
Imposing Functional Priors on Bayesian Neural Networks
Bogdan Kozyrskiy
1
, Dimitrios Milios
2
and Maurizio Filippone
1
1
Department of Data Science, EURECOM, 450 Route des Chappes, Biot, France
2
Jubile Tech Ltd., London, U.K.
Keywords:
Bayesian Inference, Markov Chain Monte-Carlo, Deep Neural Networks.
Abstract:
Specifying sensible priors for Bayesian neural networks (BNNs) is key to obtain state-of-the-art predictive
performance while obtaining sound predictive uncertainties. However, this is generally difficult because of the
complex way prior distributions induce distributions over the functions that BNNs can represent. Switching
the focus from the prior over the weights to such functional priors allows for the reasoning on what meaningful
prior information should be incorporated. We propose to enforce such meaningful functional priors through
Gaussian processes (GPs), which we view as a form of implicit prior over the weights, and we employ scalable
Markov chain Monte Carlo (MCMC) to obtain samples from an approximation to the posterior distribution
over BNN weights. Unlike previous approaches, our proposal does not require the modification of the original
BNN model, it does not require any expensive preliminary optimization, and it can use any inference tech-
niques and any functional prior that can be expressed in closed form. We illustrate the effectiveness of our
approach with an extensive experimental campaign.
1 INTRODUCTION
Artificial Neural Networks (NN) currently represent
a general class of successful models for various ma-
chine learning tasks, including computer vision, nat-
ural language processing, and many others. Bayesian
Neural Networks (BNN) combine the representation
power of NNs with Bayesian inference, making them
an attractive choice in applications where predictive
performance and accurate uncertainty quantification
is important. BNNs are difficult to use because of
the intractability of the posterior over model param-
eters, which necessitates approximations. Choosing
appropriate priors over model parameters is also cru-
cial for good performance (Fortuin, 2022; Tran et al.,
2022). In BNNs, the prior over the weights and the
network architecture determine a distribution over the
outputs of such BNNs (Sun et al., 2019), and we re-
fer to this induced prior as a functional prior. The
functional prior should encode any prior information
on the conditional distribution of the labels given the
inputs. However, it is unclear how to encode this type
of information when having to specify a prior distri-
bution over the weights.
This paper presents a framework for imposing
meaningful functional priors using scalable Markov
chain Monte Carlo (MCMC) sampling from an ap-
proximation to the posterior distribution over BNN
weights, and we specify the prior over the weights
implicitly through a prior over the induced functional
prior. Our approach is different from the literature
on Implicit Process Priors (IPPs) (Ma et al., 2019),
where the goal is to obtain an approximate frame-
work to handle the functional prior implicitly induced
by the choice of a prior distribution over the weights.
In our work, we operate in the opposite direction by
imposing a functional prior, which implicitly deter-
mines a prior over the weights; we do not know such
a prior over the weights in closed form, but we im-
plicitly determine it through the specification of the
induced functional prior.
Stochastic Processes are natural mathematical ob-
jects suitable to define distributions over functions
(Kallenberg and Kallenberg, 1997), and Gaussian
Processes (GPs) represent popular examples which
are routinely used in numerous machine learning
tasks. This type of stochastic processes is well
investigated and has strong theoretical foundations
(Williams and Rasmussen, 2006). There are theo-
retical guarantees for the generalization error of GP
regression, and this method has a strong connection
with non-Bayesian Kernel Ridge Regression (KRRs)
(Kanagawa et al., 2018). Also, it was shown in (Neal,
1996) that in the infinite width limit, shallow BNNs
are equivalent to GPs. We propose to use GPs to im-
pose functional priors over BNNs because GPs pro-
vide a flexible set of tools to encode different types of
beliefs about functions, such as periodicity or smooth-
450
Kozyrskiy, B., Milios, D. and Filippone, M.
Imposing Functional Priors on Bayesian Neural Networks.
DOI: 10.5220/0011742900003411
In Proceedings of the 12th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2023), pages 450-457
ISBN: 978-989-758-626-2; ISSN: 2184-4313
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

ness through the specification of kernels. However,
our approach is not restricted to GPs, and it can han-
dle any functional priors that can be written down in
closed form.
This paper is organized as follows. We review the
related literature in Sec. 2, and we present our method
in Sec. 3. We report results on various benchmarks
in Sec. 4 and we conclude the paper in Sec. 6, after
discussing the limitations of our work in Sec. 5.
2 RELATED WORK
A popular way of choosing prior distributions for
BNNs is to employ a Gaussian distribution over the
weight of the model (Graves, 2011; Neal, 1996). This
offers some practical advantages, for instance when
employing Variational Inference (VI) (Graves, 2011).
Mean-field VI allows for efficient calculations of the
regularization part of the VI objective function with-
out the need to resort to Monte Carlo approximations,
but the limited flexibility of the approximating distri-
butions may negatively affect performance.
Even when adopting more advanced and generally
more accurate inference techniques, such as Stochas-
tic Gradient Hamiltonian Monte Carlo (SG-HMC)
(Chen et al., 2014), the Gaussian assumption on the
prior over model parameters is still common. It was
shown by (Fortuin et al., 2021) that Gaussian priors
are problematic in terms of model performance and
the ability to detect Out-of-Domain (OOD) input ex-
amples. This work also shows how Gaussian priors
over the weights could be responsible for the cold
posterior effect described by (Wenzel et al., 2020);
this effect is characterized by the necessity of apply-
ing temperature scaling to the prior density term in
Bayes theorem in order to obtain good performance.
Flexible alternatives to Gaussian priors, such as
mixture of Gaussians (Blundell et al., 2015), Stu-
dent’s t-distribution (Fortuin et al., 2021), hierarchi-
cal Gaussian distribution (Chen et al., 2014) and many
others (Fortuin, 2022) were developed to address poor
performance of Gaussian priors. However, all these
types of priors still do not help understanding their
effect on model outputs.
An alternative to studying weight priors is to fo-
cus on their effect on NNs functional priors. A vari-
ational objective computed on a finite set of func-
tion evaluations is proposed in (Sun et al., 2019) for
finding a Bayesian posterior in the space of functions
for a functional prior defined by a stochastic process.
The authors show that the supremum of the KL di-
vergence over all sets of input points is equal to the
true KL divergence in functional space. In this set-
ting, the optimization procedure simultaneously min-
imizes the optimization objective with respect to the
parameters of the model and maximizes the KL term
with respect to the input data points, which makes the
optimization process unstable. Also, the optimization
objective requires evaluating the gradient of the ap-
proximate posterior density by the Stein gradient es-
timator (Shi et al., 2018), and this requires a careful
choice of a kernel function. The work in (Ma et al.,
2019) focuses on representing the functional prior as
a BNN and uses GPs to obtain an approximate poste-
rior over functions. The problem with this approach
is that GPs may yield a poor approximation quality
for the true functional posterior. The authors in (Sun
et al., 2019) and (Ma et al., 2019) use VI to find an
approximate posterior distribution, which means that
the optimization objective contains a functional KL
divergence term. However, in (Rudner et al., 2021) it
is claimed that the KL divergence between the func-
tional approximate posterior and the GP process func-
tional prior is problematic as it may diverge to infin-
ity. On the other hand, they acknowledge that it does
not mean that parametric models cannot approximate
GPs well.
The authors of (Tran et al., 2022) propose to im-
pose functional GP priors so as to constrain the para-
metric prior over the weights of BNNs. They propose
to optimize parameters of the prior over the weights
by minimizing the Wasserstein distance between the
BNN functional prior and the GP prior. Then, the pos-
terior over the weights is characterized by means of
MCMC.
In our work, we aim to avoid the computation of
the KL divergence or any other distance metric in
function spaces. Instead, we propose to enforce the
choice of a functional prior directly when carrying out
approximate inference of BNN weights.
3 METHODS
Consider a supervised learning task with a dataset
D{(x
i
, y
i
)}
i=1...n
of n input vectors X =
{
x
i
}
i=1...,n
and corresponding labels y =
{
y
i
}
i=1...,n
, and imag-
ine employing a NN-based model with param-
eters w to establish a parametric mapping be-
tween inputs and labels. We denote the in-
put/output mapping by f
w
(x), and for convenience
we also define f
⊤
= [ f
w
(x
1
), . . . , f
w
(x
N
)] and f
∗⊤
=
[ f
w
(x
1
), . . . , f
w
(x
N
), f
w
(
˜
x
1
), . . . , f
w
(
˜
x
M
)] as the evalu-
ation of the function f
w
(x) at the inputs X and an aug-
mented set of inputs X
∗
= [X,
˜
X], respectively. The set
X
∗
has cardinality N
∗
= N +
˜
N, and the
˜
N inputs in
˜
X
are drawn from a given p(x). Note that the sets X and
Imposing Functional Priors on Bayesian Neural Networks
451

X
∗
can be disjoint, but in order to keep the notation
uncluttered, we assume X ⊂ X
∗
3.1 Imposing Functional Priors on
BNNs
A Bayesian treatment NNs requires specifying a prior
distribution p(w) over the parameters and a likeli-
hood function for the labels given the inputs, that is
p(y|X, w). For this BNN, it is possible to write down
an expression for the posterior distribution over model
parameters as:
p(w|y, X) =
p(y|X, w)p(w)
R
p(y|X, w)p(w)dw
(1)
Carrying out inference in BNNs is extremely dif-
ficult for at least two reasons. One main difficulty
stems from the complex way in which parameters
affect the likelihood function, and this requires ap-
proximation techniques to characterize the posterior
over model parameters; popular approaches involve
MCMC and variational approximations. A second
and more subtle challenge is how to specify priors
for BNNs, because it is difficult to establish what
is the effect of prior parameters on the distribution
over the functions that BNNs can represent. In this
work, we propose a novel way to address the chal-
lenge of choosing sensible priors for BNNs by work-
ing with implicit priors over the weights induced by
the choice of functional priors, while we follow the
recent trend to employ MCMC techniques to address
the intractability of the inference process. We begin
by focusing on the distribution over the functions rep-
resented by BNNs. In particular, we consider the dis-
tribution of f
∗
, which is the distribution of f
w
(x) eval-
uated at the set of input points X
∗
, and we impose a
prior over this set of variables which encourages func-
tions to behave in a sensible way a priori. Later we
will study in particular Gaussian process priors, but
any functional prior can be incorporated as long as it
can be expressed in closed form.
We now rewrite the likelihood function in terms of
f rather than w:
p(y|X, w) → p(y|f). (2)
The main idea behind our work is to now define a
prior over f instead of w, and to perform inference
over w. With this change of variables, we should ac-
count for the change of measure through a Jacobian
term. However, such a change of variables involves
groups of variables of different dimensions in general
and even when this is not the case, computing this
term would be computationally costly. For this rea-
son, we are going to ignore the Jacobian accepting to
settle for an approximate posterior over w. With this
choice, we rewrite Bayes theorem as:
log p(f
∗
|y, X
∗
) = log p(y|f) + log p(f
∗
|X
∗
) + const.
(3)
Note that in this equation we introduced the func-
tional prior:
p(f
∗
|X
∗
) =
Z
p(f
∗
|X
∗
, w)p(w)dw, (4)
where p(f
∗
|X
∗
, w) is a Dirac’s delta placed at the eval-
uation of f
w
(x) at the inputs X
∗
due to the determin-
istic way in which inputs are mapped into outputs in
NNs. Again, we stress that while we focus on the dis-
tribution of functions represented by BNNs, we ac-
tually use the objective in eq. 3 to perform MCMC
sampling in the space of the weights w. Note that
we carry out inference over w through MCMC, but
given that we are working with an approximation to
the posterior over w, we could alternatively employ
other fast approximate inference techniques such as
VI. Here, we focus on MCMC so as to isolate the ef-
fect of the way we impose functional priors compared
with alternatives which try to characterize the exact
posterior over w (Tran et al., 2022).
Bayesian Interpretation. From a Bayesian point of
view, imposing a prior over function by specifying a
prior over f
∗
induces an implicit prior over the weights
through eq. 4. In other words, the prior over f
∗
is in
practice a prior over a deterministic transformation of
w, and this is implemented by the NN. It is interest-
ing to note that in the literature eq. 4 is usually in-
terpreted in the opposite way; that is, one uses eq. 4
starting from a prior over the weights p(w) to define
a functional prior in an implicit way (Ma et al., 2019).
The likelihood function establishes what is the like-
lihood of the labels y and it is conditioned on f or
equivalently on w and X. Therefore, the expression
in eq. 3 can be seen as an expression for the (approx-
imate) posterior over the weights w (due to the lack
of a Jacobian term), where the prior is assumed over
a transformation of such weights. In this paper, we
take this view to carry out Bayesian inference over
w using MCMC techniques. We also note that our
approach has some close similarity with the Product
of Expert approach proposed in (Wenk et al., 2019)
for inference of parameters of Ordinary Differential
Equations using Gaussian Processes.
Regularization Interpretation. While we proceed
with a Bayesian treatment of w, it is useful to interpret
eq. 3 as a regularized objective in the following way.
The first term log p(y|f) is the negative loss, which
can be equivalently be seen as a function of w and
ICPRAM 2023 - 12th International Conference on Pattern Recognition Applications and Methods
452

X
∗
, so this provides a constraint on w because the
objective promotes values of f which are compatible
with the labels y, and f depends on w and X
∗
. The
second term is a regularization term, which penalizes
functions deviating from a behavior established by the
functional prior. Because f
∗
is a function of w and X
∗
,
this translates into a regularization term for w.
3.2 Imposing Functional Priors
Through Gaussian Processes
The proposed formulation focusing on functional rep-
resentations has the advantage of putting the empha-
sis on the functions that BNNs can represent, and for
which it is possible to assume sensible priors. Here
we specify how to operate in case of Gaussian pro-
cesses (GPs), which yield a prior term in eq. 3 as:
log p(f
∗
|X
∗
) = −
1
2
f
∗⊤
C
−1
f
∗
+ const, (5)
where the covariance matrix is C = (K
X
∗
X
∗
+σ
2
n
), and
K
X
∗
X
∗
contains the evaluation of the kernel function κ
among all the inputs in X
∗
. For simplicity, we assume
a zero-mean GP, but other mean functions can be eas-
ily included. In the next subsections, we elaborate on
how to use this GP prior in practice, by proposing a
way to operate with mini-batches for scalability pur-
poses, by discussing hyper-parameter optimization,
and by discussing the properties of the proposed ap-
proach when N
∗
goes to infinity.
3.2.1 Mini-Batching
In this work, we aim to employ advanced MCMC
sampling methods based on stochastic gradients, and
in particular Stochastic Gradient Hamiltonian Monte
Carlo (SG-HMC) (Chen et al., 2014) to sample from
the weights w of BNNs. In order to do so, we need to
formulate our MCMC objective in a way that is suit-
able for mini-batching. However, extending the pre-
vious formulation to operate with mini-batches with-
out care would produce a biased estimation of the
quadratic term f
∗⊤
C
−1
f
∗
̸= E[f
∗⊤
b
C
−1
b
f
∗
b
], where f
b
and
C
b
are computed over a mini-batch X
b
.
The main difficulty of full batch training is the ne-
cessity of solving linear systems with the matrix C,
which has O(N
∗3
) complexity in the number of in-
puts in X
∗
. The literature on GPs offers many cues on
how to circumvent this problem. In particular, there
exist formulations of GPs based on random features
(Rahimi and Recht, 2007) which operate on mini-
batches (Cutajar et al., 2017). In this work, we fo-
cus on approximations based on random features, but
inducing points formulations are also possible.
Random Feature (RF) expansions of the kernel
κ(·, ·) allow one to obtain a finite-dimensional repre-
sentation for an explicit feature map which approx-
imates the true possibly infinite-dimensional feature
map. Using this expansion, we can express the Gram
matrix as a dot product of feature maps computed
over the data K ≈ ΦΦ
⊤
. We can use this property and
the Woodbury identity to rewrite the quadratic term as
follows:
f
∗⊤
C
−1
f
∗
= f
∗⊤
(ΦΦ
⊤
+ σ
2
f
I)
−1
f
∗
=
1
σ
2
f
f
∗⊤
f
∗
−
1
σ
2
f
f
∗⊤
Φ(Φ
⊤
Φ + σ
2
f
I)
−1
Φ
⊤
f
∗
.
(6)
In this case, instead of inverting a matrix of size
N
∗
× N
∗
, we invert a matrix of size D × D, where
D is the dimensionality of the RF vector. However,
this approach has two drawbacks. First, it is unsta-
ble when σ
2
f
→ 0, because after the application of the
Woodbury identity the term
1
σ
2
f
f
∗⊤
f
∗
→ ∞. Second,
this approach still does not allow mini-batching.
We can reformulate our MCMC objective by re-
placing the nonparametric term pertaining to the GP
with a parametric one based on RFs. For the set f
∗
,
we can factorize its prior probability as:
p(f
∗
|X
∗
) =
Z
p(f
∗
|β, X
∗
)p(β)dβ, (7)
where β are the parameters of RF approximation of
the GP, that is p(β) ∼ N (0, I) and p(f
∗
|β, X
∗
) ∼
N (Φβ, σ
2
f
I). In this case it is easy to verify that
p(f
∗
) = N (0, ΦΦ
⊤
+σ
2
f
I) and according to the prop-
erty of the RF approximation, the covariance ma-
trix coincides with the prior term of the objective in
eq. 6. Instead of sampling directly from the unnormal-
ized posterior p(f
∗
|X
∗
, y) marginalized over β, we can
sample from the joint density p(f
∗
, β|X, y) and discard
samples over β:
p(f
∗
, β|X
∗
, y) ∝ p(y|f)p(f
∗
|β, X
∗
)p(β). (8)
Again, when we refer to the fact that we sample f
∗
,
in practice we sample w. This RF-based approach
avoids the necessity of inverting the matrix (ΦΦ
⊤
+
σ
2
f
I) during the computation of the objective.
Resuming, the expression for the unnormalized
log-posterior in eq. 8, where the GP regularization is
approximated using RFs, is as follows:
log p(y|f) −
1
2σ
2
f
∥f
∗
− Φβ∥
2
−
||β||
2
2
+ const. (9)
It is straightforward to verify that this MCMC ob-
jective can be written as a sum of terms involving
individual input points, and it is therefore amenable
to mini-batching. It is also easy to verify that
Imposing Functional Priors on Bayesian Neural Networks
453

one can proceed with a Gibbs sampling scheme
whereby f
∗
(that is w) is sampled from the conditional
ˆp(f
∗
|β, X
∗
, y) using SG-HMC and β is sampled di-
rectly from ˆp(β|f
∗
, X
∗
, y), which has a Gaussian form.
3.2.2 Hyper-Parameter Optimization
The choice of a GP prior opens to the need to spec-
ify its kernel parameters. In the absence of any way
to determine such hyper-parameters, we propose to
optimize them by marginal log-likelihood (MLL) op-
timization, which is a popular way to proceed with
GP models. In our case, the random feature approxi-
mation lends itself to a scalable solution, avoiding the
need to invert large matrices. Again, using Woodbury
matrix identities, it is possible to rewrite the marginal
likelihood so that the cost of computing it is cubic in
the number of random features instead of cubic in the
number of input points.
3.2.3 Classification
While for regression it is natural to specify func-
tional priors through GPs and to obtain a tractable
framework to scale these through random features, for
other likelihoods things may become more involved.
For instance, in classification problems, we may wish
to specify functional priors such that the distribution
over classes is uniform a priori.
Alternatively, following an empirical Bayes ap-
proach, we could optimize the GP prior hyper-
parameters so as to maximize the marginal likeli-
hood. In this case, the random feature approximation
of GPs leads to so-called Generalized Linear Mod-
els (GLMs) and this requires approximations to be
able to compute the marginal likelihood. For classifi-
cation tasks, there exist solutions to bypass the need
to work directly with Bernoulli or Multinoulli likeli-
hoods p(y|w, X). Here we follow the idea proposed
by (Milios et al., 2018), in which labels are trans-
formed so that classification models can be replaced
by regression models with heteroskedastic observa-
tion noise. In particular, for each one-hot encoded
label y we can obtain real valued vectors
˜
y, σ
2
n
(see
(Milios et al., 2018) for details):
˜y
i
= log(α
i
) −
˜
σ
2
i
2
;
˜
σ
2
i
= log
1
α
i
+ 1
. (10)
With this transformation, we can use a Gaussian like-
lihood which is conjugate to the Gaussian prior, and
thus we can obtain a closed form solution for the
marginal likelihood of the model.
4 EXPERIMENTS
4.1 Toy Regression Dataset
We test our approach on a 1D synthetic dataset using
a two-hidden layer NN with tanh activation and 256
neurons per layer. The functional GP prior uses an
RBF kernel with length-scale l = 1 and output vari-
ance σ
2
out
= 1. Fig. 1 shows functions sampled from
the predictive posterior of the BNN with this GP prior
(GP in the figure) as well as the same GP prior ap-
proximated with 100 random features with and with-
out mini-batching (GP RFF and GP RFF mini-batch
in the figure). We also include the approach from
(Tran et al., 2022) which optimizes the Wasserstein
distance between the BNN functional prior and the
GP prior to determine the prior over BNN weights
(WDGPi-G in the figure). For the models with func-
tional prior we used a regularization set of 200 equally
spaced test points.
4.2 UCI Regression Datasets
We tested our approach on UCI datasets (Dua and
Graff, 2017) using a two-hidden layer MLP with tanh
activation and 100 neurons per layer, except for the
Protein dataset for which we used 200 neurons. We
imposed a GP prior with an RBF kernel and stan-
dardized the input vectors and labels. We used the
extended dataset X
∗
, which consists of 90% training
data and 10% of uniformly sampled vectors from the
input domain, for all experiments. Full-batch training
was used for all datasets, while mini-batch training
with a batch size of 512 was used for Kin8nm, Power,
and Protein.
As a baseline, we consider the aforementioned
WDGPi-G method with a Gaussian prior over weights
and a Hierarchical GP with a LogNormal distribution
over the GP kernel length-scale and output variance.
We compare our method to WDGPi-G and deep en-
sembles (Lakshminarayanan et al., 2017) in terms of
RMSE, as shown in Table 1. Each model in the en-
semble had the same architecture as the NN in our
method.
According to the results, our method is compet-
itive with WDGPi-G on most datasets. It is worthy
to note that WDGPi-G uses a Hierarchical Gaussian
Process as a functional prior, while our method uses a
simple GP. Hierarchical GPs represent a richer func-
tional prior, but we still achieve competitive perfor-
mance.
We tested the proposed method on the Power
dataset with deeper NN architectures featuring four
and six layers, and compared its RMSE with Deep
ICPRAM 2023 - 12th International Conference on Pattern Recognition Applications and Methods
454
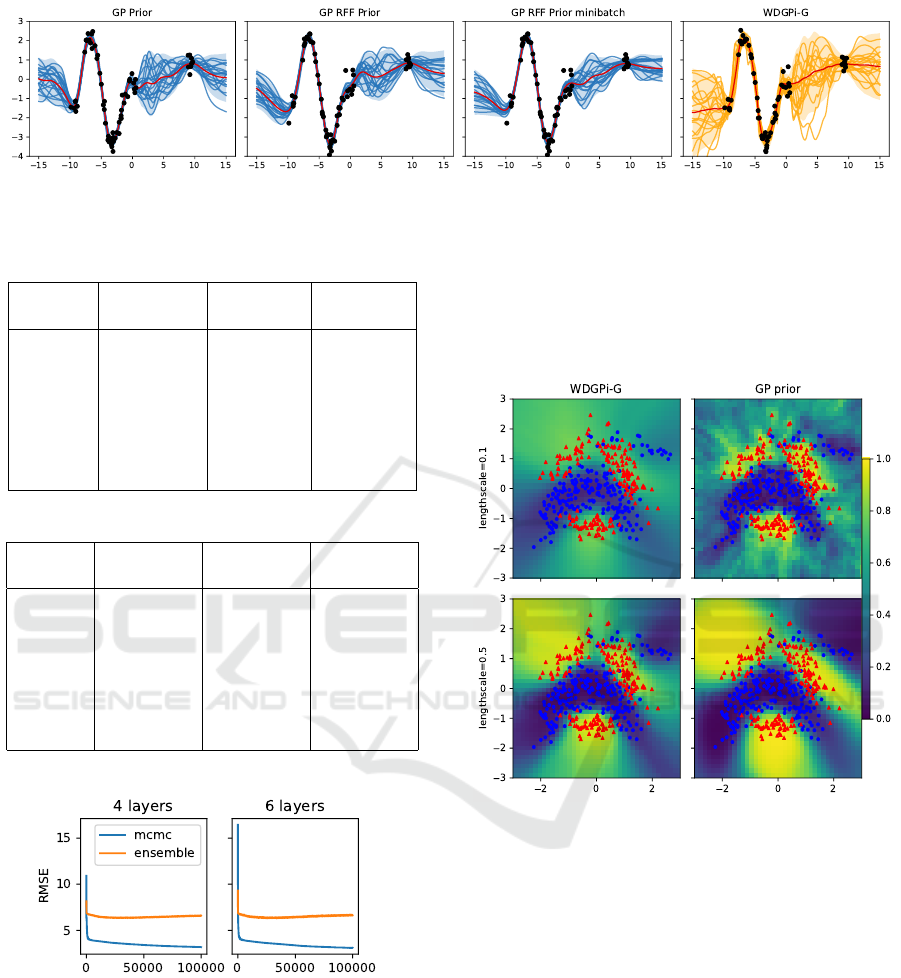
Figure 1: Sampled predictions of BNNs where the GP functional prior is imposed implicitly (our work) and by means of the
optimization of the Wasserstein distance with the functional BNN prior (WDGPi-G).
Table 1: Average RMSE for UCI regression datasets.
Dataset Functional WDGPi-G Deep
MCMC Ensembles
Boston 2.73±0.02 2.83±0.92 3.69±1.15
Concrete 4.06±0.12 4.80±0.41 5.22±0.63
Energy 0.48±0.18 0.34±0.07 1.37±0.32
Kin8nm 0.04±0.00 0.06±0.00 0.06±0.00
Power 3.24±0.06 3.72±0.18 3.86±0.21
Protein 3.61 ±0.04 3.65±0.02 4.45±0.02
Wine 0.60±0.01 0.60±0.04 0.62±0.02
Table 2: MNLL for UCI regression datasets.
Dataset Functional WDGPi-G Deep
MCMC Ensembles
Boston 2.45±0.01 2.48±0.12 3.19±1.12
Concrete 2.74±0.16 3.03±0.05 3.07±0.26
Energy 0.80±0.05 0.35±0.15 2.07±0.98
Kin8nm -1.46±0.11 -1.23±0.01 -1.32±0.08
Power 2.73±0.08 2.74±0.04 2.74±0.05
Protein 2.73±0.01 2.75±0.00 2.80±0.01
Wine 0.76±0.04 0.92±0.06 1.08±0.20
Ensembles over iterations (Fig. 2).
Figure 2: Convergence of RMSE on test data for the Power
dataset.
4.3 Toy Classification Dataset
We demonstrate the proposed approach on a 2D toy
example using the banana dataset and a two-hidden
layer NN with tanh activation and 256 neurons per
layer. We transform the labels using the method from
(Milios et al., 2018) to allow for a Gaussian likeli-
hood, as described in Sec. 3. We use an RBF kernel
with σ
out
= 5 and varying length-scales, and compare
to the WDGPi-G method from (Tran et al., 2022). We
use a grid of 40×40 points as a regularization set and
test set. The plot shows that WDGPi-G fails to incor-
porate the GP prior for a small length-scale (l = 0.1)
and the prediction function is smoother than expected.
Figure 3: Sampled predictions of the neural network with
GP prior using Mahalanobis regularization and WDGPi-G
methods.
4.4 UCI Classification Datasets
In this section, we test our approach on various UCI
classification datasets using a two-hidden layer NN
with tanh activation. We use 100 neurons in each hid-
den layer for EEG, HTRU2, Letter, and Magic, and
200 neurons for Miniboo, Drive, and Mocap. We use
the RF approximation of the functional GP prior with
D = 1000 random features and mini-batches of size
512 on all datasets. GP hyper-parameters are opti-
mized using the label transformation from Sec. 3. We
found that using this transformation with the BNN it-
self gave slightly better results than using classifica-
tion likelihoods, so we report these results in the ta-
ble. We attribute this to the optimization of GP hyper-
parameters with the transformed labels.
Imposing Functional Priors on Bayesian Neural Networks
455
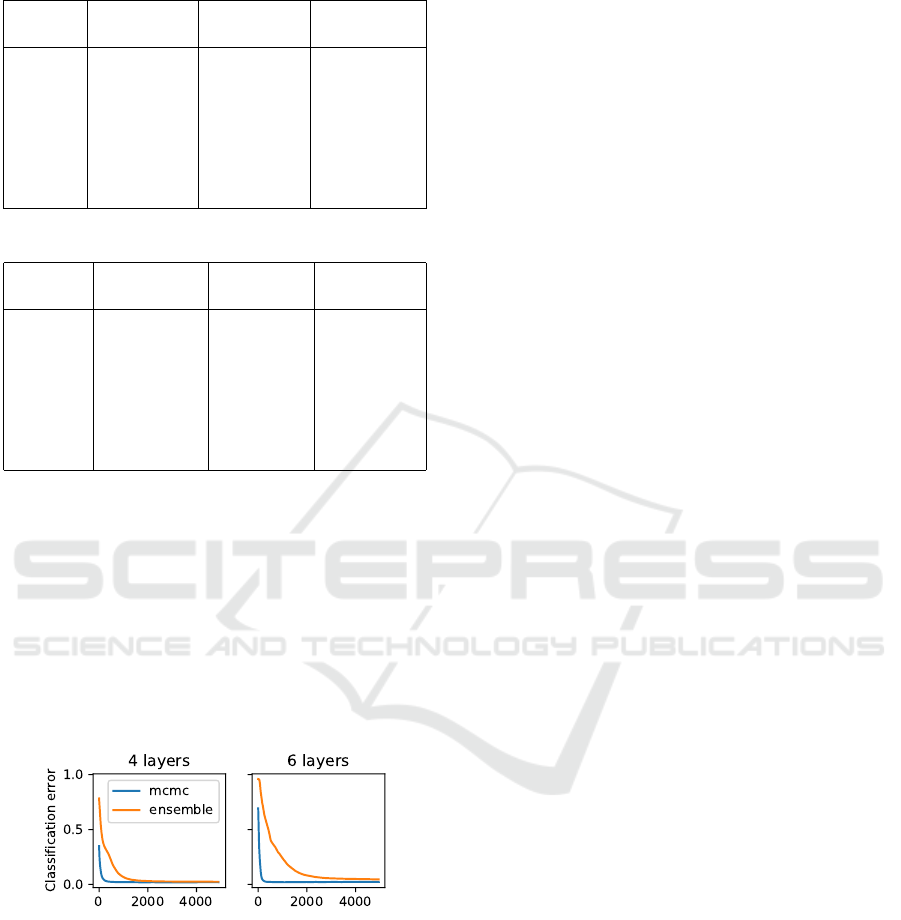
Table 3: Average classification accuracy for UCI classifica-
tion datasets.
Dataset Functional WDGPi-G Deep
MCMC Ensembles
EEG 92.51±1.82 94.13±1.96 89.04 ± 5.01
HTRU2 98.10±0.26 98.03±0.24 98.03 ± 0.20
Magic 88.16±0.33 88.37±0.29 87.90 ± 0.24
Miniboo 92.54±0.21 92.74±0.39 91.49 ± 0.19
Letter 98.22±0.18 96.90±0.29 96.38 ± 0.30
Drive 99.45±0.09 99.69±0.04 99.33 ± 0.05
Mocap 99.10±0.12 99.24±0.10 99.10 ± 0.08
Table 4: Average test NLL for UCI classification datasets.
Dataset Functional WDGPi-G Deep
MCMC Ensembles
EEG 0.33±0.04 0.18±0.04 0.24 ± 0.10
HTRU2 0.06±0.002 0.06±0.00 0.07 ± 0.01
Magic 0.31±0.00 0.29±0.00 0.30 ± 0.01
Miniboo 0.18±0.01 0.18±0.00 0.20 ± 0.01
Letter 0.09±0.01 0.17±0.00 0.15 ± 0.01
Drive 0.08±0.01 0.03±0.00 0.05 ± 0.01
Mocap 0.19±0.00 0.03±0.00 0.04 ± 0.00
We compare our approach with other classifica-
tion methods and found that it performs competitively
with the state-of-the-art, as shown in Tables 3 and 4.
Our approach does not require the Wasserstein opti-
mization phase used in WDGPi-G, while still achiev-
ing similar classification performance after optimiz-
ing GP hyperparameters.
We also tested the proposed method on the Let-
ter dataset using NNs with four and six hidden layers
and compared its convergence to the Deep Ensemble
approach in terms of classification accuracy (Fig. 4).
Figure 4: Convergence of classification error on test data
for the Letter dataset.
5 LIMITATIONS
While we consider our approach quite elegant in en-
coding prior information in the form of functional pri-
ors, we believe that it is important to point out some
limitations compared to other works.
One limitation is that the posterior distribution we
are targeting is approximate due the way we treat the
change of variables from weights to functions.
Another limitation is that the functional prior
needs to have a closed form. Even though the class
of functional priors which have this property is large,
this might be too restrictive in applications where it
is possible to sample from such priors but no closed
form is available. Prior works which perform a pre-
liminary optimization of the prior over the weights
(e.g., (Tran et al., 2022)) can operate on samples from
functional priors without the need to express these in
closed form.
Finally, the choice of a GP prior requires set-
ting its hyper-parameters. In this work, we resort
to marginal likelihood optimization, but it is possible
that this choice induces overfitting. One way around
this would be to include hyper-parameters in the set
of variables to be sampled in SG-HMC to obtain sam-
ples from their posterior at the expenses of having to
deal with a more costly MCMC sampling. Having
said that, there are situations where functional priors
are easy to elicit and express without the need to carry
out hyper-parameter optimization.
6 CONCLUSIONS
In this paper, we proposed a novel way to incorpo-
rate prior knowledge in Bayesian NNs (BNNs) in the
form of functional priors. In our view, such functional
priors implicitly determine priors over BNN weights,
and the proposed formulation yields an approximate
posterior over the weights from which it is possible
to sample through MCMC or any other approximate
inference techniques. In this paper, we studied the
scenario where functional priors are expressed in the
form of Gaussian processes (GPs), but our formula-
tion can handle any functional prior which can be ex-
pressed in closed form. We then discussed how to
scale our approach to handle large data sets by operat-
ing on mini-batches, despite the complications stem-
ming from the use of GP priors.
We tested our proposal on regression and classifi-
cation tasks and compared it with state-of-the-art ap-
proaches to carry out inference and prior optimization
for BNNs. Our results demonstrate that the proposed
approach is competitive in terms of performance and
quantification of uncertainty, while being easy to im-
plement.
We are currently investigating ways to handle GP
priors with priors over hyper-parameters for increased
flexibility, and alternative ways to specify functional
priors. Furthermore, we are investigating applica-
tions of BNNs for image classification tasks for which
BNN architectures use convolutional layers.
ICPRAM 2023 - 12th International Conference on Pattern Recognition Applications and Methods
456

ACKNOWLEDGEMENTS
MF gratefully acknowledges support from the AXA
Research Fund and the Agence Nationale de la
Recherche (grant ANR-18-CE46-0002 and ANR-19-
P3IA-0002).
REFERENCES
Blundell, C., Cornebise, J., Kavukcuoglu, K., and Wierstra,
D. (2015). Weight uncertainty in neural network. In
International conference on machine learning, pages
1613–1622. PMLR.
Chen, T., Fox, E., and Guestrin, C. (2014). Stochastic Gra-
dient Hamiltonian Monte Carlo. In Xing, E. P. and
Jebara, T., editors, Proceedings of the 31st Interna-
tional Conference on Machine Learning, volume 32
of Proceedings of Machine Learning Research, pages
1683–1691, Bejing, China. PMLR.
Cutajar, K., Bonilla, E. V., Michiardi, P., and Filippone, M.
(2017). Random feature expansions for deep Gaus-
sian processes. In Precup, D. and Teh, Y. W., editors,
Proceedings of the 34th International Conference on
Machine Learning, volume 70 of Proceedings of Ma-
chine Learning Research, pages 884–893. PMLR.
Dua, D. and Graff, C. (2017). UCI machine learning repos-
itory.
Fortuin, V. (2022). Priors in bayesian deep learning: A re-
view. International Statistical Review.
Fortuin, V., Garriga-Alonso, A., Wenzel, F., Ratsch, G.,
Turner, R. E., van der Wilk, M., and Aitchison, L.
(2021). Bayesian neural network priors revisited.
In Third Symposium on Advances in Approximate
Bayesian Inference.
Graves, A. (2011). Practical variational inference for neural
networks. In Shawe-Taylor, J., Zemel, R., Bartlett, P.,
Pereira, F., and Weinberger, K., editors, Advances in
Neural Information Processing Systems, volume 24.
Curran Associates, Inc.
Kallenberg, O. and Kallenberg, O. (1997). Foundations of
modern probability, volume 2. Springer.
Kanagawa, M., Hennig, P., Sejdinovic, D., and Sriperum-
budur, B. K. (2018). Gaussian processes and kernel
methods: A review on connections and equivalences.
arXiv preprint arXiv:1807.02582.
Lakshminarayanan, B., Pritzel, A., and Blundell, C. (2017).
Simple and scalable predictive uncertainty estimation
using deep ensembles. In Guyon, I., Luxburg, U. V.,
Bengio, S., Wallach, H., Fergus, R., Vishwanathan,
S., and Garnett, R., editors, Advances in Neural Infor-
mation Processing Systems, volume 30. Curran Asso-
ciates, Inc.
Ma, C., Li, Y., and Hernandez-Lobato, J. M. (2019). Vari-
ational implicit processes. In Chaudhuri, K. and
Salakhutdinov, R., editors, Proceedings of the 36th
International Conference on Machine Learning, vol-
ume 97 of Proceedings of Machine Learning Re-
search, pages 4222–4233. PMLR.
Milios, D., Camoriano, R., Michiardi, P., Rosasco, L.,
and Filippone, M. (2018). Dirichlet-based gaussian
processes for large-scale calibrated classification. In
Bengio, S., Wallach, H., Larochelle, H., Grauman,
K., Cesa-Bianchi, N., and Garnett, R., editors, Ad-
vances in Neural Information Processing Systems,
volume 31. Curran Associates, Inc.
Neal, R. (1996). Bayesian learning for neural networks.
Lecture Notes in Statistics.
Rahimi, A. and Recht, B. (2007). Random features for
large-scale kernel machines. In Platt, J., Koller, D.,
Singer, Y., and Roweis, S., editors, Advances in Neu-
ral Information Processing Systems, volume 20. Cur-
ran Associates, Inc.
Rudner, T. G. J., Chen, Z., and Gal, Y. (2021). Rethinking
function-space variational inference in bayesian neu-
ral networks. In Third Symposium on Advances in Ap-
proximate Bayesian Inference.
Shi, J., Sun, S., and Zhu, J. (2018). A spectral approach to
gradient estimation for implicit distributions. In Dy, J.
and Krause, A., editors, Proceedings of the 35th Inter-
national Conference on Machine Learning, volume 80
of Proceedings of Machine Learning Research, pages
4644–4653. PMLR.
Sun, S., Zhang, G., Shi, J., and Grosse, R. (2019). Func-
tional Variational Bayesian Neural Networks. In In-
ternational Conference on Learning Representations.
Tran, B.-H., Rossi, S., Milios, D., and Filippone, M. (2022).
All you need is a good functional prior for bayesian
deep learning. Journal of Machine Learning Re-
search, 23(74):1–56.
Wenk, P., Gotovos, A., Bauer, S., Gorbach, N. S., Krause,
A., and Buhmann, J. M. (2019). Fast gaussian process
based gradient matching for parameter identification
in systems of nonlinear odes. In Chaudhuri, K. and
Sugiyama, M., editors, Proceedings of the Twenty-
Second International Conference on Artificial Intelli-
gence and Statistics, volume 89 of Proceedings of Ma-
chine Learning Research, pages 1351–1360. PMLR.
Wenzel, F., Roth, K., Veeling, B., Swiatkowski, J., Tran, L.,
Mandt, S., Snoek, J., Salimans, T., Jenatton, R., and
Nowozin, S. (2020). How good is the Bayes poste-
rior in deep neural networks really? In III, H. D. and
Singh, A., editors, Proceedings of the 37th Interna-
tional Conference on Machine Learning, volume 119
of Proceedings of Machine Learning Research, pages
10248–10259. PMLR.
Williams, C. K. and Rasmussen, C. E. (2006). Gaussian
processes for machine learning, volume 2. MIT press
Cambridge, MA.
Imposing Functional Priors on Bayesian Neural Networks
457
