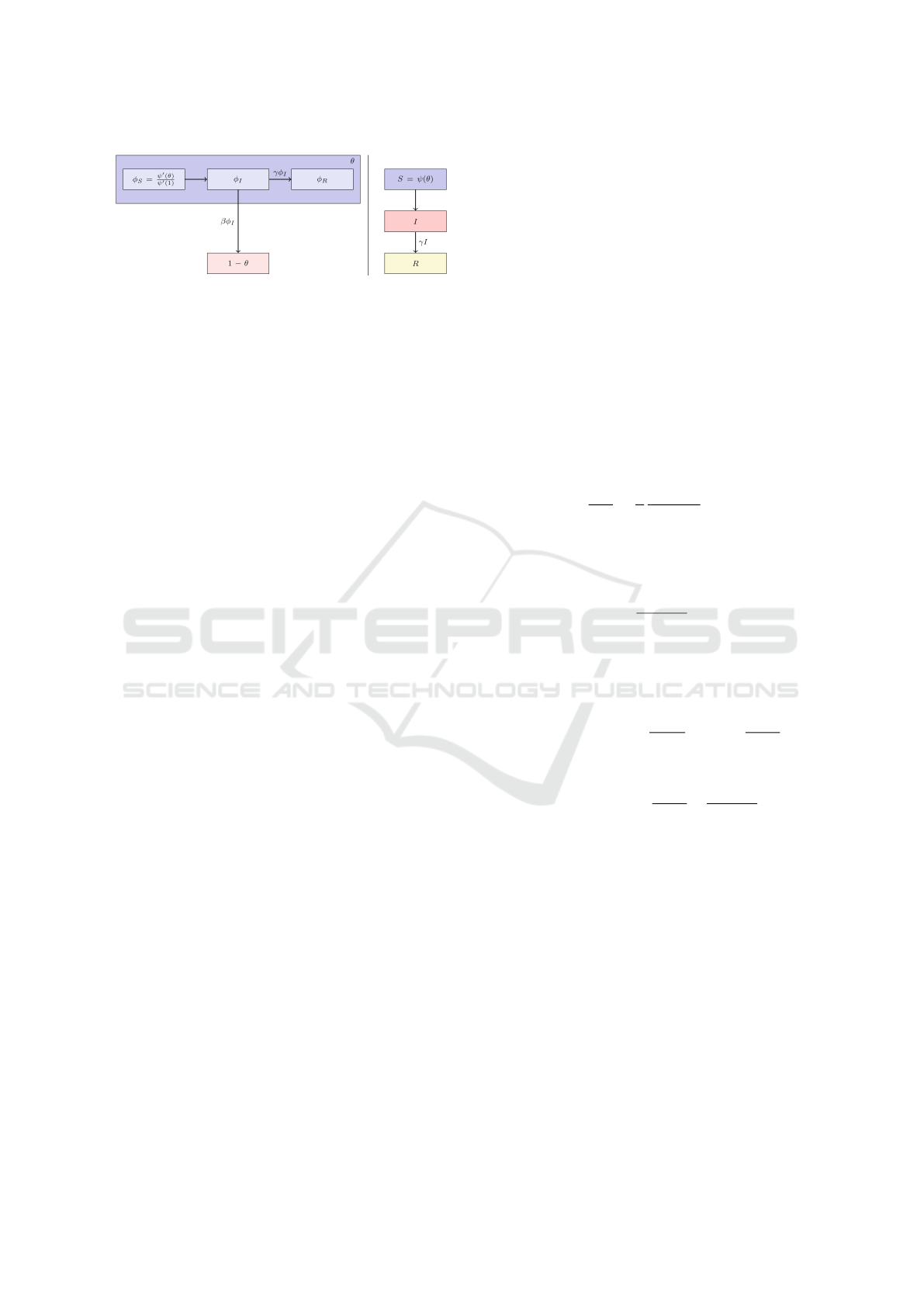
Figure 2: Flow diagram of the EBCM configuration model
of (Miller et al., 2012).
individual. Recovery occurs at the rate of γ.
S
′
= −
ˆ
βIS
I
′
=
ˆ
βIS − γI
R
′
= γI
(2)
Moreover, when building the mass action model (see
system of Equations 1), many works (Cuddington and
Beisner, 2005; Keeling and Rohani, 2011; Martcheva,
2015) interpret the transmission rate β as the product
of the contact rates and the transmission probability.
But in the specific case of the mass action model of
Miller et al.’s, the authors assume that the transmis-
sion rate per infected person
ˆ
β = β⟨k⟩ where ⟨k⟩ is
the average degree of the contact network considered.
2.2 EBCM Configuration Model
The flow diagram of the EBCM configuration model
is shown in Figure 2. The compartments S, I, and R
represent the proportions of susceptible, infected, and
recovered, respectively, as in the case of the mass ac-
tion model in section 2.1. To calculate S(t), I(t), and
R(t), the authors note that these are the probabilities
that a random test node u is in each compartment. We
calculate S(t) noting that this is also the probability
that none of u’s partners has yet transmitted to u. The
probability that a randomly selected partner v has not
yet transmitted the infection to u.
For large networks, (Miller et al., 2012; Wang
et al., 2018) assumed that the neighbors of the node
test u are independent. Given a degree k, u is sus-
ceptible at time t with probability s(k,θ(t)) = θ(t)
k
.
Thus, S(t) =
∑
k
P(k)s(k,θ(t)) = ψ(θ(t)). The ODEs
of the EBCM configuration model are obtained in (3).
S = ψ(θ)
I = 1 − S − R
(3)
The φ
S
, φ
I
and φ
R
compartments of Figure 2 repre-
sent respectively the probabilities that a partner v is
susceptible, infected and recovered but has not yet
transmitted the infection to u. The compartment 1−θ,
represents the probability that there is infection (it is
done with the rate βφ
I
between the compartment φ
I
and 1 − θ).
In order to determine θ, consider that (θ = φ
S
+
φ
I
+ φ
R
). At time t = 0, θ = φ
S
≃ 1. Because we
consider that there has not yet been an infection, so
φ
I
= φ
R
≃ 0.
The central parameter of this calculation is the de-
termination of φ
I
. Thus,
φ
I
= θ − φ
S
− φ
R
(4)
It remains to calculate φ
S
and φ
R
explicitly to obtain
φ
I
. If we finally consider that the v is infected and that
there was an infection, then
θ
′
= −βφ
I
(5)
To determine φ
R
, the authors use the fact that in Figure
2 the fluxes from φ
I
to φ
R
and from φ
I
to (1 − θ) are
proportional to one another. Both φ
R
and (1 − θ) are
equal to zero at time zero since we assume that no
infection or recovery events can occur prior to time
zero. By integrating the relation
dφ
R
dt
=
γ
β
d(1 − θ)
dt
(6)
and using the initial condition
φ
R
(0) = (1 − θ(0)) = 0 (7)
Thus,
φ
R
=
γ(1 − θ)
β
(8)
To determine φ
S
, the authors rely on the fact that
a partner v has a degree k with probability P
n
(k) =
kP(k)/⟨k⟩. Given a degree k, v is susceptible with
probability θ
(k−1)
. This allows us to obtain
φ
S
=
∑
k
P
n
(k)θ
(k−1)
=
∑
k
kP(k)
⟨k⟩
θ
(k−1)
=
ψ
′
(θ)
ψ
′
(1)
(9)
Therefore, from the relation 4, they obtain
φ
I
= θ − φ
S
− φ
R
= θ −
ψ
′
(θ)
ψ
′
(1)
−
γ(1 − θ)
β
(10)
While it is easy to recognize that the EBCM config-
uration model captures much more population struc-
ture than the mass action model, it is also important
to note that the complexity and number of differential
equations increases significantly. As we have seen,
to obtain Equation (10), the authors of (Miller et al.,
2012) had to develop new Equations 4, 5, 6, 7, 8, 9 in
addition to the one of the system of (3).
3 GENERALIZING KENDRICK’S
APPROACH
The challenge of this work is to show that Kendrick’s
approach can be extended to support aspects of con-
tact networks using an approximation of the EBCM
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
264

approach. This would improve the predictive qual-
ity of models with significant heterogeneity in the
structure of contacts while preserving the separation
of concerns. This would result in modular models,
easily extensible and reusable. The idea is to avoid
the designers/modelers/developers having to build a
model with a multitude of ordinary differential equa-
tions, having to build an explicit contact network and
having to estimate many parameters.
(Fodjo et al., 2022) showed that it was possible
to extend the compartmental framework in order to
integrate some aspects of contact network models.
Their idea was to make the force of infection a cen-
tral parameter that could be redefined from the exten-
sion points (α
gen
3
, it
gen
4
, τ
gen
5
) and according to the
concerns that one wished to model. Thus, from the
idea of (Bansal et al., 2007) and the application of
the Template Method Design Pattern (Gamma et al.,
1995), they proposed a generic definition to the force
of infection named λ (see Equation 11). (Fodjo et al.,
2022) insisted on the fact that the index ”gen” of the
extension points α
gen
, it
gen
and τ
gen
is intended to sig-
nify that they are applied to a model based on a net-
work of contacts and to indicate that they are generic
points, i.e. variable points.
λ = α
gen
it
gen
τ
gen
(11)
For our modeling and simulation purposes, we use
the same convention as the authors of (Fodjo et al.,
2022). Thus we use names with the subscript ”gen”
in the final generic definitions of the models and in
the Kendric code.
The force of infection of the mass action model
given by (2) described in section 2.1 is given by the
relation 12.
λ =
ˆ
βI = β⟨k⟩I (12)
From the relations 11 and 12 the identification of the
extension points α
gen
, it
gen
and τ
gen
gives :
α
gen
= ⟨k⟩
it
gen
= I
τ
gen
= β
(13)
In section 2.2, we have presented the EBCM config-
uration model and the array of ODEs and parameters
to be estimated from the model. This constitutes more
effort of understanding, programming for the model
designers/modelers/developers. Our goal is to cope
with the complexity of ordinary differential equations
3
It is the average number of individuals with whom a
susceptible individual is in contact or the average degree of
nodes in a contact network.
4
This is the proportion of contacts that are infectious.
5
The rate per contact at which disease is transmitted be-
tween an infectious individual and a susceptible individual.
in the EBCM framework while defining extensible
and reusable models.
In order to cope with the complexity of the or-
dinary differential equations of the EBCM frame-
work while defining extensible and reusable models,
we propose to use an approximation of each EBCM
model from the mathematical formulation of R
0
.
Indeed, (Aparicio and Pascual, 2007) showed that
it was possible to modify the simple compartmental
framework in order to integrate the parameter R
0
6
into
the model equations.
We propose to this effect to approximate the
EBCM models, by incorporating each R
0
of EBCM
model in the compartmental framework SIR (Suscep-
tible, Infected, Recovered) as shown in (Aparicio and
Pascual, 2007), so as to define these EBCM model ap-
proximations as concerns in the sense of the Kendrick
approach (i.e. a stochastic automaton that can be com-
posed) as illustrated in (Fodjo et al., 2022) and then to
be able to determine the force of infection 11, which
it would then be possible to decompose into different
points of extensions α
gen
, it
gen
and τ
gen
.
Thus, the results of this incorporation of R
0
into
the SIR compartmental framework would give (14) :
S
′
= −R
0
SI
I
′
= R
0
SI − γI
R
′
= γI
(14)
(Miller et al., 2012) gave the following formulation in
the case of the EBCM configuration model :
R
0
=
β
(β + γ)
⟨k
2
− k⟩
⟨k⟩
(15)
From (14), the force of infection of the EBCM ap-
proximation is given by the relation
λ = R
0
I =
β
(β + γ)
⟨k
2
− k⟩
⟨k⟩
I (16)
Thus from the relation 11 the identification of the ex-
tension points allows us to obtain :
α
gen
=
⟨k
2
− k⟩
⟨k⟩
I
gen
= I
τ
gen
=
β
(β + γ)
(17)
In the relationship 17, the term α
gen
=
⟨k
2
− k⟩
⟨k⟩
is ob-
tained from the python code of the EoN module
7
for
6
This parameter was an approximation of the different
contact networks considered.
7
https://epidemicsonnetworks.readthedocs.io/en/latest/
GettingStarted.html
Separation of Concerns in an Edge-Based Compartmental Modeling Framework
265

each degree distribution (Homogeneous, Poisson, Bi-
modal and PowerLaw) specified for the graph gen-
eration. For example, the code to obtain α
gen
from
the configuration model generated from a Bimodal
degree distribution
8
. We proceed in a similar way
for distributions of degree Homogeneous
9
and Power-
Law degree distribution
10
, but for the particular case
of the Poisson degree distribution, ⟨k
2
− k⟩ = ⟨k⟩
2
(Miller et al., 2012) thus, α
gen
=
⟨k
2
− k⟩
⟨k⟩
=
⟨k⟩
2
⟨k⟩
=
⟨k⟩ .
4 VALIDATION AND
DISCUSSION
To validate our approach, we replicated the EBCM
configuration model experiments of (Miller et al.,
2012) using the approximation of the EBCM config-
uration model on the distributions
11
(Homogeneous,
Poisson, Bimodal, and PowerLaw) in Kendrick (Bui
et al., 2019; Fodjo et al., 2022). In our implementa-
tions, the variable points of the λ parameter of (11) are
named ”alphagen”, ”itgen” and ”taugen”. The models
are built in different entities that are easy to define in
modules, extensible and reusable. Indeed, each model
generally includes an entity of definition of the con-
cerns then of composition of model (see Figure 6),
an entity of initialization of parameters (see Figure 7)
and finally an entity of simulation (lines 36 to 41) and
visualization (lines 43 to 49) (see Figure 8).
The challenge was to check if Kendrick’s ap-
proach to separation of concerns could capture ap-
proaches such as Miller et al.’s mass action model or
the approximation of the EBCM configuration model
while maintaining the familiar compartmental frame-
work.
The implementation of the mass action model (see
(2)) allowed us to obtain the black curve of Figure 9.
For this implementation, the code is subdivided into
two major parts. The first one is the definition of the
basic concern SIR (Susceptible, Infected and Recov-
8
https://github.com/YvanGuifo/EBCM-
ConfigurationModel/blob/main/generate heterogeneity CM
Bimodal.py
9
https://github.com/YvanGuifo/EBCM-
ConfigurationModel/blob/main/generate heterogeneity CM
Homogeneous.py
10
https://github.com/YvanGuifo/EBCM-
ConfigurationModel/blob/main/generate heterogeneity CM
PowerLaw.py
11
The Python and Kendrick code of our experi-
ments is available under https://github.com/YvanGuifo/
EBCM-ConfigurationModel
Figure 3: Definition of the classical concern SIR where λ
and the extension points are defined in lines 9 to 10.
Figure 4: Definition of the concern of mass action in lines
16 to 22. Then composition of the classic concern and the
mass action concern in line 24.
Figure 5: Composition of the basic SIR concern and the
approximation concern of the EBCM configuration model
on a Bimodal degree distribution.
Figure 6: Definition of the concern of the approximation of
the EBCM configuration model on a Bimodal degree distri-
bution.
ered) where λ is defined (see Figure 3). In this imple-
mentation, λ is defined in a general way (see lines 9 to
10). The second part of the code, is the implementa-
tion of the mass action concern named here ”maCon-
cern” as shown in Figure 4. In this definition, lines
16 to 22 allow to redefine the extension points ”al-
phagen”, ”itgen” and ”taugen”. Then in line 24 we
compose the basic concern and the mass action con-
cern.
As for the implementation of the approximation
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
266

of the EBCM configuration model in Kendrick, we
proceed in a similar way to the mass action model.
But in the case of this model (see Figure 6), the ex-
tension point ”alphagen” is obtained according to the
specified degree distribution. In the case of the Bi-
modal degree distribution for example, the value of
”alphagen” is given in line (17 to 18) of Figure 6.
Note that in our implementations, for approximation
of the EBCM configuration model what varies from
one degree distribution to another is the value of ”al-
phagen”. After having defined the concerns of the ba-
sic SIR model (see Figure 3) and the approximation
of the EBCM configuration model (see Figure 6), we
compose the different concerns as illustrated in Fig-
ure 5. The results obtained enable us to have in Fig-
ure 9 the curves of the various distributions of degrees
of the model of configuration EBCM implemented in
Kendrick.
It can be seen that, whether it is the mass action
model of Miller et al.’s. or the approximation of the
EBCM configuration model, Kendrick’s approach al-
lows us to easily define the concerns (”maConcern”
and ”cmBimodal” in the case of the approximation
of the EBCM configuration model on a Bimodal de-
gree distribution) in a way that is separate from the
basic SIR concern ”sirConcern”. Therefore, the basic
concern ”sirConcern” can be reused without ”maCon-
cern” or ”cmBimodal”. Kendrick’s approach enabled
us to define ”myConcern” and ”cmBimodal” as inde-
pendent models that can then be composed.
Regarding the results obtained while implement-
ing the EBCM configuration model on the Power-
law degree distributions (see orange curves of Fig-
ures 9 and 9), the same dynamics are observed but
with a difference regarding the infectious. This can
be explained by the fact that some individuals (super-
spreaders) have an abnormally high number of con-
tacts at the beginning of the epidemic. We often note a
strong propensity of individuals to attach themselves
preferentially to the individual with more contacts in
this type of network. We also note that the moment of
the epidemic peak is reached at the same time.
EBCM configuration model simulations on homo-
geneous, Poisson, and bimodal degree distributions
using our EBCM model approximation approach (see
Figure 9) yield improved curve heights (lower curves)
compared to a typical configuration model approach
on homogeneous, Poisson, and bimodal degree distri-
butions simulated with EoN (see Figure 9). Further-
more, we note that the curves of Figure 9 have the
same dynamics as those seen in Figure 9 and signif-
icantly improve the predictive quality that one would
expect from a mass action model (black curve). We
also note that the timing of the epidemic peak of the
Figure 7: Initialization of simulation parameters.
Figure 8: Simulation and visualization of the configuration
model.
homogeneous, Poisson and bimodal curves in Figure
9 is slightly slower compared to the respective homo-
geneous, Poisson, and bimodal curves in Figure 9.
Our approach to approximating EBCM models
can be applied to mean field social heterogeneity
(MFSH) algorithms
12
and a fixed-degree dynamic
model (DFD)
13
(Miller et al., 2012; Istvan et al.,
2019).
5 CONCLUSION
In this paper, we proposed to generalize Kendrick’s
approach to the consideration of realistic contact
structures from the edge-based compartmental mod-
eling (EBCM) framework while preserving the sepa-
ration of concerns in compartmental epidemic models
(Bui et al., 2016). To do this, we applied the solution
of (Fodjo et al., 2022) which involved defining the
usual λ parameter of epidemic models as a kind of
model method with three extension points, which al-
lowed us to easily capture aspects of contact network
models.
12
It’s model where contact rates in the population are as-
signed using a P(k) or ρ(k) distribution, while considering
that the contact duration is negligible.
13
It’s model in which each node in the dynamic network
has a constant-valued degreed, assigned using the P(k) dis-
tribution.
Separation of Concerns in an Edge-Based Compartmental Modeling Framework
267
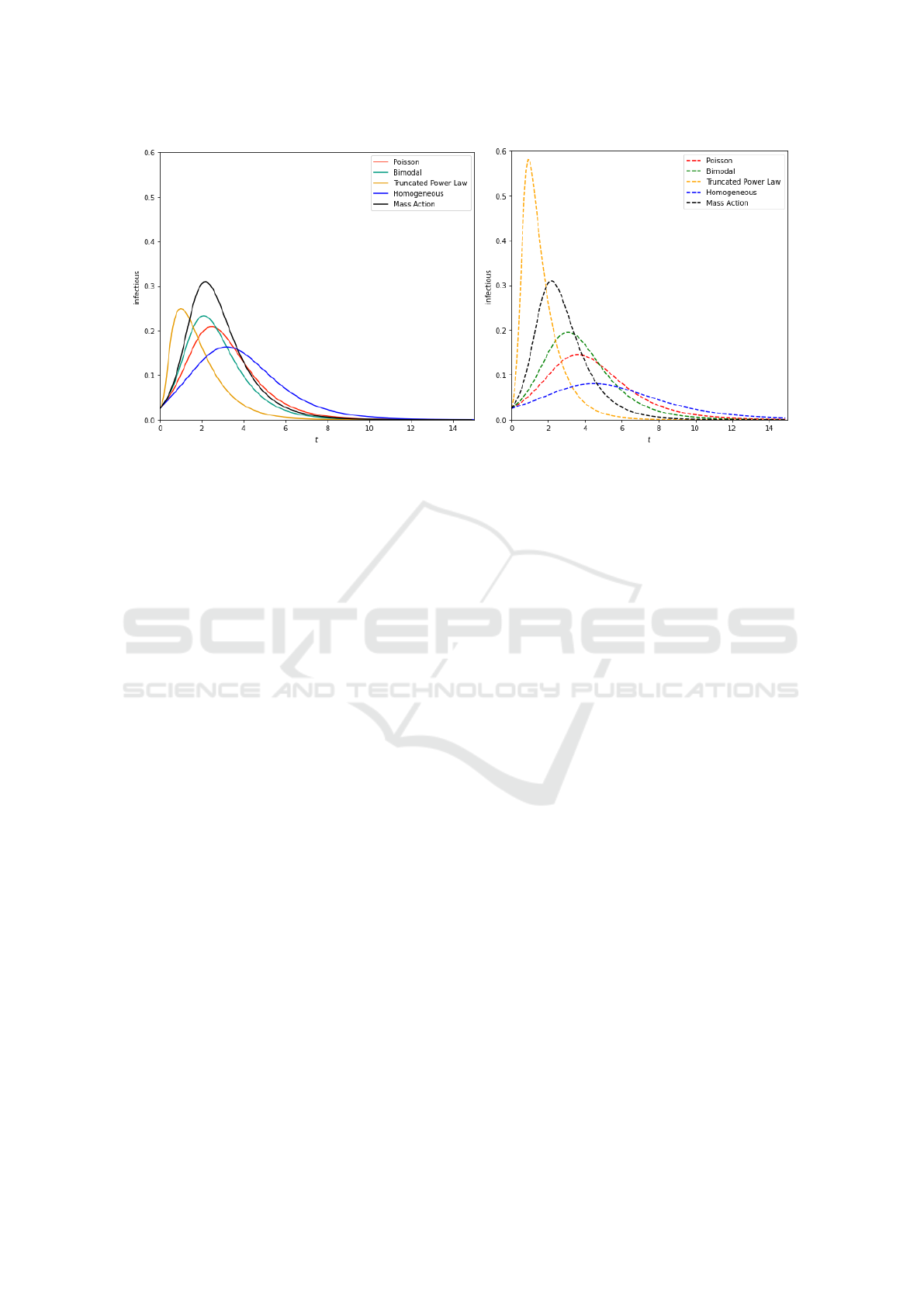
(a) EoN simulations. (b) Kendrick simulations.
Figure 9: EoN and Kendrick simulations of the mass action model (black curve) and the EBCM configuration model on four
different degree distributions : Homogeneous (blue curve), Poisson (red curve), Bimodal (green curve) and Truncated Power
Law (orange curve). Each contact network has 500,000 nodes and an average degree of 5. The Poisson degree distribution
has an average of 5, half of the nodes have a degree of 2 and the other half a degree of 8 for the bimodal distribution, and
finally a truncated Powerlaw distribution in which P(k) ∝ k
−v
e
−k/40
where v = 1.418.
In order to validate this integration, we applied the
approach of (Fodjo et al., 2022) to the mass action and
the approximation of the EBCM configuration model
of (Miller et al., 2012) and we were able to obtain
similar results close to those of (Miller et al., 2012).
Both models (mass action model and the approxima-
tion of the EBCM configuration model) were defined
as a very simple and distinct concern of the basic SIR
model in Kendrick.
If it is obvious that we were able to obtain simi-
lar results from the EBCM configuration model from
the approximation of the EBCM configuration model
in Kendrick, it is still a bit limiting. This is because
it requires to have beforehand a mathematical formu-
lation of R
0
of each EBCM model that we wish to
implement as a concern in the Kendrick sense and to
be able to find the appropriate decomposition of the
extension points of the relation 11. However, the ap-
proach we propose avoids having to build an explicit
contact network, and moreover we do not have to de-
fine a multitude of differential equations. Another en-
couraging aspect is the fact that most of the EBCM
models proposed in the literature have a mathemati-
cal formulation of R
0
, which gives us a wide range of
models to explore for future work. In the same way,
we also think that in the future, it would be interesting
to be able to implement typical EBCM models (with-
out model approximation approach) as concerns that
could be composed when adding new concerns.
REFERENCES
Aparicio, J. P. and Pascual, M. (2007). Building epidemio-
logical models from r0: an implicit treatment of trans-
mission in networks. Proceedings of the Royal Society
B: Biological Sciences, 274(1609):505–512.
Balde, C., Lam, M., and Bowong, S. (2019). Contact vacci-
nation study using edge based compartmental model
(ebcm) and stochastic simulation: An application to
oral poliovirus vaccine (opv). In International Sym-
posium on Mathematical and Computational Biology,
pages 81–96. Springer.
Bansal, S., Grenfell, B. T., and Meyers, L. A. (2007). When
individual behaviour matters: homogeneous and net-
work models in epidemiology. Journal of the Royal
Society Interface, 4(16):879–891.
Bui, T., Papoulias, N., Stinckwich, S., Ziane, M., and
Roche, B. (2019). The Kendrick modelling platform:
language abstractions and tools for epidemiology [+
correction art. no 439, 1 p.]. BMC Bioinformatics, 20.
Bui, T. M. A., Ziane, M., Stinckwich, S., Ho, T. V., Roche,
B., and Papoulias, N. (2016). Separation of concerns
in epidemiological modelling. In Companion pro-
ceedings of the 15th international conference on mod-
ularity, pages 196–200.
Cuddington, K. and Beisner, B. E. (2005). Ecological
paradigms lost: routes of theory change. Elsevier
Academic Press New York.
Danon, L., Ford, A. P., House, T., Jewell, C. P., Keeling,
M. J., Roberts, G. O., Ross, J. V., and Vernon, M. C.
(2011). Networks and the epidemiology of infectious
disease. Interdisciplinary perspectives on infectious
diseases, 2011.
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
268

Fodjo, A. Y. G., Ziane, M., Stinckwich, S., Anh, B. T. M.,
and Bowong, S. (2022). Separation of concerns in
extended epidemiological compartmental models. In
Proceedings of the 15th International Joint Confer-
ence on Biomedical Engineering Systems and Tech-
nologies - Volume 3: BIOINFORMATICS,, pages
152–159. INSTICC, SciTePress.
Gamma, E., Helm, R., Johnson, R., Vlissides, J., and Pat-
terns, D. (1995). Elements of reusable object-oriented
software, volume 99. Addison-Wesley Reading, Mas-
sachusetts.
Heesterbeek, H., Anderson, R. M., Andreasen, V., Bansal,
S., De Angelis, D., Dye, C., Eames, K. T., Edmunds,
W. J., Frost, S. D., Funk, S., et al. (2015). Modeling
infectious disease dynamics in the complex landscape
of global health. Science, 347(6227):aaa4339.
Istvan, Z., Miller, K., Joel, C. S., and Peter, L. (2019). Math-
ematics of Epidemics on Networks: From Exact to Ap-
proximate Models. Springer.
Keeling, M. J. and Rohani, P. (2011). Modeling infectious
diseases in humans and animals. Princeton university
press.
Kermark, M. and Mckendrick, A. (1927). Contributions to
the mathematical theory of epidemics. part i. Proc. r.
soc. a, 115(5):700–721.
Kiss, I. Z., Miller, J. C., Simon, P. L., et al. (2017). Math-
ematics of epidemics on networks. Cham: Springer,
598.
Levin, S. A. and Durrett, R. (1996). From individu-
als to epidemics. Philosophical Transactions of the
Royal Society of London. Series B: Biological Sci-
ences, 351(1347):1615–1621.
Martcheva, M. (2015). An introduction to mathematical epi-
demiology, volume 61. Springer.
Meyers, L. A., Pourbohloul, B., Newman, M. E., Skowron-
ski, D. M., and Brunham, R. C. (2005). Network the-
ory and sars: predicting outbreak diversity. Journal of
theoretical biology, 232(1):71–81.
Miller, J. C., Slim, A. C., and Volz, E. M. (2012). Edge-
based compartmental modelling for infectious dis-
ease spread. Journal of the Royal Society Interface,
9(70):890–906.
Molina, C. and Stone, L. (2012). Modelling the spread of
diseases in clustered networks. Journal of theoretical
biology, 315:110–118.
Plateau, B. and Stewart, W. J. (2000). Stochastic automata
networks. In Computational Probability, pages 113–
151. Springer.
Trapman, P., Ball, F., Dhersin, J.-S., Tran, V. C., Wallinga,
J., and Britton, T. (2016). Inferring r 0 in emerging
epidemics—the effect of common population struc-
ture is small. Journal of The Royal Society Interface,
13(121):20160288.
Wang, Y., Cao, J., Li, X., and Alsaedi, A. (2018). Edge-
based epidemic dynamics with multiple routes of
transmission on random networks. Nonlinear Dynam-
ics, 91(1):403–420.
Yang, J. and Xu, F. (2019). The computational approach for
the basic reproduction number of epidemic models on
complex networks. IEEE Access, 7:26474–26479.
Zhang, Z., Wang, H., Wang, C., and Fang, H. (2015). Mod-
eling epidemics spreading on social contact networks.
IEEE transactions on emerging topics in computing,
3(3):410–419.
Separation of Concerns in an Edge-Based Compartmental Modeling Framework
269
