
GASTon: A Graph-Exploration System for Indexing, Annotating and
Visualizing PubMed Articles to Enhance the Analysis of Social
deTerminants of Health
Simone Bottoni
1
, Alberto Trombetta
1
, Flavio Bertini
2
, Danilo Montesi
3
, Francesca Bonin
4
,
Alessandra Pascale
4
, Martin Gleize
4
and Pierpaolo Tommasi
4
1
Department of Theoretical and Applied Sciences, University of Insubria, Varese, Italy
2
Department of Mathematical, Physical and Computer Sciences, University of Parma, Parma, Italy
3
Department of Computer Science and Engineering, University of Bologna, Bologna, Italy
4
IBM Research Europe - Dublin, Ireland
fl
{fbonin, apascale, martin.gleize, ptommasi}@ie.ibm.com
Keywords:
Graph-Based Knowledge Visualisation, Graph-Exploration Tool, PubMed Knowledge Base, Social Determi-
nants of Health.
Abstract:
Many works have shown associations between social determinants of health (SDoH) –the social circumstances
in which people live– and health-related outcomes. However, the lack of SDoH data increases the challenges
in measuring and understanding their effect on people’s health and health systems. In this paper, we present
GASTon, a system for the indexing, annotation, and graph-based rendering of PubMed information to enable
the search and retrieval of SDoHs in scientific literature. Our work provides a way to associate specific
concepts with peer-reviewed articles to simplify the search for social factors. It builds a knowledge graph based
on PubMed publications and associates them with concepts extracted from the Unified Medical Language
System (UMLS) Metathesaurus. GASTon allows a full-text search and graph-based navigation and supports
an overview of the concepts and related publications. Moreover, the architecture allows scale-up thanks to its
containerized nature and parallelization capabilities. The system is open-source under the Apache V2 license.
1 INTRODUCTION
The World Health Organisation (WHO) defines the
Social Determinants of Health (SDoH) as the circum-
stances in which people grow, live, and work that af-
fect their health (Wilkinson et al., 2003). Examples
of SDoH include socioeconomic status, education, or
transportation, and addressing these is important to
improve an individual’s health and reduce disparities
across communities (Artiga and Hinton, 2018). Pre-
vious works have shown associations between SDoH
and health-related outcomes, such as wealth linked to
risk of hospital admissions or costs (Artiga and Hin-
ton, 2018)(Meddings et al., 2017). The global SARS-
COV-2 pandemic has further revealed the stark in-
equity and inequality in healthcare provision and re-
sources, often associated with different dimensions of
SDoH such as race, income, education level, or job
security (Bettencourt-Silva et al., 2020). Therefore,
timely and effective interventions that address social
and healthcare needs are now more critical than ever.
In recent years, there has been a growing number
of initiatives that tackle SDoH, including nutritional
programs, addressing food insecurity (i.e., availability
and access to healthy foods), transportation programs
boosting access to employment, or housing interven-
tions tackling homelessness issue (Artiga and Hinton,
2018). However, more effort is needed to provide
combined or coordinated approaches. On one side,
it is essential to measure the impact of SDoH across
the health continuum. On the other, there is a need to
identify gaps and inconsistencies in data, especially
since electronic health record systems have not tradi-
tionally been designed to capture SDoH-related data.
Furthermore, healthcare terminologies such as ICD-
10 or SNOMED-CT do not extensively or adequately
cover social concepts (Bettencourt-Silva et al., 2020).
Therefore, it is not straightforward to consistently col-
lect data about SDoH across datasets. Lack of SDoH
data is, in fact, one of the main challenges when it
424
Bottoni, S., Trombetta, A., Bertini, F., Montesi, D., Bonin, F., Pascale, A., Gleize, M. and Tommasi, P.
GASTon: A Graph-Exploration System for Indexing, Annotating and Visualizing PubMed Articles to Enhance the Analysis of Social deTerminants of Health.
DOI: 10.5220/0011714900003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 5: HEALTHINF, pages 424-431
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

comes to measuring and understanding their effects.
Previous work has explored the connection be-
tween SDoH and health concepts based on pub-
lished literature (Park et al., 2021)(Gleize et al.,
2021)(Bettencourt-Silva et al., 2020)(Hatef et al.,
2019)(Meddings et al., 2017). However, the typical
bottleneck of these efforts lies in acquiring scientific
articles in a scalable, constant, and up-to-date manner,
as well as in being able to perform natural language
searches for SDoH related terms.
In this work, we present GASTon, a system that
provides a graph exploration rendering of the PubMed
knowledge base augmented with medical concept an-
notations. Specifically, GASTon consists of the fol-
lowing capabilities:
1. processes and indexes the whole PubMed
1
;
2. annotates the article with medical concepts
(UMLS concepts (Bodenreider, 2004));
3. updates the repository every day;
4. exposes an interface to allow searching and graph-
based rendering of data.
5. searches and identifies SDoH in the processed
data by exploiting the association between articles
and UMLS concepts.
GASTon aims to facilitate access to published ev-
idence and improve the discovery of social deter-
minants of health factors within scientific articles.
Thanks to its containerized nature and parallelization
capabilities, it can easily scale up to handle heavier
workloads. The system has been made open source
under Apache V2 license
2
The paper is organized as follows. Section 2
presents an overview of the related works. Section 3
and Section 4 describe the dataset and the UMLS tax-
onomy used for annotation, respectively. In Section 5,
we present the overall workflow of GASTon, and in
Section 6 the detailed description of the architecture.
Finally, conclusions and future works are reported in
Section 7.
2 RELATED WORK
A few tools have been developed as PubMed deriva-
tives. Among these we mention: GoPubMed (Doms
and Schroeder, 2005), SemanticMedline (Kilicoglu
1
We refer here only to the publicly available informa-
tion provided by PubMed, i.e., abstract and available infor-
mation about authors, etc.
2
The system is open source and accessible for de-
ployment at the following links: https://github.com/
SimoneBottoni/PubMedKnowledgeGraph
Figure 1: Workflow of the system.
et al., 2007), MeshNet (Yang and Lee, 2018), and
MeSHy (Theodosiou et al., 2011).
GoPubMed (Doms and Schroeder, 2005) extracts
Gene Ontology (GO) (Harris et al., 2004) terms from
the abstracts of the publications in PubMed search re-
sult, and groups the publications according to the GO
terms. The users are given the PubMed search result
as a list in which the publications are categorized ac-
cording to the GO terms. Such a grouping can provide
the users with an easy way to identify publications
with research themes (or concepts), and the search can
be refined using the sub-theme.
Semantic MEDLINE extracts predictions based
on UMLS concepts from the publications in the
PubMed search result (Kilicoglu et al., 2007). The list
of predictions is entered into the automatic summa-
rizer and then assorted into the list of semantic con-
densates (list of UMLS concepts), which is provided
to the users.
MeSHNet (Yang and Lee, 2018) is a prototype
application to explore the research trend of a re-
search area using a network graph. The authors ap-
ply a methodology to visualize a social network com-
posed of medical keywords in PubMed literature, so-
called MeSH terms; they select only the ”noteworthy”
MeSH terms from those extracted from the publica-
tions in a research area defined by a search query and
represent them as a graph. The graph is handy for ex-
ploring research trends and under-investigated areas.
However, to our understanding, no information from
the original article is maintained in the graph.
MeSHy (Theodosiou et al., 2011) is specifically
designed to provide random knowledge domains that
might have implications for the users so that they can
explore novel and promising research areas. The sys-
tem calculated the rarity of MeSH terms associated
with an article.
GASTon annotates the articles with UMLS med-
ical concepts. In addition to previous work, it of-
fers a graph-based visualization of the articles and
the concepts with a free text search capability. Un-
like MeSH Net, GASTon uses UMLS concepts and
visualizes the complete information of the articles. In
addition, thanks to the annotation process with UMLS
concepts, Gaston provides experts, through full-text
search, a way to visualize the association between ar-
GASTon: A Graph-Exploration System for Indexing, Annotating and Visualizing PubMed Articles to Enhance the Analysis of Social
deTerminants of Health
425
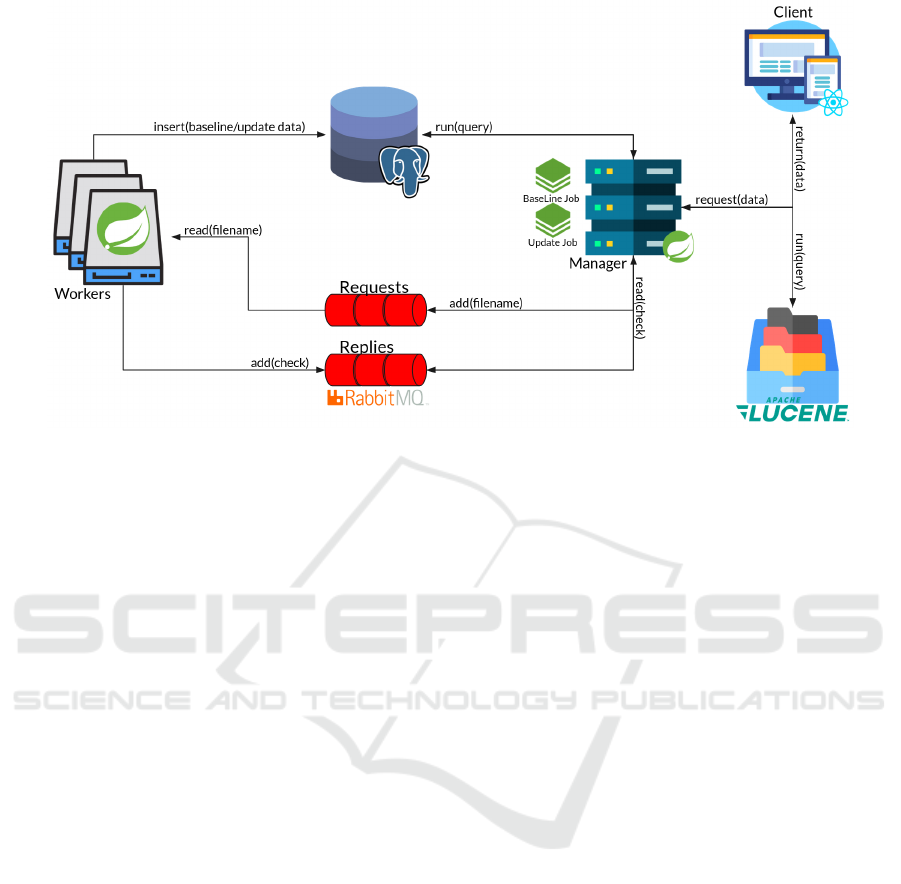
Figure 2: Architecture of the system.
ticles and concepts that help them to find new possi-
ble associations between social determinants of health
and health-related issues.
3 PubMed KNOWLEDGE BASE
PubMed is a free resource repository of biomedical
and life sciences literature. It has the purpose of im-
proving the search and retrieval of health-related in-
formation. The PubMed database includes more than
34 million citations and abstracts of biomedical liter-
ature.
PubMed comprises three primary sources, Med-
line, PubMed Central, and Bookshelf. Medline is a
bibliographic database containing references to arti-
cles from life science journals, newspapers, maga-
zines, and newsletters, concentrating on biomedicine;
it is the online version of MEDical Literature Analy-
sis and Retrieval System (MEDLARS). PubMed Cen-
tral (PMC) is an archive of biomedical and life science
full-text journal literature. The Bookshelf contains
citations, full-text books, and individual chapters re-
lated to biomedical, health, and life science.
PubMed is maintained by the National Center for
Biotechnology Information (NCBI), which publishes
a baseline annually that contains the whole PubMed
citation records (i.e., articles) in XML format. Ev-
ery day the NCBI also publishes updated files daily,
including new, revised, or deleted articles. Each
XML file contains articles with a specific structure
described in the PubMed Document Type Definition
(DTD). The article information includes the id, called
PubMed ID (PMID), the title, the abstract, the journal,
and the list of authors. It also contains information re-
garding an article’s history in the PubMed repository,
such as the date the record was created and the last
date the article was revised. PubMed does not include
full-text articles, but links to the full-text are usually
present.
4 UNIFIED MEDICAL
LANGUAGE SYSTEM (UMLS)
The UMLS is a set of files and software that com-
bines many health and biomedical vocabularies and
standards to create more effective and interoperable
biomedical information systems and services (Boden-
reider, 2004)(McCray et al., 2001). It includes differ-
ent knowledge sources, such as a Metathesaurus, a set
of hierarchies, definitions, and other relationships and
attributes.
UMLS Metathesaurus (Schuyler et al., 1993) is
a large, multi-lingual biomedical thesaurus organized
by concepts that contain information about biomedi-
cal and health-related concepts, their various names
and relationships. It is organized by concept or
meaning, connecting different names over a unique
and permanent Concept Unique Identifier (CUI).
The UMLS Metathesaurus is used by different tools
to extract specific terminologies or ontologies from
text. One of the most used is MetaMap (Aronson,
2006)(Aronson and Lang, 2010) and its lite versions
MetaMapLite, a customizable and faster version of
MetaMap (Demner-Fushman et al., 2017). These
HEALTHINF 2023 - 16th International Conference on Health Informatics
426
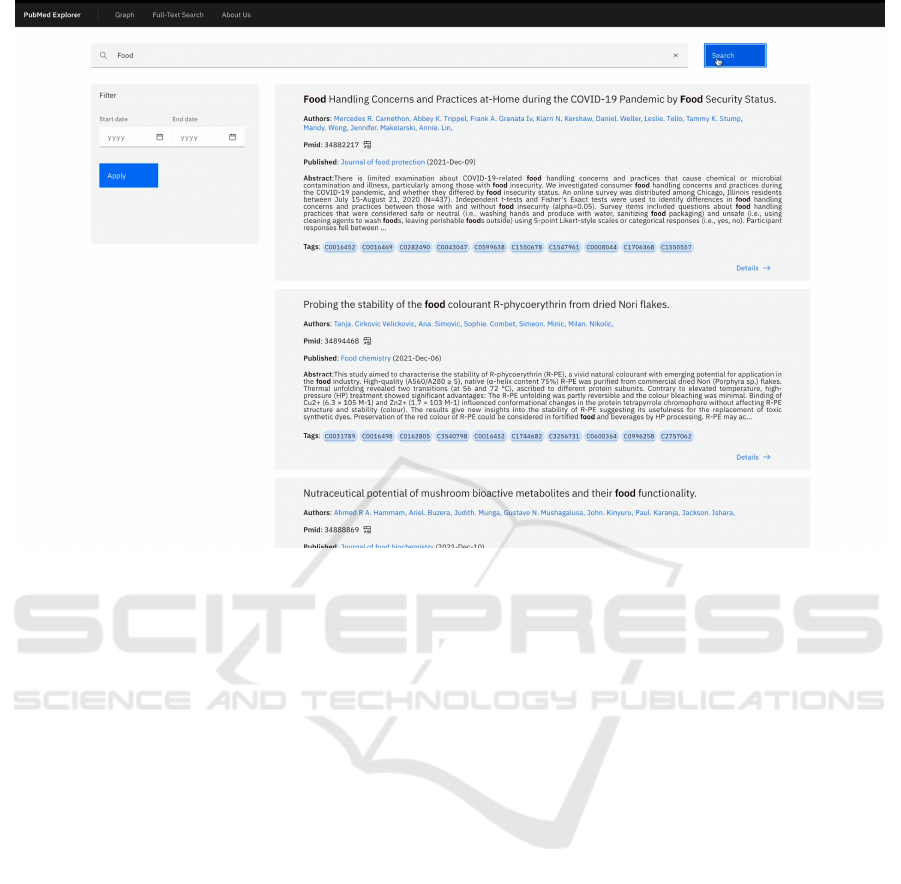
Figure 3: Web UI. Example of the Full-Text search view.
named entity recognition tools use NLP techniques
to map biomedical text to the UMLS Metathesaurus
concepts.
5 GASTon WORKFLOW
The primary purpose of our tool is to provide an easier
and faster way to explore SDoHs in clinical literature.
We first need to index the PubMed repository and en-
sure that the index stays updated daily. Then we need
to expose all the available information externally and
allow to query them in a simple way.
This section provides a high-level overview of
how PubMed data are indexed, processed, and anno-
tated. The workflow and its main phases are shown in
Figure 1. GASTon downloads the PubMed XML files
from the baseline repository and processes a user-
defined amount of files in parallel. The process work-
flow consists of three phases: parsing, annotating, and
collecting/indexing.
In the Parsing phase, an XML parser extracts all
the available information from an XML PubMed file,
converting them into a more convenient format for
GASTon. Then it starts the annotation phase, analyz-
ing the text (title and abstract) of the extracted articles
and associating them with specific UMLS Metathe-
saurus concepts that describe their specific character-
istics. The parsed and annotated PubMed baseline
data is stored in a relational database. To improve
the system’s performance, an index –optimized for
textual data– is built upon a subset of interest (e.g.,
we currently focus on data regarding authors, articles,
and concepts rather than on the history of articles).
Every day updated information on articles is re-
leased. GASTon looking for updates on the articles
and processing them through the previously described
phases. We store all the updated information in the
database incrementally, keeping track of history.
6 GASTon ARCHITECTURE
In Figure 2 we present the GASTon architecture. It
has three main components: a Manager that orches-
trates the interactions, a set of workers that concur-
rently processes the PubMed files, and a Web User
Interface (UI) to query and visualize the data.
6.1 Manager
The core component of our system is the Manager. It
is written using the Spring Boot framework (spring
projects, 2018)(Walls, 2015), and it has the purpose
of managing the system. In particular, it initializes
the system, manages the analysis of the PubMed data,
GASTon: A Graph-Exploration System for Indexing, Annotating and Visualizing PubMed Articles to Enhance the Analysis of Social
deTerminants of Health
427
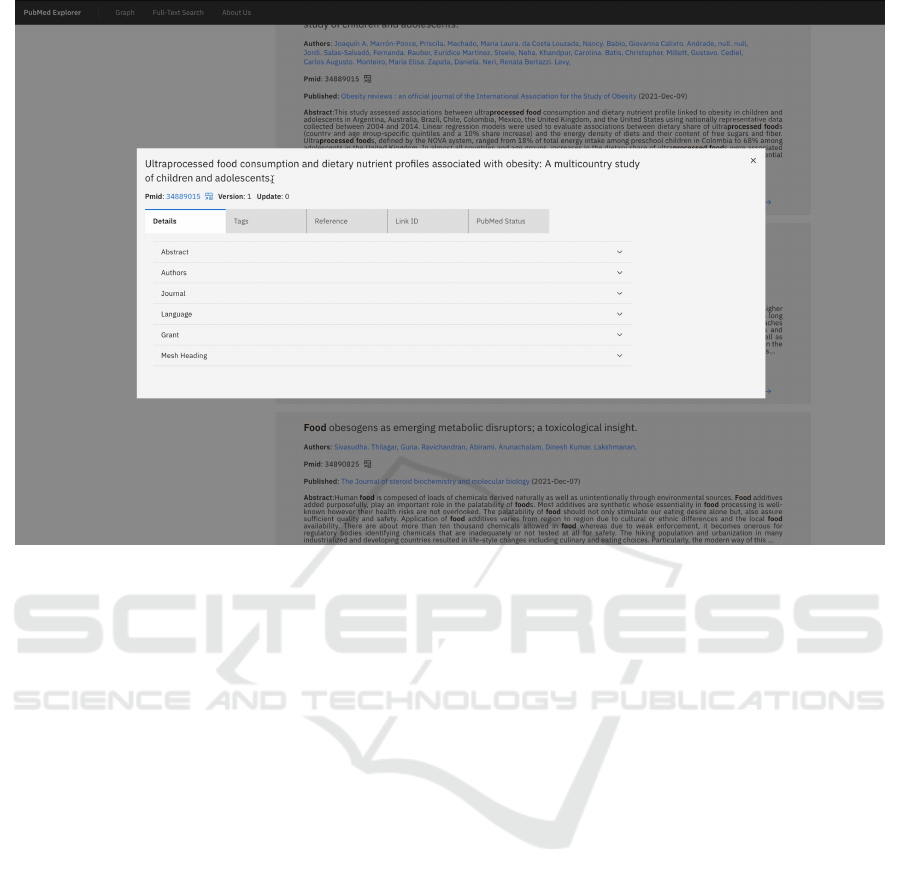
Figure 4: Web UI. Example of the Full-Text search view details.
generates the index, and answers the users’ requests.
System Initialization. The Manager creates, if they
do not exist, a PostgreSQL relational database and
two RabbitMQ messaging queues for communica-
tions with the Workers (a requests queue and a replies
queue).
Job Orchestration. After the initialization, the
Manager distributes the baseline PubMed files, to be
processed by the workers through a remote partition-
ing job (spring projects, 2008)(Lui et al., 2011) called
BaseLine job. The Manager keeps track of the Base-
Line job progress, and once completed, it enables a
second remote partitioning job, the Update job. Every
day, at a given user-defined time, the Manager awakes
to handle the Update job to elaborate on the daily up-
dates released by PubMed.
Index Maintenance. The Manager keeps an up-
dated index of the most relevant data. This in-
dex is built using Hibernate Search (hibernate,
2010)(Bernard and Griffin, 2008) and Apache
Lucene (Białecki et al., 2012).
Fulfilling Users’ Queries. The Manager also han-
dles queries coming from web clients. A set of REST-
ful Application Programming Interface (API)s are ex-
posed to fulfil the requests.
6.2 Worker(s)
GASTon allows starting a user-defined number of
workers. Each worker processes a subset of the
PubMed repository concurrently for higher through-
put. Each worker reads a message from the request
queue that points to a specific PubMed file and pro-
cesses it. The process consists of the parsing, annota-
tion, and collection phases described in Section 5.
During the parsing phase, a worker parses the in-
formation regarding the articles using Java Sax. The
articles’ data extracted by the parser, are sent, in the
annotation phase, by a worker to the MetaMapLite’s
APIs (Demner-Fushman et al., 2017) to generate a list
of keywords for each article. A worker passes as in-
put to the APIs the title and the abstract of each article
in the form of raw text. Then, MetaMapLite’s APIs
matches the input text with the UMLS Metathesaurus
and identifies a list of relevant concepts. Each con-
cept is retrieved in the MetaMap Indexing (MMI) for-
mat and includes the CUI that is the keyword itself,
the preferred term that is the keyword correspond-
ing name in natural language, a score, and trigger in-
formation. The score represents the relevance of the
UMLS concept according to the MMI ranking func-
HEALTHINF 2023 - 16th International Conference on Health Informatics
428
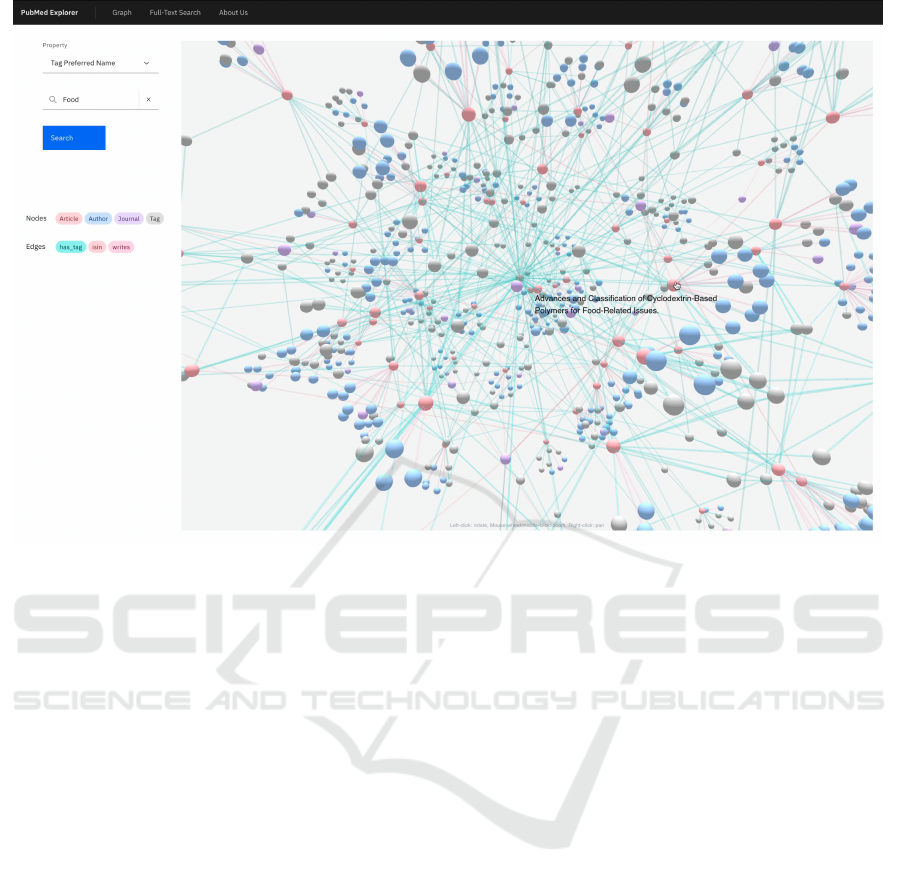
Figure 5: Web UI. Example of an explorable 3D graph view.
tion (Aronson, 1997); instead, the trigger includes the
input text word that triggered the keyword and its
location in the input text. Finally, in the collection
phase, a worker stores the extracted articles’ data and
the keywords in the database.
Once a worker processes a file, it notifies the Man-
ager with a message in the replies queue containing
the status of the workflow.
6.3 Web UI
We developed a modern multi-page React web appli-
cation that exploits the REST APIs provided by the
Manager to provide a convenient and easy way to ac-
cess and query the processed PubMed data. Our web
UI comprises two tools. The first is a full-text search
that allows one to search between the indexed data;
the second is a 3D Graph that explores the relation-
ships among concepts and articles.
Full-Text Search Tool. We show the Full-Text
search tool in Figure 3. The Full-Text Search enables
scanning the processed PubMed repository to find de-
tailed information that matches a specific topic. The
tool allows users to insert a query using natural lan-
guage and output a list of articles that match the query.
Each result represents a single article, showing essen-
tial information such as the title, the author(s), the
journal, the abstract, and the list of matched UMLS
concepts. Each article has a detailed view, shown in
Figure 4, that includes all the available information
in the PubMed repository regarding the article. The
detailed view includes the basic information, the list
of references, links where it is possible to find the ar-
ticle, and the article’s history in the PubMed reposi-
tory. The detailed view also includes the complete list
of UMLS concepts associated with the article, with
the score and triggers, as presented in Section 6.2.
Through this concepts list and the score related to
each concept, it is possible to identify social deter-
minants of health. This association can allow experts
to discover new relations between social determinants
of health and health issues.
3 D Graph Tool. The 3D exploration graph tool,
shown in Figure 5, provides a graphical overview of
the concepts, the relationships among them, and the
related publications. This tool helps understand the
connections between different articles and the UMLS
concepts, with additional explanations about discov-
ered relationships or information about concepts.
The tool allows searching data by PMID, CUI, or
UMLS concept names. The result list is visualized
as a 3D graph. For the graph visualization, we used
react-force-graph, a React library (Asturiano, 2018)
displaying 3-dimensional space graph data structure
rendered using a force-directed iterative layout.
Each node represents a different type of entity, dis-
GASTon: A Graph-Exploration System for Indexing, Annotating and Visualizing PubMed Articles to Enhance the Analysis of Social
deTerminants of Health
429
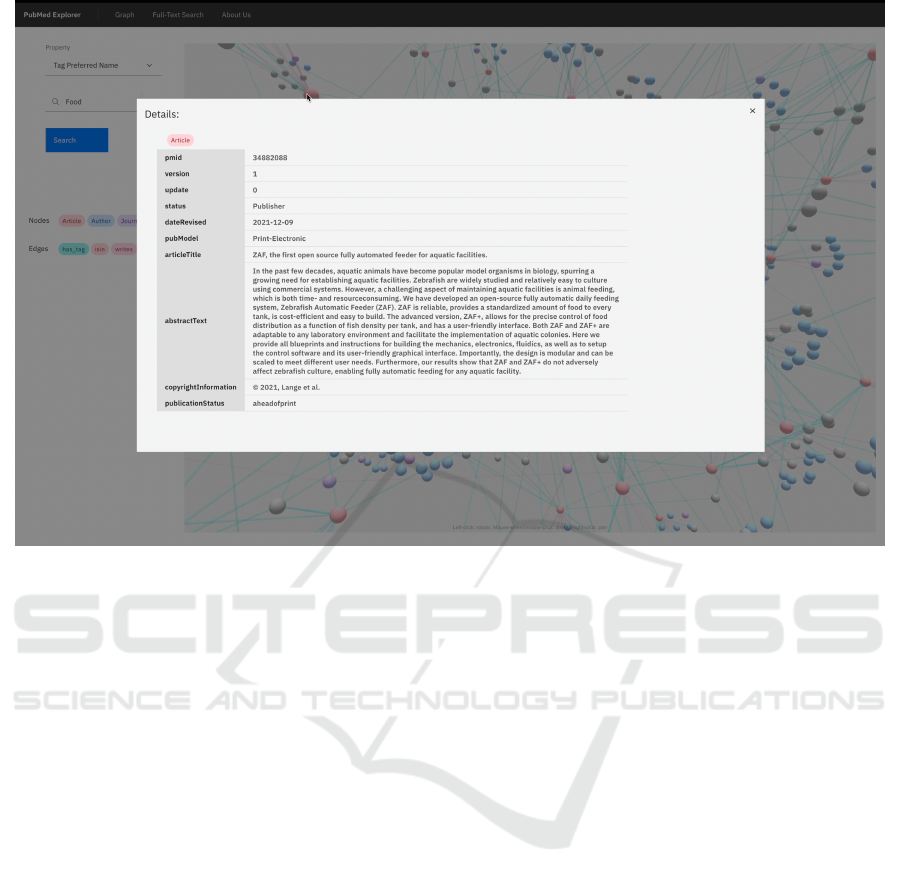
Figure 6: Web UI. Example of an explorable 3D graph view details.
tinguishable by colour. It is possible to find nodes
representing articles, authors, journals, or UMLS con-
cepts. The centre of the graph is always the searched
node. Every node has a detailed view, shown in Fig-
ure 6, where it is possible to find other data regards the
node. Edges also have a detailed view that displays in-
formation regards the relationship between the nodes
they connect.
7 CONCLUSIONS AND FUTURE
WORK
In this paper, we presented a novel system for the
indexing, annotation, and graph-based rendering of
PubMed articles that facilitates the retrieval of SDoH
from PubMed articles. Our work provides a way to
associate specific UMLS Metathesaurus concepts to
the PubMed scientific literature to simplify the search
for social factors and their possible links to health is-
sues. The solution consists of a pipeline that down-
loads and parses PubMed articles’ records and then
annotates them using MetaMap UMLS concepts. In
addition, we provide a set of accessible APIs to query
the processed data and a modern multi-page web ap-
plication to search and visualize articles information.
Specifically, we developed a Full-Text Search inter-
face to search articles by keyword(s) and an interac-
tive 3D graph tool that allows us to have an overview
of the concepts and the related publications and to un-
derstand the relationships between different articles
and the UMLS concepts.
Further analysis can be conducted to improve the
search and visualization tools, integrating new ways
and functionalities to visualize and explore data (e.g.,
a dynamic list of relevant fields). This analysis must
be conducted with medical experts that can provide
solid feedback regards the accuracy and the utility of
the retrieved results and the benefit of the proposed
tools that allow to visualization of that results. These
improvements may give users a better and more ac-
cessible overview of the relationships between arti-
cles and concepts to help identify social factors that
can impact people’s health.
REFERENCES
Aronson, A. R. (1997). The mmi ranking func-
tion. Available in the website: https://ii. nlm. nih.
gov/MTI/Details/mmi. shtml.
Aronson, A. R. (2006). Metamap: Mapping text to the umls
metathesaurus. Bethesda, MD: NLM, NIH, DHHS,
1:26.
Aronson, A. R. and Lang, F.-M. (2010). An overview of
MetaMap: historical perspective and recent advances.
HEALTHINF 2023 - 16th International Conference on Health Informatics
430

Journal of the American Medical Informatics Associ-
ation, 17(3):229–236.
Artiga, S. and Hinton, E. (2018). Beyond health care: the
role of social determinants in promoting health and
health equity. Health, 20(10):1–13.
Asturiano, V. (2018). react-force-graph. https://github.com/
vasturiano/react-force-graph.
Bernard, E. and Griffin, J. (2008). Hibernate search in ac-
tion. Simon and Schuster.
Bettencourt-Silva, J., Mulligan, N., Sbodio, M., Segrave-
Daly, J., Williams, R., Lopez, V., and Alzate, C.
(2020). Discovering New Social Determinants of
Health Concepts from Unstructured Data: Frame-
work and Evaluation. Stud Health Technol Inform,
270:173–177.
Białecki, A., Muir, R., and Ingersoll, G. (2012). Apache
lucene 4. In OSIR@SIGIR.
Bodenreider, O. (2004). The unified medical language sys-
tem (umls): integrating biomedical terminology. Nu-
cleic acids research, 32(suppl 1):D267–D270.
Demner-Fushman, D., Rogers, W. J., and Aronson, A. R.
(2017). MetaMap Lite: an evaluation of a new Java
implementation of MetaMap. Journal of the American
Medical Informatics Association, 24(4):841–844.
Doms, A. and Schroeder, M. (2005). Gopubmed: Explor-
ing pubmed with the gene ontology. Nucleic acids
research, 33:W783–6.
Gleize, M., Mulligan, N., Di Bari, A., and Bettencourt-
Silva, J. H. (2021). Social determinant trends of covid-
19: An analysis using knowledge graphs from pub-
lished evidence and online trends. In MIE, pages 744–
748.
Harris, M., Deegan, J., Ireland, A., Lomax, J., Ashburner,
M., Foulger, R., Eilbeck, K., Lewis, S., Marshall, B.,
Mungall, C., Richter, J., Rubin, G., Blake, J., Bult, C.,
Dolan, M., Drabkin, H., Eppig, J., Hill, D., Ni, L., and
White, R. (2004). The gene ontology (go) database
and informatics resource. Nucleic Acids Res, 32:258–
261.
Hatef, E., Kharrazi, H., Nelson, K., Sylling, P., Ma, X.,
Lasser, E. C., Searle, K. M., Predmore, Z., Bat-
ten, A. J., Curtis, I., Fihn, S. D., and Weiner, J. P.
(2019). The association between neighborhood so-
cioeconomic and housing characteristics with hospi-
talization: Results of a national study of veterans. The
Journal of the American Board of Family Medicine,
32:890 – 903.
hibernate (2010). hibernate-search. https://github.com/
hibernate/hibernate-search.
Kilicoglu, H., Fiszman, M., Rodriguez, A., Shin, D., Rip-
ple, A., and Rindflesch, T. (2007). Semantic medline:
A web application for managing the results of pubmed
searches. Proceedings of the Third International Sym-
posium on Semantic Mining in Biomedicine (SMBM
2008).
Lui, M., Gray, M., Chan, A., and Long, J. (2011). Spring in-
tegration and spring batch. In Pro Spring Integration,
pages 561–589. Springer.
McCray, A. T., Burgun, A., and Bodenreider, O. (2001).
Aggregating umls semantic types for reducing con-
ceptual complexity. Studies in health technology and
informatics, 84(0 1):216.
Meddings, J., Reichert, H., Smith, S. N., Iwashyna, T. J.,
Langa, K. M., Hofer, T. P., and McMahon, L. F.
(2017). The impact of disability and social determi-
nants of health on condition-specific readmissions be-
yond medicare risk adjustments: a cohort study. Jour-
nal of general internal medicine, 32(1):71–80.
Park, Y., Mulligan, N., amd Morten Kristiansen, M. G., and
Bettencourt-Silva, J. H. (2021). Discovering asso-
ciations between social determinants and health out-
comes: Merging knowledge graphs from literature
and electronic health data. In AMIA 2021, American
Medical Informatics Association Annual Symposium,
Virtual Event, USA. AMIA.
Schuyler, P. L., Hole, W. T., Tuttle, M. S., and Sherertz,
D. D. (1993). The umls metathesaurus: representing
different views of biomedical concepts. Bulletin of the
Medical Library Association, 81(2):217.
spring projects (2008). spring-batch. https://github.com/
spring-projects/spring-batch.
spring projects (2018). spring-boot. https://github.com/
spring-projects/spring-boot.
Theodosiou, T., Vizirianakis, I., Angelis, L., Tsaftaris, A.,
and Darzentas, N. (2011). Meshy: Mining unantic-
ipated pubmed information using frequencies of oc-
currences and concurrences of mesh terms. Journal of
biomedical informatics, 44:919–26.
Walls, C. (2015). Spring Boot in action. Simon and Schus-
ter.
Wilkinson, R., Marmot, M., for Europe, W. H. O. R. O.,
Project, W. H. C., for Health, W. I. C., and Society
(2003). Social Determinants of Health: The Solid
Facts. Academic Search Complete. World Health Or-
ganization, Regional Office for Europe.
Yang, H. and Lee, H. (2018). Research trend visualization
by mesh terms from pubmed. International Journal of
Environmental Research and Public Health, 15:1113.
GASTon: A Graph-Exploration System for Indexing, Annotating and Visualizing PubMed Articles to Enhance the Analysis of Social
deTerminants of Health
431
