
Wearable Data Generation Using Time-Series Generative Adversarial
Networks for Hydration Monitoring
Farida Sabry
1 a
, Wadha Labda
1 b
, Tamer Eltaras
1 c
, Fatima Hamza
1 d
, Khawla Alzoubi
2 e
and Qutaibah Malluhi
1 f
1
Qatar University, Doha, Qatar
2
Engineering Technology Department, Community College of Qatar, Doha, Qatar
Keywords:
Data Generation, Synthetic Data, GAN, Wearable Devices, Biosignals.
Abstract:
Collection of biosignals data from wearable devices for machine learning tasks can sometimes be expensive
and time-consuming and may violate privacy policies and regulations. Successful and accurate generation
of these signals can help in many wearable devices applications as well as overcoming the privacy concerns
accompanied with healthcare data. Generative adversarial networks (GANs) have been used successfully in
generating images in data-limited situations. Using GANs for generating other types of data has been actively
researched in the last few years. In this paper, we investigate the possibility of using a time-series GAN
(TimeGAN) to generate wearable devices data for a hydration monitoring task to predict the last drinking
time of a user. Challenges encountered in the case of biosignals generation and state-of-the-art methods for
evaluation of the generated signals are discussed. Results have shown the applicability of using TimeGAN
for this task based on quantitative and visual qualitative metrics. Limitations on the quality of the generated
signals were highlighted with suggesting ways for improvement.
1 INTRODUCTION
The recent advances in wearable technology have mo-
tivated researchers and industry to investigate many
machine learning based applications that can learn
from the vast amount of signals from wearable sen-
sors (Sabry et al., 2022a). There are a variety of non-
invasive signals that can be captured from the human
body using different sensors to learn about the health
condition of the user for a wide variety of machine
learning healthcare applications (Sabry et al., 2022a).
Some of the problems that arise with these types of
applications are the cost and time associated with the
data collection phase, the limited size of datasets used
for training (Sabry et al., 2022b; Delmastro et al.,
2020). Additionally, the collection of biosignals data
is erroneous which increases the cost and time even
more. Data collection of health data is subjected to
a
https://orcid.org/0000-0001-5639-983X
b
https://orcid.org/0000-0001-6097-2395
c
https://orcid.org/0000-0002-8664-9091
d
https://orcid.org/0000-0002-3689-7003
e
https://orcid.org/0000-0002-2797-2673
f
https://orcid.org/0000-0003-2849-0569
many regulations to ensure the safety of subjects and
preserve the privacy of their sensitive data. Though
anonymizing this sensitive data by removing identi-
fying features or adding noise and grouping individu-
als into broader categories is sometimes done to over-
come this privacy problem, it is usually not effective
with small dataset sizes used in research from wear-
able devices.
One way to overcome these problems is to gener-
ate synthetic training data that follows the same dis-
tribution for real world data (Piacentino et al., 2021;
Zhou et al., 2019; Yoon et al., 2019). Synthetic data
can be used with various objectives such as data un-
derstanding, data imputation (Luo et al., 2018), data
correction and noise removal (Kiranyaz et al., 2022;
Lu et al., 2021; Zargari et al., 2021), data augmenta-
tion (Um et al., 2017; Kiyasseh et al., 2020; Lo et al.,
2021; Montero et al., 2021), and data privacy (Xin
et al., 2020; Nguyen et al., 2020; Piacentino et al.,
2021).
A synthetically generated dataset must have the
same mathematical and statistical properties as the
real-world dataset. For biosignals generation, there
are additional constraints on the shape and repeated
pattern of some signals which must be preserved in
94
Sabry, F., Labda, W., Eltaras, T., Hamza, F., Alzoubi, K. and Malluhi, Q.
Wearable Data Generation Using Time-Series Generative Adversarial Networks for Hydration Monitoring.
DOI: 10.5220/0011757200003414
In Proceedings of the 16th Inter national Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 4: BIOSIGNALS, pages 94-105
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

synthetic data. The generated data has to be realis-
tic enough to gain true insights from it when used in
different machine learning tasks (Sabry et al., 2022a).
Synthetic data is less subjected to data privacy con-
cerns or missing values problems. Synthetic data gen-
eration has been approached using many techniques
that either change the real data to guarantee its pri-
vacy such as differential privacy (Ping et al., 2017;
Xin et al., 2020; Um et al., 2017) or learn from the
real data using a variety of machine-learning tech-
niques (Reiter, 2005; Zhang et al., 2017; Xu et al.,
2019; Yoon et al., 2019) to learn the distribution of
the data and then sample the distribution to generate
the synthetic data.
Generative adversarial networks (GANs) are
among the machine learning techniques that can be
used to generate synthetic data that is similar to the
actual data in terms of data distribution and statisti-
cal features. The authors in (Goodfellow et al., 2014)
were the first to introduce GAN to generate new syn-
thetic image data that are difficult to be distinguished
from real data images but at the same time not memo-
rized from the training set. With the potential of GAN
and development of research for its variants, GAN has
been used to generate various other types of data such
as tabular data with discrete and continuous values
(Xu et al., 2019). Although there are some similar-
ities in the used techniques, there are differences in
implementation for every type of data which requires
some adjustments in the GAN architecture. In this
study, we investigate the use of a variant of GAN, spe-
cific for time-series data (Yoon et al., 2019), to gener-
ate synthetic data of different biosignals which have
special characteristics such as amplitude thresholds,
repetitive pattern, positions of peaks and troughs to
be used for hydration monitoring based on the real
dataset we collected in (Sabry et al., 2022b). Though
there are some studies (Furdui et al., 2021; Belo et al.,
2017; Zhou et al., 2019) that used other types of
GANs to generate electrocardiogram (ECG) or gal-
vanic skin response (GSR) signals for other classifi-
cation tasks, but few studied using GANs for a regres-
sion problem (Ning et al., 2018) and for the best of our
knowledge this is the first study to use GAN for hy-
dration monitoring data generation to predict the last
drinking time.
The rest of the paper is organized as follows. Sec-
tion 2 reviews briefly the background for this re-
search giving a summarized overview for GAN and
its variants. Related work for using GANs for biosig-
nals generation and challenges for biosignals data are
discussed in this section as well. Section 3 briefly
describes the dataset (Sabry et al., 2022b) used in
this research. In section 4, the methodology for us-
ing times-series GAN for synthetic data generation
for hydration monitoring is presented, together with
the used evaluation metrics. Results and discussion
are presented in section 5. The paper is then con-
cluded in section 6 with a summary of the research,
its results, its limitations and future improvements are
highlighted.
2 BACKGROUND AND RELATED
WORK
Generative adversary network (GAN) is a machine
learning model that is simply based on two deep neu-
ral networks; one is called the generator network (G)
and the other is called discriminator network (D) and
both of them compete in a zero-sum game to reach
Nash equilibrium with a value function V (D, G) given
in Equation 1 where x represents the input real sam-
ples to the discriminator and z represents the input
noise samples to the generator from which it starts
to generate fake data G(z). While G works on gener-
ating realistic data by minimizing V, D is a classifier
that is used to differentiate the real and fake data as
genuinely as possible by maximizing V . A typical
GAN model is shown in Figure 1, both the generator
and discriminator typically have multiple convolution
layers to capture the details of the input data. GAN
was first introduced by Goodfellow et al (Goodfellow
et al., 2014) in 2014 and since then research has been
developing different GAN variants that were able to
generate images, videos, music notes, etc with high
quality. The authors in (Jabbar et al., 2022) review the
different variants of GANs, their wide range of appli-
cations, shortcomings affecting the training stability
and different training solutions.
G
min
D
max
V (D, G) =
E
x∼p
data
(x)
logD (x)
+
E
z∼p
z
(z)
log (1 − D (G(z)))
(1)
The basics for generative modeling in GAN can be
seen as improvements to autoencoders. Autoencoder
and its probabilistic version, variational autoencoder,
are used to represent high-dimensionality data and
compress it into a small representation with simple
neural networks (encoder and decoder) without mas-
sive data loss (Jabbar et al., 2022). Generative models
were then used to to improve the quality of the gener-
ated data and guarantee better privacy protection mea-
sures. Here we list some examples for GAN variants
used in literature for generation of different biosignals
which have special characteristics and challenges dif-
Wearable Data Generation Using Time-Series Generative Adversarial Networks for Hydration Monitoring
95
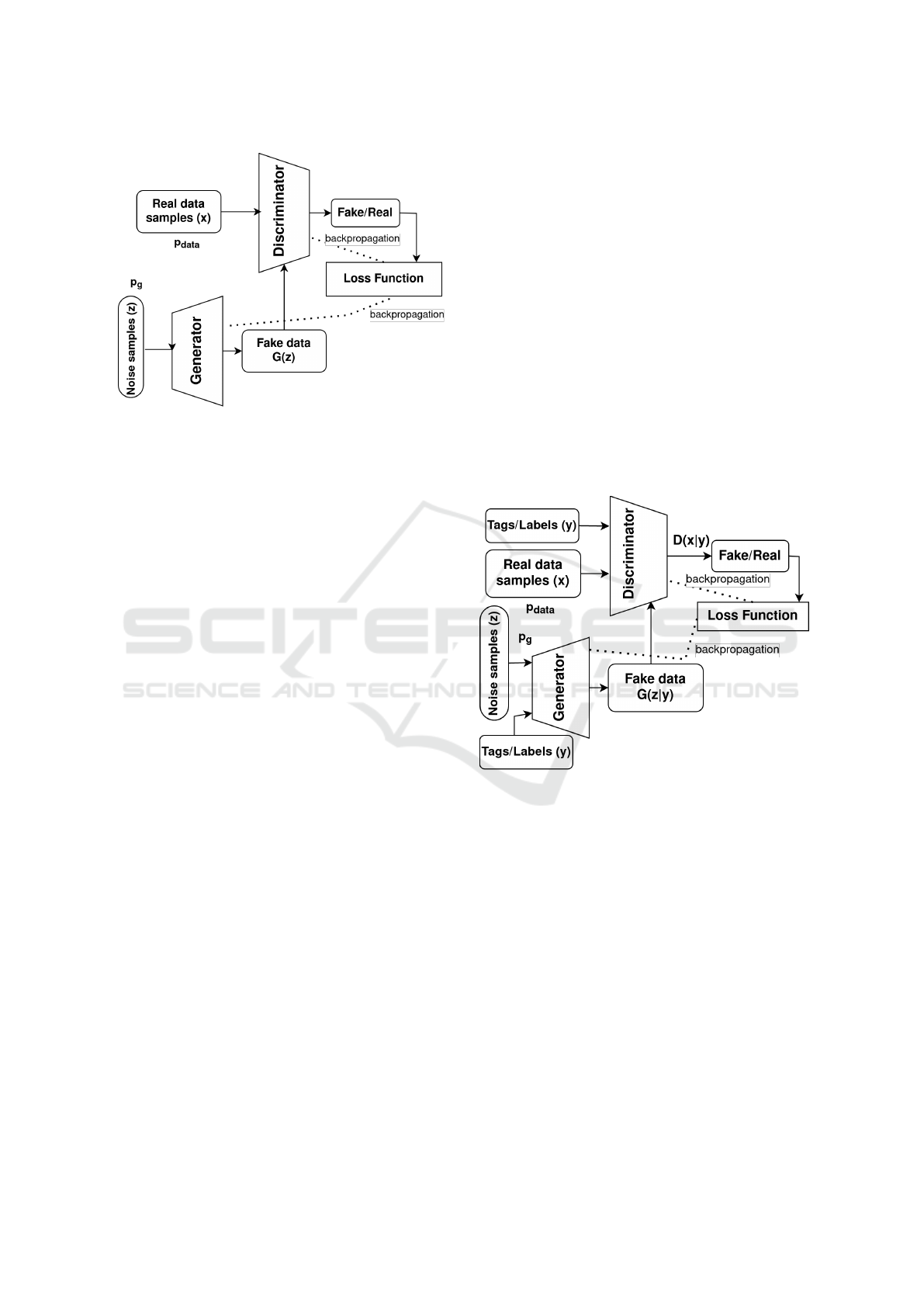
Figure 1: General GAN architecture.
ferent from images that will be discussed in the next
subsection.
• Conditional GAN: It is very similar to the sim-
ple GAN in Figure 1 but with the output of the
generator and discriminator conditioning on la-
bels y or any additional information in general for
both the real samples and the noise samples as
shown in Figure 2 with modified objective func-
tion in Equation 2 to improve the diversity of the
generated dataset. The first term in the objec-
tive function is the expected log likelihood of the
real data samples under the discriminator, given
the label or context y that the discriminator is
trying to predict. The second term is the ex-
pected log likelihood of the synthetic data sam-
ples under the discriminator, given the label or
context y. It has been used in (Kiyasseh et al.,
2020) for the generation of pathological photo-
plethysmogram (PPG) signals in order to boost
medical classification performance for different
cardiac classes. PPG signals were downsampled
and split into overlapping 5sec-frames and fed to
three different CGANs which may not handle the
temporal behavior of the data and produce un-
realistic data. They modified the loss functions
with different terms to improve inter-class diver-
sity and penalize the network for generating unre-
alistic samples. In (Harada et al., 2018), they pro-
posed forming each neural network in GAN based
on a recurrent neural network (RNN) using long
short-term memories (LSTM) for its hidden lay-
ers to deal with the time-series data generation for
ECG and EEG signals. An ICU dataset record-
ing oxygen saturation, heart rate, respiratory rate
and mean arterial pressure has been used in (Es-
teban et al., 2017) to generate synthetic data for
various ICU prediction tasks of whether a patient
will go in a critical condition defined by thresh-
olds for these variables. They showed that the
prediction accuracy decreases when training with
synthetic data by a maximum of 12%. Generating
synthetic signals for four kinds of biomedical sig-
nals (electrocardiogram (ECG), electroencephalo-
gram (EEG), electromyography (EMG), photo-
plethysmography (PPG)) was done in (Hazra and
Byun, 2020). They used a CGAN with bidirec-
tional grid long short-term memory for the gener-
ator network and convolutional neural network for
the discriminator network to capture time-series
dependency.
G
min
D
max
V (D, G) =
E
x∼p
data
(x)
logD (x|y)
+
E
z∼p
z
(z)
log (1 − D (G(z|y)))
(2)
Figure 2: Conditional GAN architecture.
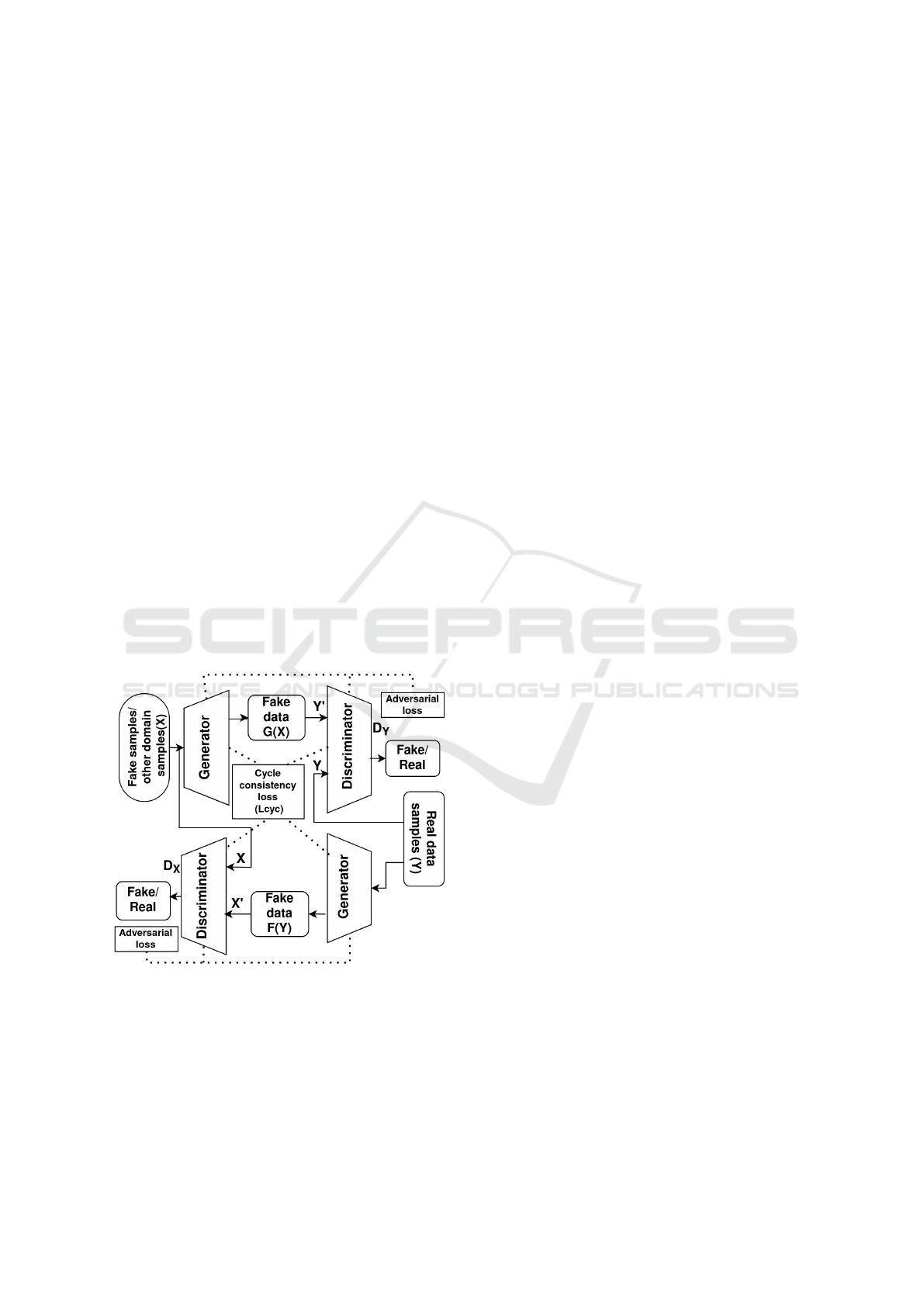
• Cycle GAN (Zhu et al., 2017): It is a special kind
of conditional GAN where the condition is not a
label or tag but rather a sample data from a dif-
ferent domain, in the case of data generation it
can be mapping of a sample from synthetic data
to real data or vice versa, one GAN works in the
forward cycle and the other in the backward, or
it can be to transform one raw signal to a de-
rived signal. General architecture for a cycle GAN
can be modeled as shown in Figure 3 and its full
objective function is given in Equation 3 where
adversarial losses for the forward and backward
GANs are calculated as in Equation 1 and L
cyc
is
calculated according to Equation 4 to ensure cy-
cle consistency by reducing the possibilities for
the mapping function so that an individual input
x
i
can be mapped to a desired output y
i
. How-
ever, cycle GANs require a relatively big dataset
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
96
to reduce the replication percentage of the gener-
ated data. For biosignals generation, Cycle GAN
has been used in (Aqajari et al., 2021) for respi-
ratory rate estimation through learning a mapping
of PPG signals to respiratory signals and updat-
ing the objective function to include a weighted
term that takes into account the respiratory rate of
the generated respiratory signals. It has been used
in (Zargari et al., 2021) too, for noise artifacts re-
moval without depending on accelerometer data
through PPG signal to image transformation. The
authors in (Kiranyaz et al., 2022) also used oper-
ational cycle-GANs for ECG signals restoration,
not whole signal generation. They used quanti-
tative evaluation by measuring the performance
gain for peak detection as well as the visual qual-
itative evaluation.
L (G, F, D
X
, D
Y
) = L
GAN
(G, D
Y
, X, Y )+
L
GAN
(F, D
X
, Y, X )+
λL
cyc
(G, F)
(3)
L
cyc
(G, F) =
E
x∼p
data
(x)
∥(F (G (x))) − x∥
1
+
E
y∼p
data
(y)
∥(G(F (y))) − y∥
1
(4)
Figure 3: Cycle GAN architecture.
• Wasserstein GAN (W-GAN) (Arjovsky et al.,
2017): It is very similar to the traditional GAN
but with calculating the loss function based on
Wasserstein distance between the real data distri-
bution and the generated data distribution rather
than depending on Jensen–Shannon divergence.
This GAN variant was introduced to solve mode
collapse problem which is a GAN training chal-
lenge. In the context of biosignal generation, an
auxiliary conditioned WGAN has been used in
(Furdui et al., 2021) to generate galvanic skin re-
sponse (GSR) and electocardiogram (ECG) sig-
nals that can be used for arousal classification by
combining the Wasserstein loss with the gradi-
ent penalty and classification loss to discard sig-
nals that doesn’t improve the classification to en-
hance the generalizability of emotion recognition
algorithms. WGAN was also used with modified
Gate Recurrent Units (GRUs) in (Luo et al., 2018)
for imputation of data in physionet dataset, au-
thors were able to model the temporal irregularity
of the incomplete time series. Their results out-
performed the baselines in terms of accuracy of
imputation for the missing data during recording
process.
• Style Generator Architecture for GAN (Style
GAN): It is a modified architecture to GAN where
more complexity is added to the generator to have
26.2M trainable parameters (Karras et al., 2021).
This is done through adding a mapping network of
fully connected layers to map the input to a latent
representation, followed by a synthesis network
which is controlled by this representation through
adaptive instance normalization (AdaIN) at each
convolution layer. This aims to achieve better in-
terpolation and also model the factors of variation
better, its usage has vastly improved the gener-
ation of images that feel-like real. A variant of
style GAN was used in (Montero et al., 2021) for
fetal brain ultrasound plane classification. It was
used in (Thambawita et al., 2021) for generation
of gastroenterology data images. To the best of
our knowledge, styleGAN hasn’t been applied to
biosignals with time-series patterns.
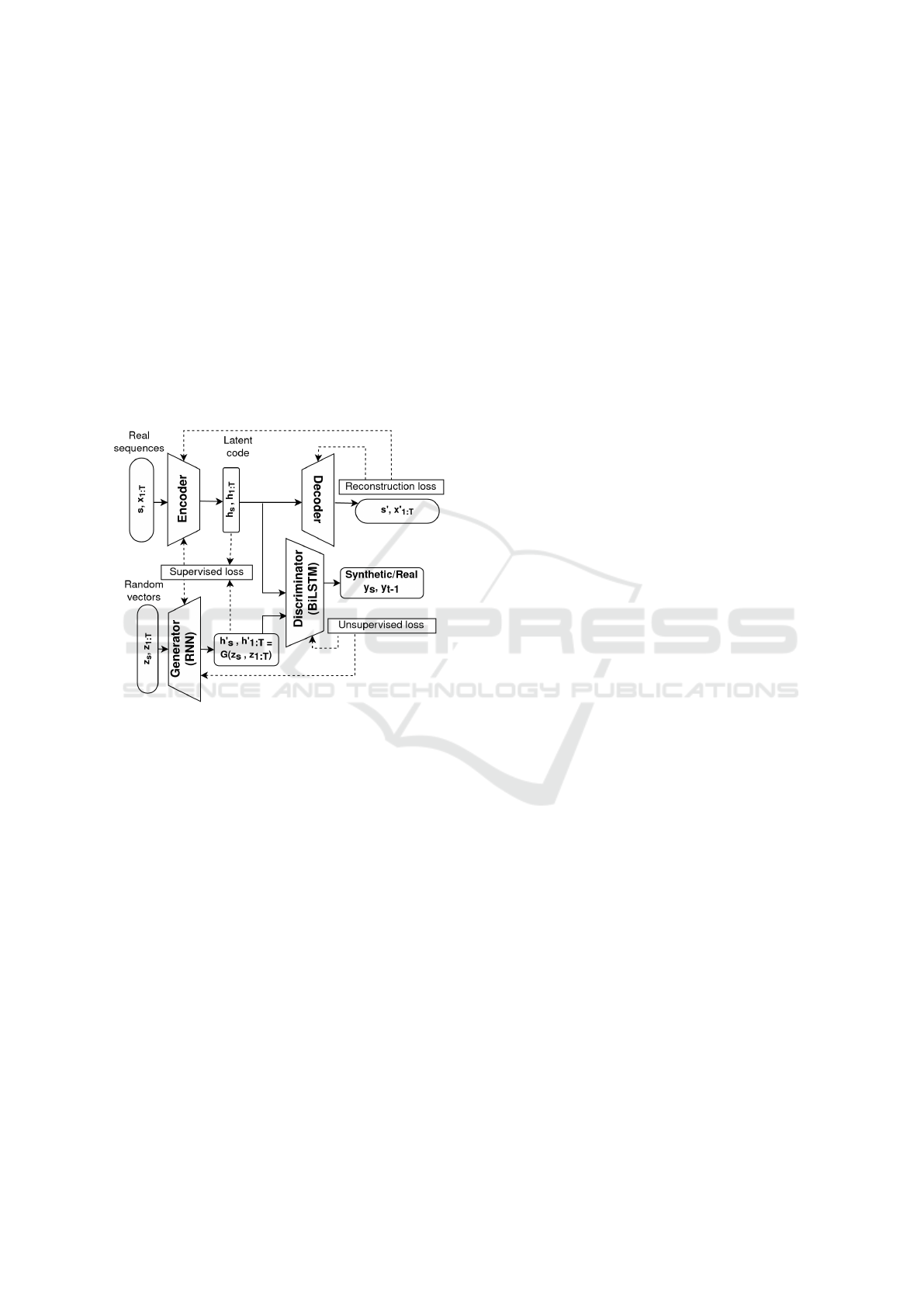
• Time-series GAN (Yoon et al., 2019) is another
type of GAN that combines the traditional un-
supervised GAN network with a supervised au-
toregressive model to model the temporal dynam-
ics of a time-series. A typical architecture of the
TimeGAN is shown in Figure 4. Weights are up-
dated based on three losses; the supervised loss,
the unsupervised loss and the reconstruction loss
of the autoregressive model defined as in Equa-
tions 5, 6 and 7 respectively where the expected
log likelihood is calculated over all time steps for
both real and synthetic data. It has been used to
generate stock and energy prediction data and re-
ported improvement over other CGANs with units
that capture temporal dynamics such as RNN and
LSTM (Brophy et al., 2021). It hasn’t been ap-
plied before for biosignals data generation as we
Wearable Data Generation Using Time-Series Generative Adversarial Networks for Hydration Monitoring
97
propose in this paper.
L
supervised
=
E
s,x
1:T
∼p
∑
t
∥h
t
− G
X
(h
s
, h
t−1
, z
t
)∥
2
(5)
L
unsupervised
=
E
s,x
1:T
∼p
log(y
s
) +
∑
t
log(y
t
)
+
E
s,x
1:T
∼p
′
log(1 − y
′
s
) +
∑
t
log(1 − y
′
t
)
(6)
L
reconstruction
=
E
s,x
1:T
∼p
∥s − s
′
∥
2
+
∑
t
∥x
t
− x
′
t
∥
2
(7)
Figure 4: Time-series GAN architecture.
2.1 Challenges of Biosignals Generation
Biosignals dataset collection is considered very chal-
lenging for many reasons. Most of the time, the real
dataset is with a limited number of subjects due to
the cost involved and the privacy constraints. The
collected data may suffer from missing/incorrect data
due to errors in sensors’ attachment or noisy sig-
nals due to motion artifacts (Elgendi, 2016). There
are other inadequacies that might appear in the col-
lected data which include insufficient measurement
time, different sensors’ configurations used by dif-
ferent subjects and inconsistent measurement times
(Hernang
´
omez et al., 2022). Generative modeling to
generate synthetic data based on the real biosignals
dataset collected consequently becomes challenging.
In addition to the paucity of the collected data and
its aforementioned inadequacies, class imbalance data
(Kiyasseh et al., 2020) for infected or abnormal sam-
ples pose other difficulties for synthetic data genera-
tion.
Real-world tabular data collected for biosignals
usually consists of mixed types; continuous data for
signals from sensors such as photoplethysmography
(PPG), ECG, GSR, accelerometer, magnetometer, gy-
roscope and discrete data such as sex, age, medicine
intake, etc. To generate a mix of these discrete and
continuous data simultaneously, GANs must apply
both softmax and tanh on the output (Xu et al., 2019).
Modeling this kind of mixed tabular data in general
to generate realistic data is a non-trivial task (Xu
et al., 2019). A review for some approaches to han-
dle discrete data is presented in (Jabbar et al., 2022).
Continuous data for biosignals usually follow a non-
Gaussian distribution unlike the images pixel values
which follow a Gaussian-like distribution. In GANs,
the last layer has a tanh function which can success-
fully output a value in the range [-1,1] for images data
after normalization using a min-max transformation
whereas continuous non-Gaussian data will lead to
vanishing gradient problem.
As well known in the machine learning field,
highly imbalanced data poses many challenges for
learning in general and for learning a GAN model
in particular as minority classes are underrepresented
making a GAN susceptible to severe mode collapse.
In this case, the generator produce outputs with small
diversity leading to only slight changes to the data dis-
tribution that may be hardly detected by the discrim-
inator. To decrease this effect and prevent the gener-
ator from optimizing for a single fixed discriminator,
Wasserstein loss is used to avoid the discriminator be-
ing stuck in local minima. Unrolled GAN (Metz et al.,
2017) is another way to face this problem which uses
a generator loss function based on the current and fu-
ture discriminator’s classifications so that the genera-
tor don’t optimize for a single discriminator.
In the next subsection, we review the sate-of-art
evaluation metrics to evaluate GANs.
2.2 Evaluation Metrics
Evaluation of GAN models is an active research area
regarding unstable training (Jabbar et al., 2022) as
there is no standard function for evaluation. In this
section, we briefly review the different evaluation
metrics used in literature to evaluate the GAN and
outputs generated by it.
Evaluation metrics can be classified as qualitative
or quantitative. Qualitative evaluation refers to human
visual assessment for the generated samples from the
GAN but it lacks a suitable objective evaluation met-
ric. Quantitative evaluation refers to objective metrics
that either assess the similarity of the generated syn-
thetic data to the real data, calculate the distance be-
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
98

tween the two distributions or evaluate the accuracy
for the classification task or regression error when
replacing the real dataset with synthetic dataset for
training the machine learning model. Most of the pro-
posed metrics in literature are applicable to the image
data (Brophy et al., 2021).
For image data, inception-score (IS) and Fr
´
echet
inception distance (FID) are most commonly used to
evaluate the diversity and quality of the generated
samples for its correlation with human findings of the
generated samples (Jabbar et al., 2022).
Pearson correlation coefficient was used in (Hazra
and Byun, 2020) to verify the quality of synthetic
data when compared to the original data as well as
root mean square error (RMSE), percent root mean
square difference (PRD), mean absolute error (MAE),
and Frechet distance (FD) for statistical analysis.
However, they were used as one-to-one measure be-
tween synthetic data and original data used to gener-
ate it with assumption of having the same sequence
length as the original data (aligned sequences of fixed
length).
Propensity score which represents the probabil-
ities of whether a record is real or synthetic, has
been used for many record data (Dankar and Ibrahim,
2021). It involves building a binary classification
model for classification but not formulated for time-
series data. To deal with time-series data, authors in
(Yoon et al., 2019) introduced a discriminative score
as a quantitative measure for similarity using a 2-
layer LSTM to classify sequences as either synthetic
or original. The classification error is the score and
the less error means the generated sequences are simi-
lar to the original data. They also introduced a predic-
tive score which is similar to the Train on Synthetic,
Test on Real where a sequence-prediction model with
2-layer LSTM is trained to predict next-step tem-
poral vectors over each input sequence. Then, the
trained model is tested on the original dataset. For
visualizing how closely the distribution of generated
data represents that of the original, authors in (Yoon
et al., 2019) used t-Stochastic Neighbor Embedding
(t-SNE) (van der Maaten and Hinton, 2008) and prin-
cipal component analysis (PCA) (Bryant and Yarnold,
2001) by flattening the temporal dimension on both
the original and synthetic datasets and results are as-
sessed visually.
As can be deduced from the above brief review,
there are no standard evaluation metric for fair model-
to-model comparison and specially when it comes
to time-series data. The development of a quantita-
tive evaluation metric for the synthetic data genera-
tion still requires future research, especially for time-
series data. As this is not the main focus of the pa-
per, we chose to use the scores used by (Yoon et al.,
2019) as they are the only ones developed for time
sequences. Additionally, we used Train on Synthetic
Test on Real (TSTR) metric as this is the main goal
for the generation of synthetic data to use more data
for learning a model that can predict the output value
in the regression problem of hydration monitoring for
the last drinking time.
3 DATASET
For generating wearable device data for the task of
hydration monitoring, we used the real dataset we col-
lected in (Sabry et al., 2022b) using sensors from the
calibrated Shimmer Galvanic Skin Response (GSR)
unit
1
. The last time for water and food intake was
recorded for subjects wearing the GSR unit who
fasted during Ramadan or were voluntary fasting with
no restrictions on movement or the time of wear-
ing the device and also in non-fasting conditions, i.e.
the last drinking time is within 1 hour. The signals
are recorded from the shimmer device at a frequency
of 512 Hz and include PPG, GSR Skin Resistance,
GSR Skin Conductance, accelerometer in the three
directions (X,Y,Z), magnetometer (X,Y,Z), gyroscope
(X,Y,Z), ambient temperature and pressure. A total of
3386 min (56.4 h) data were collected from 11 healthy
subjects (9 females and 2 males). All data collection
was subject to Qatar University Institutional Review
Board (IRB) approval procedures covered by the IRB
approval: QU-IRB 1538-EA/21. The raw dataset is
available at Zenodo
2
.
4 WEARABLE DATA
GENERATION
The approach followed for generating wearable data
that can be used for hydration monitoring task in
(Sabry et al., 2022b) can be presented as shown in
Figure 5. First, a real dataset was built of the biosig-
nals from the wearable device (PPG, GSR) as well
as that of the movement sensors (accelerometer, mag-
netometer and gyroscope) and the ambient tempera-
ture and pressure sensors as discussed in the previous
section. Preprocessing of the dataset included down-
sampling the signals at 1 min intervals and extract-
ing features from signals aggregating the mean val-
1
http://www.shimmersensing.com/products/gsr-optical
-pulse-development-kit (accessed on 16 Nov. 2022)
2
https://zenodo.org/record/6299964 (accessed on 16
Nov. 2022)
Wearable Data Generation Using Time-Series Generative Adversarial Networks for Hydration Monitoring
99

ues and calculating the accumulated changes in the
magnitude for accelerometer, magnetometer and gy-
roscope data (Sabry et al., 2022b) as in Equations
8, 9, 10, 11, 12 and 13. |ACC| refers to the mag-
nitude of the accelerometer from the components in
the three directions (acc
x
t
, acc
y
t
, acc
z
t
). cumAcc is
the accumulative change in accelerometer magnitude
over time. |MAG| refers to the magnitude of the
magnetometer from the components in the three di-
rections (mag
x
t
, mag
y
t
, mag
z
t
). cumMag is the ac-
cumulative change in magnetometer magnitude over
time. |GY RO| refers to the magnitude of the gyro-
scope from the components in the three directions
(gyro
x
t
, gyro
y
t
, gyro
z
t
). cumGyro is the accumulative
change in gyroscope magnitude over time. These
magnitude values for accelerometer, magnetometer
and gyroscope as well as the accumulative changes
in them that represent the motion jerk can reflect the
effort and state of activity of the subject. For the hy-
dration monitoring task, the fine details of the signals
is not as relevant as the big changes of it through
time so downsampling and aggregation can be seen
feasible. It also helps in speeding up the training of
the generative adversary network for time-series data
(TimeGAN).
The output signals together with the column for
the recorded last drinking time for each of the fasting
and non-fasting samples for each subject are then in-
put to the TimeGAN module and are considered the
real sequences. The TimeGAN generator uses gated
recurrent units in its recurrent neural network with 20
hidden nodes in each of its three layers and the dis-
criminator uses bidirectional long short-term memory
units to increase the amount of information available
to the network. The generated synthetic signals are
then updated with the objective to minimize the three
loss functions in Equations 5, 6 and 7 for 500 iter-
ations. The TimeGAN is then evaluated using dis-
criminative score (Yoon et al., 2019) mentioned in
section 2.2 with lower value means better. It repre-
sents the post-hoc classification error for a supervised
classifier fed with original signals labeled as real ex-
amples and generated signals labeled as synthetic ex-
amples and tested with a held-out test set. A predic-
tive score is also calculated to evaluate the effective-
ness of TimeGAN to capture the conditional distribu-
tions over time. It represents the mean absolute error
(MAE) in predicting the next-step temporal vectors
over each input sequence in the original dataset using
an LSTM model trained with the synthetic generated
dataset.
Training on synthetic data and testing on real was
done for the main problem of hydration monitoring
where we trained the three models that achieved the
Table 1: Mean discriminative and predictive scores for gen-
erated fasting and non-fasting data.
Discriminative score Predictive score
fasting 0.319 3.9
non-fasting 0.225 0.276
best performance in (Sabry et al., 2022b); random for-
est, gradient boosted regression and extra trees.
|ACC| =
q
(acc
x
t
2
+ acc
y
t
2
+ acc
z
t
2
) (8)
cumAcc =
t−1
∑
j=1
|ACC|
j+1
− |ACC|
j
(9)
|MAG| =
q
(mag
x
t
2
+ mag
y
t
2
+ mag
z
t
2
) (10)
cumMag =
t−1
∑
j=1
|MAG|
j+1
− |MAG|
j
(11)
|GY RO| =
q
(gyro
x
t
2
+ gyro
y
t
2
+ gyro
z
t
2
) (12)
cumGyro =
t−1
∑
j=1
|GY RO|
j+1
− |GY RO|
j
(13)
5 RESULTS
5.1 Quantitative Evaluation
The synthetic output of the TimeGAN is evaluated
quantitatively using the discriminative and predic-
tive scores for the output of TimeGAN obtained after
training for 500 iterations as shown in Table 1. The re-
sults show that the average predictive score for gener-
ating fasting samples is relatively high, this can be at-
tributed to the smaller number of samples and shorter
sequences used in training the TimeGAN with fasting
sequences as well as the missing of some other fea-
tures in the original dataset we used in (Sabry et al.,
2022b) such as height, weight, sex and age which af-
fect these signals differently during fasting.
The generation with different fasting and nonfast-
ing samples in the dataset from different subjects was
used to check for the relation between the discrimina-
tive and predictive scores and the length of the gen-
erated sequences. The results are shown in Figure 6
and no relation can be inferred for the dependence of
theses scores on the length of the generated sequence.
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
100
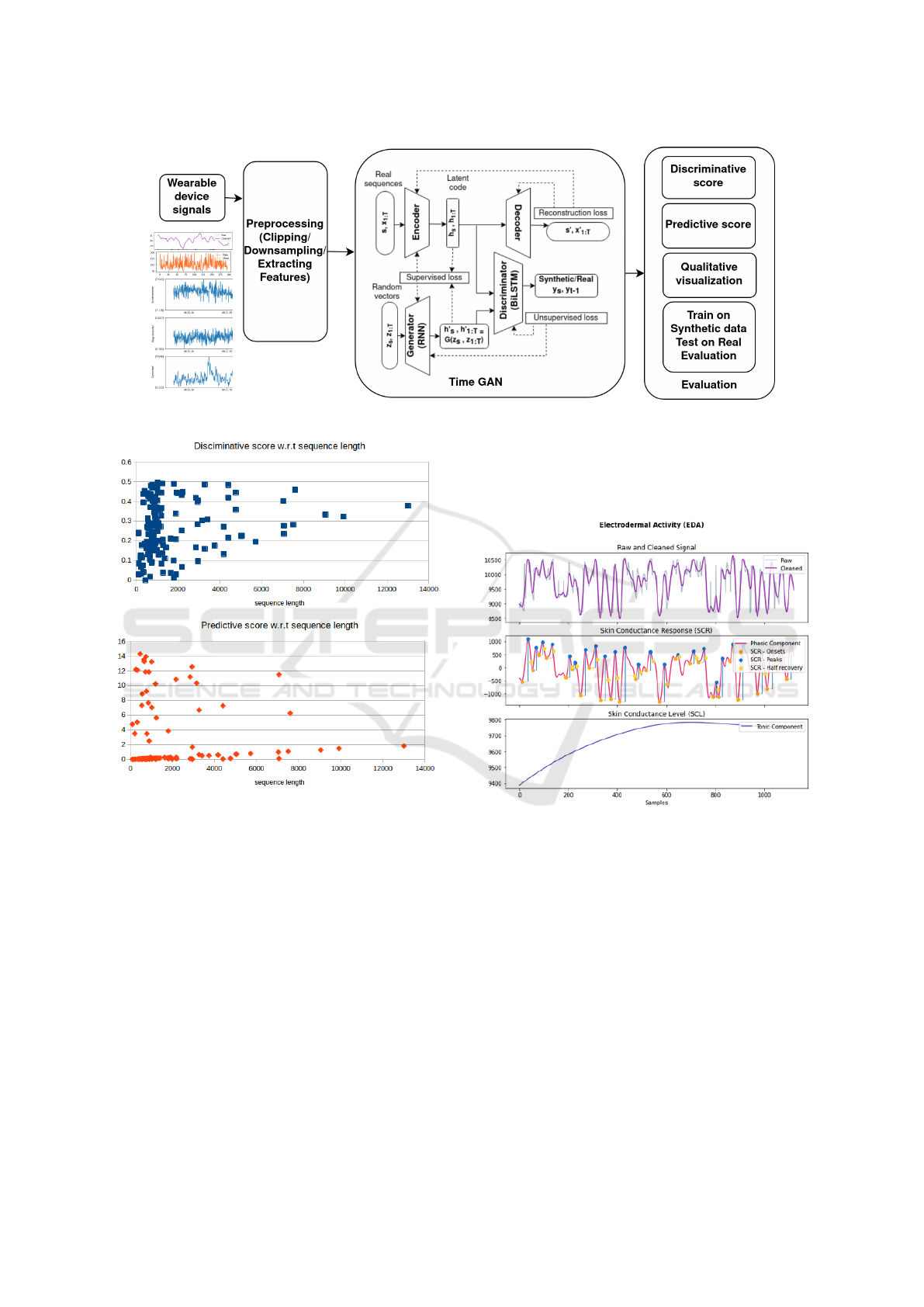
Figure 5: Overview of the wearable data generation steps.
Figure 6: Discriminative score (a) and predictive score (b)
vs generated sequence length.
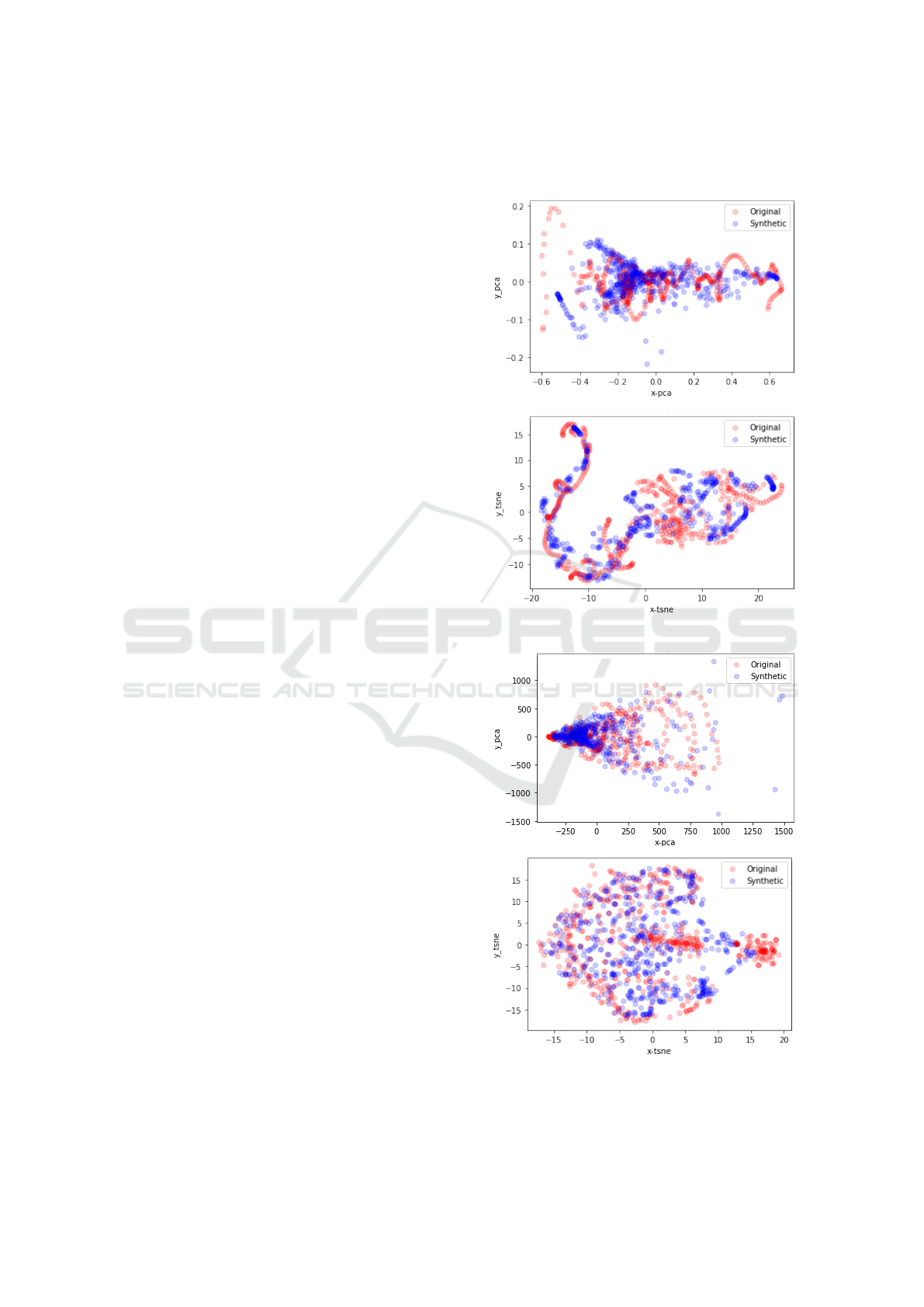
5.2 Qualitative Visualization
As for the qualitative visualization to assess how the
distribution of the generated signals is close to that of
the real signal, flattening of the time-series sequence
for all signals was done using the functions provided
by the authors of (Yoon et al., 2019). PCA and t-SNE
visualizations are shown in Figure 8 for both fasting
and non-fasting sequences. The figure shows the syn-
thetic data projections are closely following that of
the original data for these groups of sequences and
the rest of sequences show similar closeness with few
exceptions. A collective quantitative metric is prefer-
ably to be introduced to evaluate the overall distance
from the original distribution for all sequences.
Plotting samples of the generated signals sepa-
rately however doesn’t show good quality signals
most of the time e.g. an electrodermal activity signal
in Figure 7 plotted from a GSR generated signal.
Figure 7: Galvanic skin response generated sample.
5.3 Training on Synthetic Test on Real
The results of the hydration monitoring task for pre-
dicting the last drinking time were evaluated by gen-
erating balanced synthetic data for fasting and non-
fasting since the original data was having more non-
fasting samples. Different synthetic data sizes were
generated and used for training the best performing
models in (Sabry et al., 2022b), namely random for-
est, gradient boosted regression and extra trees. The
average results for 10 runs are shown in Figure 9. It
can be shown in the three graphs in Figure 9 that us-
ing synthetic data has decreased the root-mean-square
error for prediction with respect to training with the
small unbalanced real dataset (TRTR) represented by
Wearable Data Generation Using Time-Series Generative Adversarial Networks for Hydration Monitoring
101
the red line on top of the graphs with a best case of
approximately 1 hour difference in the case of using
Gradient Boosted Regressor to predict the last drink-
ing time. The best RMSEs are achieved especially for
cases when the real dataset sizes used for training are
less than 1500 samples, after that the differences in
the accuracy in prediction between using generated
data and real data in training start to shrink. This
suggests the feasibility of generation of wearable data
biosignals using TimeGAN to predict the last drink-
ing time in a hydration monitoring application espe-
cially in cases of small real dataset sizes collected. As
pointed out in (Sabry et al., 2022b), predicting the last
drinking time can provide the flexibility of adjusting
the alerting threshold on the wearable device based
on the health conditions, needs, and activity of the
user. The alerted user/caregiver can then choose to in-
put the correct drinking time if the prediction was not
accurate for his/her situation to enable a closed-loop
solution with online training to take place to improve
the model’s accuracy.
6 CONCLUSION
In this paper, we proposed using a generative adver-
sarial networks model, specifically designed for time-
series data, known as TimeGAN (Yoon et al., 2019)
to test its applicability for generating wearable biosig-
nals data. It was able to generate the synthetic wear-
able data signals with low discriminative and predic-
tive scores. Low discriminative score means a clas-
sifier can’t distinguish between original and synthetic
dataset samples. Low predictive score means that the
post-hoc sequence-prediction model is able to pre-
dict next-step temporal data signals for each input se-
quence. Visual evaluation also showed that most of
the generated signals are following the same distri-
bution of the original data. Training with synthetic
data and testing on real (TSTR) also showed that us-
ing synthetic data has decreased the root mean square
error (RMSE) for the regression task of predicting the
last drinking time compared to the training on real
testing on real (TRTR) for using different synthetic
data sizes.
One of the main objectives of biosignal wear-
able data generation is to protect the privacy of the
users’ data used in training, another evaluation met-
ric needs to be added to assess TimeGAN’s privacy
and its resistance to leak real data that participated in
the training by just the GAN memorising it (Boun-
liphone et al., 2016; Esteban et al., 2017). Another
metric for every type of synthetic biosignal generated
could be evaluated to test the quality of the generated
signals and possibly categorize them like classifying
PPG signals quality in (Elgendi, 2016).
(a) For fasting sequences.
(b) For non fasting sequences.
Figure 8: Qualitative PCA and tSNE visualization for fast-
ing and non-fasting sequences.
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
102
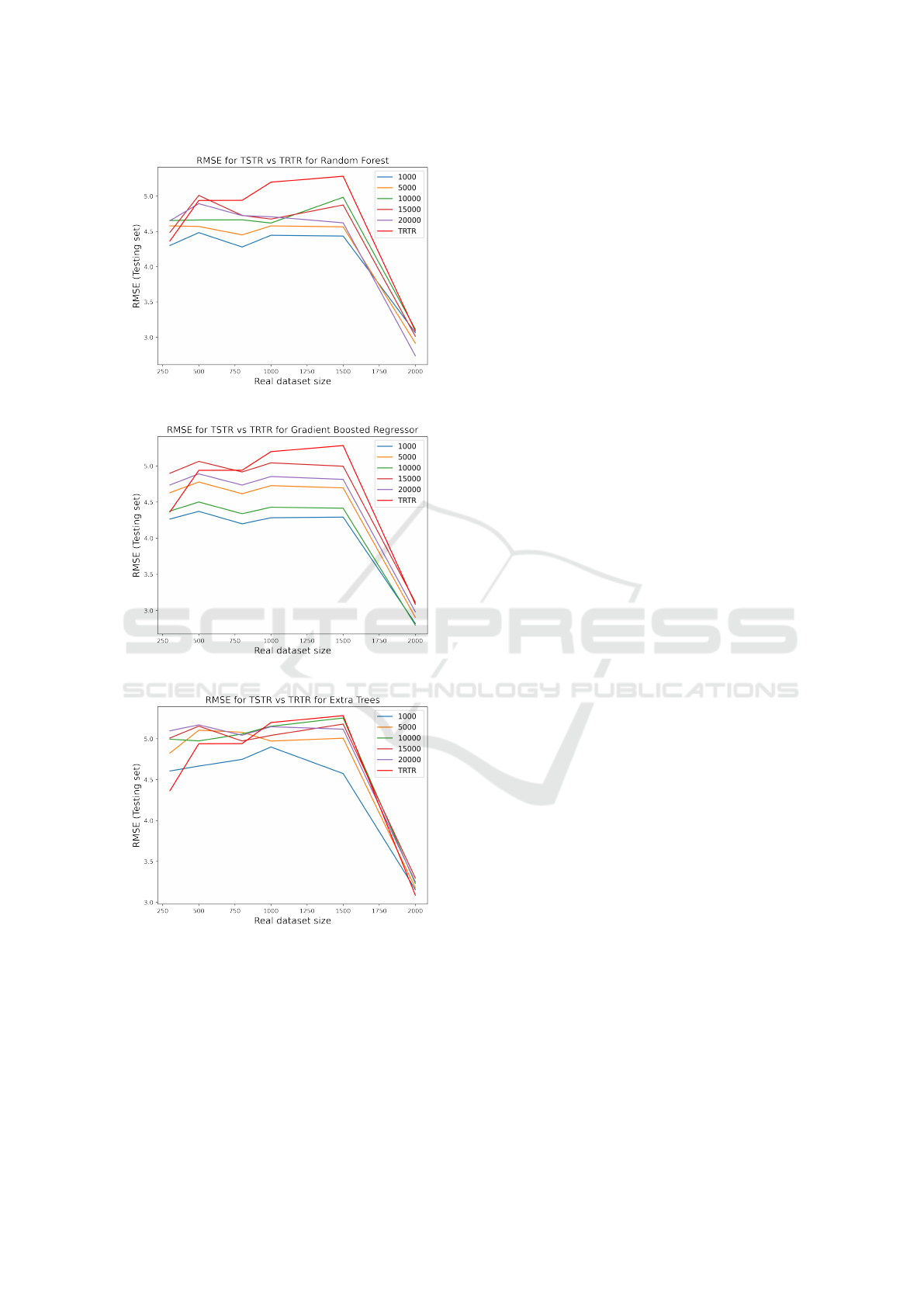
(a) Random Forest.
(b) Gradient Boosted Regressor.
(c) Extra trees.
Figure 9: Train Synthetic Test Real vs Train Real Test Real
for different models.
ACKNOWLEDGEMENTS
This publication was made possible by a grant from
the Qatar National Research Fund (QNRF), Project
Number ECRA 01-006-1-001. The contents of this
research are solely the responsibility of the authors
and do not necessarily represent the official views of
the Qatar National Research Fund (QNRF).
REFERENCES
Aqajari, S. A. H., Cao, R., Zargari, A. H. A., and
Rahmani, A. M. (2021). An End-to-End and Ac-
curate PPG-based Respiratory Rate Estimation Ap-
proach Using Cycle Generative Adversarial Net-
works. arXiv:2105.00594 [cs, eess]. arXiv:
2105.00594.
Arjovsky, M., Chintala, S., and Bottou, L. (2017). Wasser-
stein gan.
Belo, D., Rodrigues, J., and Vaz, J. (2017). Biosignals
learning and synthesis using deep neural networks.
BioMed Eng Online, 16.
Bounliphone, W., Belilovsky, E., Blaschko, M. B.,
Antonoglou, I., and Gretton, A. (2016). A test of rela-
tive similarity for model selection in generative mod-
els. In Bengio, Y. and LeCun, Y., editors, 4th Interna-
tional Conference on Learning Representations, ICLR
2016, San Juan, Puerto Rico, May 2-4, 2016, Confer-
ence Track Proceedings.
Brophy, E., Wang, Z., She, Q., and Ward, T. (2021). Gen-
erative adversarial networks in time series: A survey
and taxonomy. arXiv.
Bryant, F. and Yarnold, P. (2001). Principal-component
analysis and exploratory and confirmatory factor
analysis. American Psychological Association.
Dankar, F. K. and Ibrahim, M. (2021). Fake it till you make
it: Guidelines for effective synthetic data generation.
Applied Sciences, 11(5).
Delmastro, F., Martino, F. D., and Dolciotti, C. (2020).
Cognitive Training and Stress Detection in MCI Frail
Older People Through Wearable Sensors and Machine
Learning. IEEE Access, 8:65573–65590.
Elgendi, M. (2016). Optimal signal quality index for
photoplethysmogram signals. Bioengineering (Basel,
Switzerland), 3(21).
Esteban, C., Hyland, S. L., and R
¨
atsch, G. (2017). Real-
valued (Medical) Time Series Generation with Recur-
rent Conditional GANs.
Furdui, A., Zhang, T., Worring, M., Cesar, P., and El Ali,
A. (2021). Ac-wgan-gp: Augmenting ecg and gsr sig-
nals using conditional generative models for arousal
classification. In Adjunct Proceedings of the 2021
ACM International Joint Conference on Pervasive and
Ubiquitous Computing and Proceedings of the 2021
ACM International Symposium on Wearable Comput-
ers, UbiComp ’21, page 21–22, New York, NY, USA.
Association for Computing Machinery.
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B.,
Warde-Farley, D., Ozair, S., Courville, A., and Ben-
gio, Y. (2014). Generative adversarial nets. In Ghahra-
mani, Z., Welling, M., Cortes, C., Lawrence, N., and
Weinberger, K., editors, Advances in Neural Infor-
mation Processing Systems, volume 27. Curran Asso-
ciates, Inc.
Wearable Data Generation Using Time-Series Generative Adversarial Networks for Hydration Monitoring
103

Harada, S., Hayashi, H., and Uchida, S. (2018). Biosig-
nal data augmentation based on generative adversarial
networks. volume 2018, pages 368–371.
Hazra, D. and Byun, Y.-C. (2020). SynSigGAN: Gener-
ative Adversarial Networks for Synthetic Biomedical
Signal Generation. Biology, 9(12):441.
Hernang
´
omez, R., Visentin, T., Servadei, L., Khod-
abakhshandeh, H., and Sta
´
nczak, S. (2022). Im-
proving Radar Human Activity Classification Using
Synthetic Data with Image Transformation. Sensors,
22(4):1519.
Jabbar, A., Li, X., and Omar, B. (2022). A Survey on Gen-
erative Adversarial Networks: Variants, Applications,
and Training. ACM Computing Surveys, 54(8):1–49.
Karras, T., Laine, S., and Aila, T. (2021). A style-based
generator architecture for generative adversarial net-
works. IEEE Transactions on Pattern Analysis and
Machine Intelligence, 43(12):4217–4228.
Kiranyaz, S., Devecioglu, O., Ince, T., Malik, J., Chowd-
hury, M., Hamid, T., Mazhar, R., Khandakar, A.,
Tahir, A., Rahman, T., and Gabbouj, M. (2022). Blind
ecg restoration by operational cycle-gans. IEEE Trans
Biomed Eng.
Kiyasseh, D., Tadesse, G. A., Nhan, L. N. T., Van Tan,
L., Thwaites, L., Zhu, T., and Clifton, D. (2020).
PlethAugment: GAN-Based PPG Augmentation
for Medical Diagnosis in Low-Resource Settings.
IEEE Journal of Biomedical and Health Informatics,
24(11):3226–3235.
Lo, J., Cardinell, J., Costanzo, A., and Sussman, D. (2021).
Medical Augmentation (Med-Aug) for Optimal Data
Augmentation in Medical Deep Learning Networks.
Sensors, 21(21):7018.
Lu, H., Han, H., and Zhou, S. K. (2021). Dual-GAN:
Joint BVP and Noise Modeling for Remote Physio-
logical Measurement. In 2021 IEEE/CVF Conference
on Computer Vision and Pattern Recognition (CVPR),
pages 12399–12408, Nashville, TN, USA. IEEE.
Luo, Y., Cai, X., Zhang, Y., and Xu, J. (2018). Multivariate
Time Series Imputation with Generative Adversarial
Networks. page 12.
Metz, L., Poole, B., Pfau, D., and Sohl-Dickstein, J. (2017).
Unrolled generative adversarial networks. In Interna-
tional Conference on Learning Representations.
Montero, A., Bonet-Carne, E., and Burgos-Artizzu, X. P.
(2021). Generative Adversarial Networks to Improve
Fetal Brain Fine-Grained Plane Classification. Sen-
sors, 21(23):7975.
Nguyen, H., Zhuang, D., Wu, P.-Y., and Chang, M.
(2020). AutoGAN-based dimension reduction for pri-
vacy preservation. Neurocomputing, 384:94–103.
Ning, X., Yao, L., Wang, X., Benatallah, B., Zhang, S.,
and Zhang, X. (2018). Data-Augmented Regression
with Generative Convolutional Network. In Hacid,
H., Cellary, W., Wang, H., Paik, H.-Y., and Zhou,
R., editors, Web Information Systems Engineering –
WISE 2018, volume 11234, pages 301–311. Springer
International Publishing, Cham. Series Title: Lecture
Notes in Computer Science.
Piacentino, E., Guarner, A., and Angulo, C. (2021). Gen-
erating synthetic ecgs using gans for anonymizing
healthcare data. Electronics, 10(4):389.
Ping, H., Stoyanovich, J., and Howe, B. (2017). Data-
synthesizer: Privacy-preserving synthetic datasets. In
Proceedings of the 29th International Conference on
Scientific and Statistical Database Management, SS-
DBM ’17, New York, NY, USA. Association for Com-
puting Machinery.
Reiter, J. P. (2005). Using cart to generate partially synthetic
public use microdata. Journal of Official Statistics,
21:441–462.
Sabry, F., Eltaras, T., Labda, W., Alzoubi, K., and Malluhi,
Q. (2022a). Machine learning for healthcare wearable
devices: The big picture. Journal of Healthcare Engi-
neering.
Sabry, F., Eltaras, T., Labda, W., Hamza, F., Alzoubi, K.,
and Malluhi, Q. (2022b). Towards on-device dehy-
dration monitoring using machine learning from wear-
able device’s data. Sensors, 22(5).
Thambawita, V., Hicks, S., Isaksen, J., Stensen, M., Hau-
gen, T., Kanters, J., Parasa, S., de Lange, T., Johansen,
H., Johansen, D., Hammer, H., Halvorsen, P., and
Riegler, M. (2021). Deepsynthbody: the beginning
of the end for data deficiency in medicine. pages 1–8.
Um, T. T., Pfister, F. M. J., Pichler, D., Endo, S., Lang, M.,
Hirche, S., Fietzek, U., and Kuli
´
c, D. (2017). Data
Augmentation of Wearable Sensor Data for Parkin-
son’s Disease Monitoring using Convolutional Neural
Networks. In Proceedings of the 19th ACM Interna-
tional Conference on Multimodal Interaction, pages
216–220. arXiv:1706.00527 [cs].
van der Maaten, L. and Hinton, G. (2008). Visualizing data
using t-sne. Journal of Machine Learning Research,
9(86):2579–2605.
Xin, B., Yang, W., Geng, Y., Chen, S., Wang, S., and
Huang, L. (2020). Private FL-GAN: Differential Pri-
vacy Synthetic Data Generation Based on Federated
Learning. In ICASSP 2020 - 2020 IEEE International
Conference on Acoustics, Speech and Signal Process-
ing (ICASSP), pages 2927–2931, Barcelona, Spain.
IEEE.
Xu, L., Skoularidou, M., Cuesta-Infante, A., and Veera-
machaneni, K. (2019). Modeling Tabular data using
Conditional GAN. arXiv, page 11.
Yoon, J., Jarrett, D., and Schaar, M. (2019). Time-
series generative adversarial networks. In Wallach,
H., Larochelle, H., Beygelzimer, A., d'Alch
´
e-Buc, F.,
Fox, E., and Garnett, R., editors, Advances in Neural
Information Processing Systems, volume 32. Curran
Associates, Inc.
Zargari, A. H. A., Aqajari, S. A. H., Khodabandeh,
H., Rahmani, A. M., and Kurdahi, F. (2021).
An Accurate Non-accelerometer-based PPG Mo-
tion Artifact Removal Technique using CycleGAN.
arXiv:2106.11512 [cs]. arXiv: 2106.11512.
Zhang, J., Cormode, G., Procopiuc, C. M., Srivastava, D.,
and Xiao, X. (2017). Privbayes: Private data release
via bayesian networks. ACM Trans. Database Syst.,
42(4).
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
104

Zhou, B., Liu, S., Hooi, B., Cheng, X., and Ye, J. (2019).
BeatGAN: Anomalous Rhythm Detection using Ad-
versarially Generated Time Series. In Proceedings
of the Twenty-Eighth International Joint Conference
on Artificial Intelligence, pages 4433–4439, Macao,
China. International Joint Conferences on Artificial
Intelligence Organization.
Zhu, J.-Y., Park, T., Isola, P., and Efros, A. A. (2017).
Unpaired image-to-image translation using cycle-
consistent adversarial networks.
Wearable Data Generation Using Time-Series Generative Adversarial Networks for Hydration Monitoring
105
