
System Modeling and Machine Learning in Prediction of Metastases
in Lung Cancer
Andrzej Swierniak
1a
, Emilia Kozłowska
1b
, Krzysztof Fujarewicz
1c
, Damian Borys
1d
,
Agata Wilk
1e
, Jaroslaw Smieja
1f
and Rafal Suwinski
2g
1
Department of Systems Biology and Engineering, Silesian University of Technology, Akademicka 16,
44-100 Gliwice, Poland
2
The 2nd Radiotherapy and Chemotherapy Clinic, M. Sklodowska-Curie National Research Institute of Oncology,
Gliwice Branch, Wybrzeze Armii Krajowej 15, Gliwice, Poland
rafal.suwinski@io.gliwice.pl
Keywords: Medical Image Processing, NSCLC, AI Based Models, Metastases, Survival Analysis.
Abstract: The aim of this paper is to present goals and preliminary results of our project devoted to system engineering
approach in prediction of metastases in lung cancer. More specifically we consider existing and develop new
methods of system modeling, machine learning, signal processing and intelligent control to find biomarkers
enabling prediction of risk of tumor spread and colonization of distant organs in non-small-cell lung
carcinoma basing on clinical data and medical images. The results could bring us knowledge about the
dynamics and origin of metastatic dissemination of lung cancer. By dynamics, we understand when and where
a tumor will disseminate, and by origin we mean dissemination path (directly from original tumor or through
lymphatic nodes). This information is very valuable for clinicians, as it could guide the personalized treatment
of lung cancer patients. The results will elucidate important issues concerning prediction of individual
progress of cancer and treatment outcome in oncology. They will provide both theoretical and simulation
tools to support decision making and diagnostics in oncology, on the basis of individual patient state.
1 INTRODUCTION
In this paper we describe main goals and methods
used in a project in which we combine system
engineering methodology with clinical data to predict
metastases in lung cancer. The interest of proposing
original models and methods is to support analysis of
clinical and imaging data and aim at better prediction
of spread and colonization of tumor cells to distant
organs, with emphasis on the most common subtype
of lung cancer - non-small-cell lung carcinoma
(NSCLC). Since the metastatic tumor is mainly
incurable, due to its resistance to treatment, our
research is directed to answer the following urgent
a
https://orcid.org/0000-0002-5698-5721
b
https://orcid.org/0000-0002-3069-3085
c
https://orcid.org/0000-0002-1837-6466
d
https://orcid.org/0000-0003-0229-2601
e
https://orcid.org/0000-0001-7554-1803
f
https://orcid.org/0000-0002-6120-4424
g
https://orcid.org/0000-0002-3895-7938
biological and clinical question: how, when, and
where the primary tumor will spread to distant
locations. The proposal is focused on reducing
probability of metastasis and evolution of cancer in
distant sites and emergence of evolving resistance to
therapies. To verify applicability of these methods, all
theoretical considerations are related to clinical data
and radiological images, to which we have access. To
reach that goal, both experimental and system
modeling methods are employed in order to meet the
following intermediate objectives:
• Analysis of available radiomic data
incorporated in Positron Emission Tomography/
Computed Tomography (PET/CT) images, and
220
Swierniak, A., Kozłowska, E., Fujarewicz, K., Borys, D., Wilk, A., Smieja, J. and Suwinski, R.
System Modeling and Machine Learning in Prediction of Metastases in Lung Cancer.
DOI: 10.5220/0011705300003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 3: BIOINFORMATICS, pages 220-227
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

application of signal processing and statistical
inference tools to develop original estimators
supporting prognosis of tumor spread to local
lymph nodes and distant organs.
• Development of stochastic compartmental
models based on branching birth-death
processes in which the primary tumors can
metastasize to local lymph nodes and next,
distant metastases can emerge in liver, brain,
and bones. Inter-patient heterogeneity is
accounted by assuming statistical distributions
of model parameters, using the mixed-effects
statistical framework.
• Modification of existing mathematical models
based on ordinary and/or partial differential
equations describing cancer growth and therapy
aimed at taking into account both local and
distant metastases. Estimation of model
parameters is based on available clinical data.
• Development of machine learning tools required
for integration of radiomic and clinical data with
mathematical models mentioned above.
• Modification of models based on evolutionary
game theory supporting analysis of interaction
of different cancer cells phenotypes leading to
emergency of metastasis and resistance to
treatment.
2 BIOMEDICAL BACKGROUND
AND JUSTIFICATION OF
TACKLING SCIENTIFIC
PROBLEM
Lung cancer is one of the most commonly diagnosed
cancer and is the leading cause of cancer-related
deaths. The most common histological subtype is
non-small-cell lung carcinoma (NSCLC), accounting
for 85% of all lung cancer cases (Inamura, 2017).
Advanced NSCLC is more likely to metastasize,
leading to severe symptoms and a decrease in overall
survival. The presence of distant metastases is one of
the most predictive factors of poor prognosis (Popper,
2016). Distant metastases (distant cancer) refer to
cancers that have spread via blood or lymphatic
vessels from the original location (the primary tumor)
to distant organs or lymph nodes. The main cause of
cancer death is associated with metastases, which are
mainly incurable. Thus, distant cancer is resistant to
treatment intervention. Even though cancer
researchers have made a lot of effort to understand the
appearance of metastases, only few preclinical studies
about metastases were translated to clinical practice.
The proposal aims at tackling metastases in the
most common type of lung cancer. If successful, the
project outcome will be information about the
dynamics of tumor metastases in lung cancer, i.e.,
when, where, and how the primary tumor will
metastasize. Information is extracted using a non-
invasive PET/CT imaging techniques. This
information is incorporated in different types of
known and original models using machine learning
tools. The results could bring us knowledge about the
dynamics and origin of metastatic dissemination of
lung cancer. By dynamics, we understand when and
where a tumor will disseminate, and by origin we
mean dissemination path (directly from original
tumor or through lymphatic nodes). This information
is very valuable for clinicians, as it could guide the
personalized treatment of lung cancer patients.
3 MODELING METASTASIS –
METHODS AND TOOLS
Metastasis is a complex process that involves the
spread of a cancer to distant parts of the body from its
original site. In order to become clinically detectable
lesions, it must complete a series of steps at multiple
temporal and spatial scales. The deterministic
description of this process is based on either ODE or
PDE modes. Saidel et al. (Saidel et al, 1976) proposed
a compartmentalized translational ODE model of
metastasis distribution over the time. An important
contribution, used as a basis of many subsequent
works, in the field of modeling metastasis was
introduced by K. Iwata et al. (Iwata et al, 2000). Their
model for the colony size distribution of multiple
metastatic tumors raising from untreated tumor is
represented by the hyperbolic PDE. Model by Iwata
et al. was further analyzed and extended by Barbolosi
et al.(Barbolosi et al, 2009), Devys et al. (Devys et al,
2009), and Benzekry (Benzekry E., 2011). In (Iwata
et al, 2000, Barbolosi et al, 2009, Devys et al, 2009)
the primary tumor is subject to the Gompertz law,
while in (Benzekry E., 2011) the primary tumor is
described by model of tumor growth including
angiogenesis (Hahnfeldt et al, 1999). This model was
developed further by the same group (Benzekry E. et
al, 2016) in conjunction with clinical data and the
mathematical formulation of a metastatic
dissemination. A different, hybrid approach to the
problem of modeling of invasive cancer and
metastases was introduced by Franssen et al.
(Franssen et al, 2019). The authors presented the
general spatial modelling framework of the metastatic
System Modeling and Machine Learning in Prediction of Metastases in Lung Cancer
221
spread of cancer. Their model was then simulated
using clinical data from breast cancer patients and
data of metastatic sites (bones, lung and liver). The
model (Franssen et al, 2019) was further used by a
group of Benzekry in (Bilous et al, 2019, Nicolo et al,
2000). In (Nicolo, 2000) the authors compare
predictions of the metastatic relapse given by a
machine learning and mechanistic modeling
techniques. In (Bilous et al, 2019) a model of the
dynamics of brain metastasis in NSCLCS is
discussed. To our knowledge, this is the only work in
which explicit metastasis in non-small cell lung
cancer is taken into consideration in tumor dynamics
modeling. In (Smieja et al, 2022) we have proposed
probably the simplest model of tumor progression
including metastasis. At the one hand, it contains the
minimum number of compartments and parameters.
At the other hand, it is able to represent
heterogeneous response treatment in a population of
patients and provide a good fit to clinical survival
curves or progression including metastasis which
enables estimation of parameters based on clinical
data.
Stochastic modeling has been used in
mathematical oncology for a relatively short time,
and includes various techniques to take into account
randomness in the process such as tumor progression
or metastasis. Indeed, tumor growth is a random
process as each tumor cell have different cell cycle
length due to internal (process of DNA repair) or
external factors (competition for space and
resources). Thus, stochastic mathematical models
provide a powerful toolbox for mechanistic modeling
of cancer. We have developed a model of NSCLC
progression and dissemination to local lymph nodes
and distant sites (Kozłowska and Swierniak, 2022).
The model is in the form of stochastic
multicompartmental birth-death branching process
model. The branching process is powerful tool in
modeling various processes in biology, especially in
cancer (see e.g. (Kimmel and Axelrod, 2015)). This
mathematical framework is also useful for
mathematical modeling of local metastases, as shown
in (Haeno et al, 2012) by modeling metastases in
pancreatic cancer. The structure of the proposed
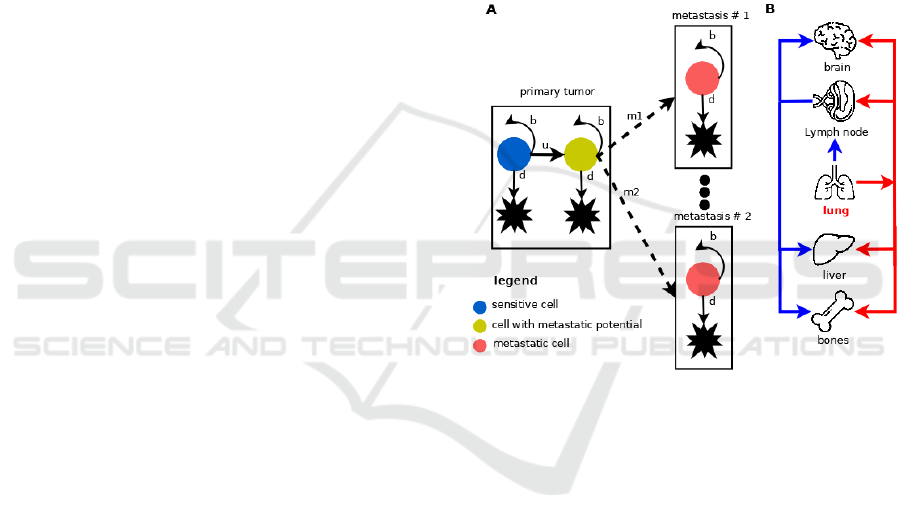
model is presented in Figure 1.
The model considers Gompertzian growth of
lung cancer cells from a single cell of Type I, which
does not have the ability to metastasize. The cell,
however, has accumulated all necessary aberrations
needed for proliferation and has fitness advantage
over healthy cells. Type I cell is also treatment-naïve
and thus sensitive to chemo- and radiotherapy. At
each discrete time point (representing the moment of
cell division), the cell can divide or die, and the
population of cells grows according to Gompertzian
growth law. In addition, a cell has small probability
to mutate to Type II cell, which has metastatic
potential, during division. Type II cell is an
aggressive type of cells, thus its growth dynamics is
exponential. This new type of cell appears with
probability u per cell division, as shown in Figure 1
A. Type II lung cancer cell can undergo a process of
dissemination with probability m per cell division,
leading to appearance of a new lung cancer cell in one
of metastasis sites. We assume that the cells in
metastasis sites are resistant to standard treatment,
which is composed of chemotherapy combined with
radiotherapy.
Figure 1: Mathematical model of NSCLC progression and
dissemination. A. Each cell in primary tumor compartment
(lung) can undergo one of three processes division, death,
and dissemination to local/distant site. Each cell in
metastasis compartment can divide or die. We do not
consider secondary metastases. B. Two paths of metastatic
dissemination. Blue arrows indicate dissemination through
lymphatic vessels and red ones through blood vessels.
The model considers two ways of metastatic
dissemination: through blood vessels (hematogenous
route) and through lymphatic vessels (lymphatic
route), as shown in Figure 1 B. The lymphatic route
is shown in blue color, whereas hematogenous route
is depicted with red color. In the first route of
dissemination, lung cancer cells disseminate first
through lymph nodes (local metastases), and next to
one of three distant sites: brain, liver or bones. Those
three distant sites characteristic for NSCLC
metastasis. The hematogenous route of tumor
dissemination is modelled as a single step process
where a Type II lung cancer cell colonizes one of
three distant site brain, liver or bones with probability
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
222

m. Global sensitivity analysis of a preliminary version
of this model was performed in (Kozłowska and
Swierniak, 2022). We found four parameters that
affect MFS: the growth rate of the primary tumor, the
growth rate of distant metastases, dissemination rate
from the primary tumor to distant metastases, and
carrying capacity. From all four parameters, we can
control two of them (to some extent): the growth rate
of the primary tumor (using chemotherapy) and
carrying capacity (using antiangiogenic therapy).
Yet another possibility for modeling invading
and colonization of distant organs is given by
evolutionary games. Evolutionary game theory
(EGT) combines mathematical tools of theory of
games with Darwinian adaptation and species
evolution and may be applied to analysis and
simulation of evolutionary changes within different
subpopulations due to interactions between them. The
result of these interactions (and, possibly, the effect
of environment) is a change of the degree of
evolutionary adjustment which, in turn, may cause
stabilization of the population structure. Using EGT,
it is possible to foresee, whether a population tends to
be heterogeneous or rather only one phenotype
survives and dominates. Introducing changes of the
replicator equations (RE) describing the behavior in
the population in time allows to follow dynamics of
changes. EGT has also been applied to study
development of cellular populations since cells, like
whole organisms, compete for space and nutrients,
exchange signals, cooperate, and show kinds of
“altruism” resembling animals in evolution. Starting
from the pioneering work of Tomlinson and Bodmer
(Tomlinson and Bodmer, 1997) this machinery was
used to model different tumor related phenomena.
Basanta et al. (Basanta et al, 2008) were probably the
first to use this machinery in modeling phenomena
leading to tumor cell invasion and migration. The
authors assume that at initial stage cancer cells are
specified by autonomous growth and then they can
switch to anaerobic glycolysis or become
increasingly motile and invasive. It allows to study
the circumstances, under which mutations confer
increased motility to cells needed for invasion of
other tissues and metastasis. In their next paper
(Basanta et al, 2010), the authors extended their
model by adding phenotype which could switch to
anaerobic glycolysis and be motile. Their model is
directed to glioblastomas. EGT is based on the
assumption of perfect mixing inside the population
(mean field approach) and interaction of each pair of
strategies.
To overcome this simplification and enable
analysis of local arrangement and internal
interactions in the neighborhood, the evolutionary
games have been transferred into spatial lattice by
application of cellular automata techniques, leading
to the so called spatial evolutionary game theory. In
(Swierniak and Krzeslak, 2013) the analysis of all
these three models is appended by RE and SEGT
tools (if absent in original study) which allows to give
an approximate answer on questions regarding time
and place of the switch, leading to tumor migration.
In the project we propose more complex EGT models
of tumor- tumor cells interactions containing different
strategies of dissemination of NSCLC which will take
into account results of other tasks in the project.
Moreover, we apply new tools of spatial evolutionary
tools, proposed recently. These tools take into
account heterogeneity at the cell level (the so called
Mixed Spatial Evolutionary Games – MSEG) and
varying in time (and possibly also in space) effects of
environment (Evolutionary Games with Resources
and Spatial Evolutionary Games with Resources,
respectively). In the former case it leads to multilayer
structure of the game (Swierniak and Krzeslak, 2016)
and in the latter case to time varying pay-off tables
(Swierniak et al, 2018). Moreover, we propose new
algorithms which enable modeling of 3D structure in
spatial games.
4 IMAGE PROCESSING,
FEATURE EXTRACTION AND
SELECTION, MACHINE
LEARNING BASED MODEL
Positron Emission Tomography/Computed
Tomography (PET/CT) examination is currently
routinely used in radiation treatment planning and
staging of patients with NSCLC. It allows for
relatively precise assessment of primary tumor
volume and volume of involved mediastinal lymph
nodes. Retrospective data (including PET/CT images
and history of the treatment) for at least 100 patients
with stage IIIAN2-IIIB NSCLC, who had pre-
treatment PET/CT imaging and underwent curative
radio-chemotherapy have been selected and acquired
from the database. For all those patients all clinical
information will be extracted. Images acquired from
PET/CT device and stored in a standard DICOM
format (Digital Imaging and Communications in
Medicine) will be processed to obtain a set of
radiomics features. Target lesions, for primary tumor,
as well as for nodes or metastatic lesions, are prepared
manually by an experienced specialist, using medical
image viewer software and/or automatically extracted
System Modeling and Machine Learning in Prediction of Metastases in Lung Cancer
223
directly from image, if needed. All regions of interest
(ROI) are stored for subsequent analyses. With high-
throughput computing, it is now possible to rapidly
extract a vast number of quantitative features from
tomographic images (computed tomography (CT),
magnetic resonance (MR), positron emission
tomography (PET)).
The main concept behind this process was that
biomedical images contain information reflecting
underlying pathophysiology and that these
relationships can be revealed via quantitative image
analyses. The conversion of digital medical images
into mineable high-dimensional data is known as
radiomics (d’Amico et al, 2020, van Griethuysen et
al, 2017, Gillies et al, 2016, Kumar et al, 2012,
Lambin et al, 2012). Radiomics is designed to
develop decision support tools; therefore, it involves
combining radiomic data with other patient
characteristics (clinical, molecular etc.), if available,
to increase the power of the decision support models.
It has been proven that features not perceptible to the
eye of the reporting physician — such as intra-tumor
heterogeneity, distribution of signal values within the
tumor area and more — can be indicative of certain
biological characteristics of the tissue, such as
proliferation, hypoxia, necrosis, angiogenesis and
even tumoral genotype (d’Amico et al, 2020).
Quantitative image features based on intensity, shape,
size or volume, and texture offer information on
tumor phenotype and microenvironment that is
distinct from that provided by clinical reports,
laboratory test results, and genomic or proteomic
assays. These features, in conjunction with other
information, can be correlated with clinical treatment
outcomes data and used for clinical decision support
(Figure 2). Radiomics provides imaging biomarkers
that could potentially aid cancer detection, diagnosis,
assessment of prognosis, prediction of response to
treatment, and monitoring of disease status etc.
Acquired pre-treatment PET/CT images are
preprocessed in order to save the data in appropriate
format for subsequent radiomics analysis. Manually
or automatically generated ROIs are preprocessed in
the similar way. Radiomic features are extracted from
the target lesions (described by ROIs) using the
program based on PyRadiomics (https://
pyradiomics.readthedocs.io) package for Python (van
Griethuysen et al, 2017), including: (i) First order
features (energy, entropy, minimum, percentiles,
maximum, mean, median, interquartile range, range,
standard deviation, skewness, kurtosis, among
others); (ii) Shape Features (volume, surface area,
sphericity, among others); (iii) higher order statistics
texture analysis, including: Gray-Level Co-
occurrence Matrix (GLCM), Grey-Level Dependence
Matrix (GLDM), Grey-Level Run Length Matrix
(GLRLM), Grey-Level Size Zone Matrix (GLSZM)
and Neighboring-Gray Tone Difference Matrix
(NGTDM). Using additional filters (for example
Local Binary Patters, wavelets etc.) on the original
image is also considered. This allows to multiply the
resulting radiomic features, which can potentially
highlight features invisible in ROIs.
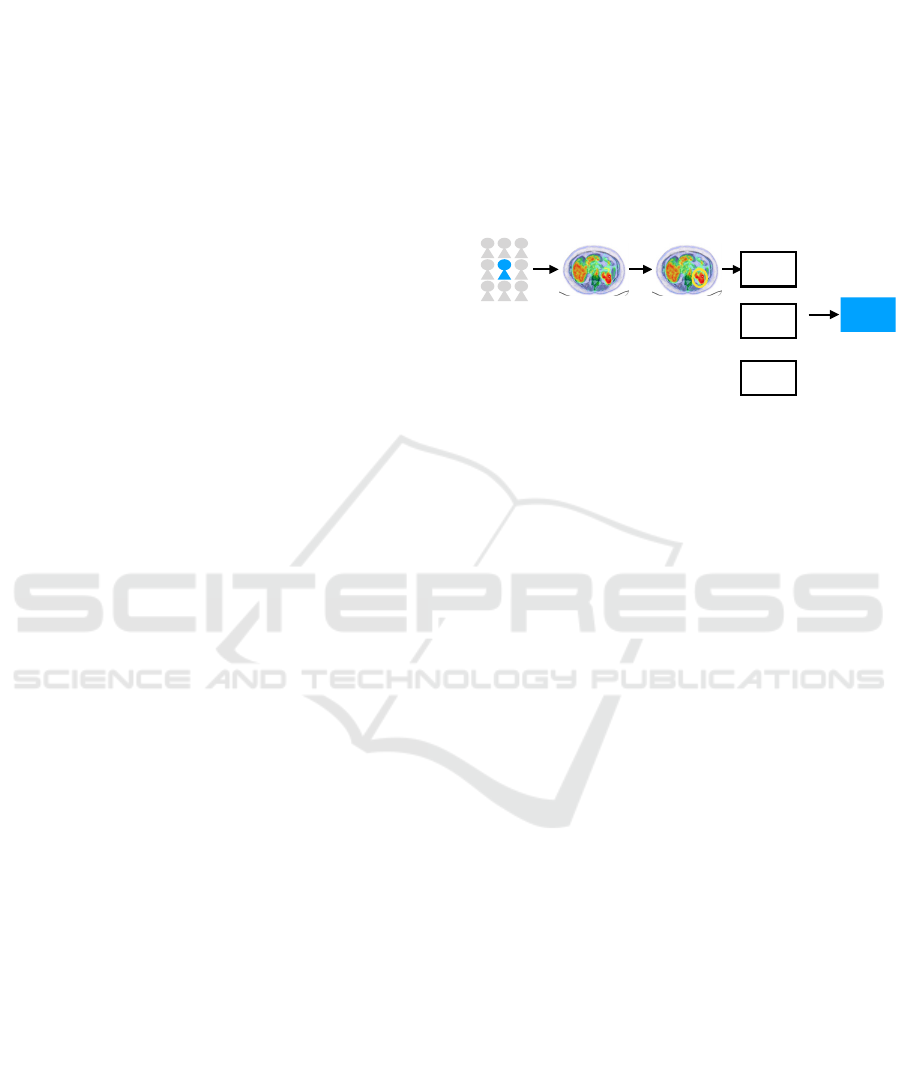
Figure 2: Illustration of a typical workflow for radiomics
signature development: I group selection, II Data
acquisition, III Image segmentation, IV Feature extraction,
V model validation. Model can be build using not only
radiomic features.
Machine learning algorithms could be applied in two
ways:
a. Classical approach - as a separate algorithm /
predictor whose output is the predicted time and place
of cancer metastasis.
b. Non-classical approach - as part of a larger
model, part of which is one of the dynamic models
developed in the project.
In the first case, the problem is formulated in a
typical way for supervised learning, in which we have
a learning set in the form of PET / CT images,
radiomic features extracted on their basis, and
additional clinical data for a given patient cohort. This
data is accompanied by the set outputs of the model
in the form of information about the time and places
of metastasis. In this situation, the classic division of
the model structure into (i) feature selection and (ii)
training the classifier (predictor) may apply. In
addition, the problem of simultaneous use of radiomic
data and clinical data whose nature is different may
be interesting. In this context, we have tested different
ways of integrating this data and choose the best one.
For this purpose, we use the software, which allows
testing various methods of data integration, while
protecting against information leakage, which may
result in an optimistic bias of prediction quality
assessment. In (Fujarewicz et al, 2022) we have
presented the attempt to use the radiomics features to
predict the metastasis for lung cancer patients. The
I II III IV V
PET/ CT ROI
.csv
file
model
radiomic s
features
ML / AI
clinical
information
+
+
molecular
data
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
224

obtained accuracy of the best classifier confirms the
potential of such prediction of metastasis.
The non-classical approach (b) to the application
of machine learning algorithms and feature selection
relies on the construction of a combined model, part
of which is the dynamic models developed in the
project. In the combined model, a task of the machine
learning algorithm (instead of metastasis prediction)
determines the values of parameters (different for
each patient) of dynamic models. It seems that this
approach, although more difficult than the classical
approach, has a chance of better prediction, because
it combines the advantages of machine learning and
modeling of dynamic systems. The difficulty in
building such a model lies in the fact that the
described approach cannot build a typical supervised
learning data set. While we have input data (radiomic
and clinical), we do not have the set values of these
parameters, but only the set (observed) responses of
patients to therapy. The model learning process must
therefore take account of this fact and requires
development of new learning / adaptation methods.
In the case of the classic approach to the use of
artificial intelligence algorithms (in which the output
of the artificial intelligence algorithm is the predicted
time and place of metastasis) the model learning task
can be formulated as a regression / approximation
task, with continuous output variables, or as a
classification task, in which the model output is the
label of the appropriate class. It is also possible to
create a hybrid model that has both continuous (time
to metastasis) and discrete (place of metastasis)
outputs. In all these cases, we test various regression
and classification techniques such as: support vector
machines (SVMs) with different kernels, linear and
logistic regression, convolution neural networks,
linear (LDA) and quadratic (QDA) discriminant
analysis, classifier ensembles (bagging, boosting,
random forests) and others.
An important element of building the machine
learning model is the selection of features
(radiometric, clinical). In this case, we use various
approaches, ranging from the simplest filter methods
to more complex wrapped methods and embedded
methods. It is also possible to use methods for
transforming feature spaces such as Principal
Components Analysis (PCA) or Independent
Components Analysis (ICA). Nevertheless SVM is
our first choice in extraction and selection of radiomic
features.
In the case of a non-classical approach, in which
the outputs of the machine learning algorithm are the
parameters of the dynamic model, in general case, it
is impossible to use available methods of supervised
learning because (desired) parameters of the dynamic
model are not known and only desired response of the
dynamic model (i.e. patient’s response) is given. In
this case, we present the machine learning task as a
mathematical programming task – optimization of the
performance index depending on the prediction error
in the parameters space of the particular machine
learning method.
In the special case, when the dynamic model has
a form enabling to build based on it the sensitivity
model, we develop an original gradient algorithm
based on the backpropagation of the prediction error
(through the model adjoint to the dynamic model)
enabling the determination of the gradient of the
performance index with respect to the parameters of
the machine learning algorithm. In some respects,
such an algorithm is similar to the algorithms
developed earlier, which involve the use of adjoint
sensitivity analysis for complex systems (Fujarewicz
and Galuszka, 2004, Fujarewicz et al, 2007).
Selection of features, as in the case of the classical
approach, is possible using filter or wrapped methods.
5 CONCLUSIONS
Although the first attempt to use mathematical
modeling to study quantitatively metastases of
untreated lung cancer had more than sixty years of
history (see, (Colins, 1956)), there are currently no
mechanistic models incorporating biomarkers, which
could play a role of prognosis tools that could inform
when and where NSCLC may metastasize. Such tools
could be of great interest to clinicians, supporting
treatment decisions, such as whether to use systemic
therapy or not and with what intensity and duration.
We hope that by extraction of radiomic features from
PET/CT images, their selection and incorporation in
existing and newly built mechanistic models,
predicting NLSCLC spread and metastatic
dissemination will become possible.
On the other hand, there exist many studies in
which empirical models based on statistical data are
used to predict the risk of metastases taking into
account different genomic or proteomic features of
patients. Those studies are related to genotyping and
genomic profiling (e.g. (Li et al, 2013)), expression
of multiple mRNA markers in bronchoscopy (e.g.
(Suwinski et al, 2012)), gene polymorphisms (e.g.
(Butkiewicz et al, 2015)), blood serum proteins (e.g.
(Suwinski et al, 2019)) or more general serum
comparative analysis (e.g. (Pietrowska et al, 2014)),
to mention only a few of them. The approach
proposed in the paper is the first step in construction
System Modeling and Machine Learning in Prediction of Metastases in Lung Cancer
225

of mixed models, which combine mechanistic
models of tumor dynamics with machine learning
models and using data from diagnostic investigations
(in this case biomedical images).
Dynamical models of cancer growth based on
ordinary differential equations (ODE), partial
differential (PDE) and other structured or agent-based
models (see, (Swierniak st al, 2016, Ledzewicz and
Schaettler, 2015, Clairambaut, 2014), for survey),
usually concentrate only at local tumor eradication,
some additionally take into account the surrounding
tissue. Those models do not take into account cancer
reappearance in distant sites after treatment.
Moreover, there is no available mathematical model
taking into account an intermediate step of metastasis
dissemination, which is spread of tumor cells to local
lymph nodes. The mixed machine learning and
mechanistic model proposed by us could be applied
also to other types of solid cancers such as rectal, head
and neck or breast cancer, which also have high
metastasis potential. Thus, in the future, we plan to
extend the method to other types of solid cancers.
In addition, we plan to incorporate molecular
data from liquid biopsy (see e.g. (Suwinski et al,
2019)). Proteomics profiling of blood serum from
about 100 non-small cell lung cancers will be
performed, which will allow to incorporate molecular
features into prediction of distant metastases. In
(Jaksik and Smieja, 2022) we have presented an
attempt to identify which -omics dataset or
combination of them, provide the most relevant
information for the prognosis of lung cancer survival.
This, will enable integration of biomedical images
with molecular data. However, this work is beyond
the scope of this paper.
ACKNOWLEDGEMENTS
This work was supported by Polish National Science
Centre, grant number: UMO-2020/37/B/ST6/01959
and Silesian University of Technology statutory
research funds.
REFERENCES
d’Amico, A., Borys, D., Gorczewska, I. (2020). Radiomics
and artificial Intelligence for PET imaging analysis.
Nuclear Medicine Review, 23, 1: 36–39
Barbolosi, D., Benabdallah, A., Hubert, F., Verga, F.
(2009). Mathematical and numerical analysis for a
model of growing metastatic tumors. Math Biosci
218(1):1-14
Basanta, D., Hatzikirou, H., Simon M, and Deutsch, A.
(2008). Evolutionary game theory elucidates the role of
glycolisys in glioma progression and invasion, Cell
Prolif., 41:980-987.
Basanta, D., Scott, J. G., Rockne R., Swanson, K.R., and
Anderson, A. R. A. (2010) The role of IDH1 mutated
tumor cells in secondary glioblastomas: an evolutionary
game theoretical view, Phys. Biol. 8: 015016.
Benzekry, S. (2011). Mathematical analysis of a two-
dimensional population model of metastatic growth
including angiogenesis. J Evol Equ 11(1):187-213.
Benzekry, S., Tracz, A., Mastri, M., Corbelli, R., Barbolosi,
D., Ebos, J.M. (2016). Modeling spontaneous
metastasis following surgery: an in vivo-in silico
approach. Cancer Res 76(3):535-547.
Bilous, M., Serdjebi, C., Boyer, A., Tomasini, P.,
Pouypoudat, C., Barbolosi, D., Barlesi, F., Chomy, F.,
Benzekry, S. (2019). Quantitative mathematical
modeling of clinical brain metastasis dynamics in non-
small cell lung cancer. Sci. Rep. 9:13018.
Butkiewicz, D., Krzesniak, M., Drosik, A., Giglok, M.,
Gdowicz-Kłosok, A., Kosarewicz, A., Rusin, M.,
Masłyk, B., Gawkowska-Suwinska, M., Suwinski, R.
(2015). The VEGFR2, COX-2 and MMP-2
polymorphisms are associated with clinical outcome of
patients with inoperable non-small cell lung cancer, Int.
J.Cancer, 137: 2332–2342.
Clairambault, J. (2014). Deterministic mathematical
modelling for cancer chronotherapeutics: cell
population dynamics and treatment optimisation. In
Mathematical Oncology 2013, A. d'Onofrio, A.
Gandolfi Eds., Part III:265-294, Birkhäuser, New York.
Collins, V.P., Loeffley, K.R., and Tivey, H (1956).
Observations on growth rates of human tumors, Am. J.
Roentgen, 76, 988-1002
Devys, A., Goudon, T., Lafitte, P. (2009). A model
describing the growth and the size distribution of
multiple metastatic tumors. Discrete Cont Dyn-B
12:731-767.
Franssen, L. C., Lorenzi, T., Burgess, A. E. F. & Chaplain,
M. A. J. (2019). A Mathematical Framework for
Modelling the Metastatic Spread of Cancer. Bull. Math.
Biol. 81:1965
Fujarewicz, K., Galuszka, A. (2004). Generalized
backpropagation through time for continuous time
neural networks and discrete time measurements., In:
Lecture Notes in Computer Science (eds: L. Rutkowski,
J. Siekmann, R. Tadeusiewicz and L. A. Zadeh), 3070:
190-196, Springer. 2004.
Fujarewicz, K., Kimmel, M., Lipniacki, T., Swierniak, A.
(2007). Adjoint systems for models of cell signaling
pathways and their application to parameter fitting.
IEEE-ACM Transactions on Computational Biology
and Bioinformatics, 4(3): 322-335.
Fujarewicz, K., Wilk, A., Borys, D., d’Amico, A.,
Suwiński, R., Świerniak, A. (2022). Machine Learning
Approach to Predict Metastasis in Lung Cancer Based
on Radiomic Features. In: Intelligent Information and
Database
Systems, Nguyen, N.T. et al. (eds): 40-50,
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
226

Lecture Notes in Computer Science, ACIIDS 2022,
Springer, Cham.
Gillies, R.J., Kinahan, P.E. and Hricak, H. (2016).
Radiomics: images are more than pictures, they are
data. Radiology, 278(2),563-577.
van Griethuysen, J. J. M., Fedorov, A., Parmar, C., Hosny,
A., Aucoin, N., Narayan, V., Beets-Tan, R. G. H.,
Fillon-Robin, J. C., Pieper, S., Aerts, H. J. W. L. (2017).
Computational Radiomics System to Decode the
Radiographic Phenotype. Cancer Research, 77(21),
e104–e107.
Haeno, H. et al. (2012). Computational modeling of
pancreatic cancer reveals kinetics of metastasis
suggesting optimum treatment strategies. Cell 148,
362–375.
Hahnfeldt, P., Panigraphy, D., Folkman, J., and Hlatky, L.
(1999). Tumor development under angiogenic
signaling: a dynamical theory of tumor growth,
treatment, response and postvascular dormancy.
Cancer Research, 59:4770–4775.
Inamura, K. (2017). Lung cancer: understanding its
molecular pathology and the 2015 WHO classification.
Front Oncol. Aug 28; 7.
Iwata, K., Kawasaki, K., Shigesada, N. (2000). A
dynamical model for the growth and size distribution of
multiple metastatic tumors. J Theor Biol 203(2):177-
186.
Jaksik, R., Smieja, J. (2022). Prediction of lung cancer
survival basing on -omic data, In: Intelligent
Information and Database Systems, Nguyen, N.T. et al.
(eds):116-127. Lecture Notes in Computer Science, vol
13758. ACIIDS 2022, Springer, Cham.
Kimmel, M., Axelrod, D.E. (2015). Branching Processes in
Biology, Springer, New York, Heidelberg Dordrecht
London.
Kozłowska, E., Świerniak, A. (2022). The Stochastic
Mathematical Model Predicts Angio-Therapy Could
Delay the Emergence of Metastases in Lung Cancer. In:
Biocybernetics and Biomedical Engineering – Current
Trends and Challenges, D.G., Zieliński K., Liebert A.,
Kacprzyk J. (eds). Lecture Notes in Networks and
Systems, vol 293. Springer, Cham.
Kumar, V., Gu, Y., Basu, S., Berglund, A., Eschrich, S.A.,
Schabath, M.B., Forster, K., Aerts, H.J., Dekker, A.,
Fenstermacher, D. and Goldgof, D.B. (2012).
Radiomics: the process and the challenges. Magnetic
resonance imaging, 30(9).1234-1248.
Lambin, P., Rios-Velazquez, E., Leijenaar, R., Carvalho,
S., Van Stiphout, R.G., Granton, P., Zegers, C.M.,
Gillies, R., Boellard, R., Dekker, A. and Aerts, H.J.
(2012). Radiomics: extracting more information from
medical images using advanced feature
analysis. European Journal of Cancer,48(4),.441-446.
Ledzewicz, U., Schaettler, H. (2015). Optimal Control for
Mathematical Models of Cancer Therapies, Springer,
New York.
Li, T., Kung, H.J., Mack, P.H., Gandara, D.R. (2013).
Genotyping and Genomic Profiling of Non–Small-Cell
Lung Cancer: Implications for Current and Future
Therapies, Journal of Clinical Oncology, 31 (8), 1039-
1043.
Nicolo, C., Perier, C., Prague, M., Bellera, C., MacGrogan,
G., Saut, O., Benzekry, S. (2000) Machine Learning
and Mechanistic Modeling for Prediction of Metastatic
Relapse in Early-Stage Breast Cancer. JCO Clin
Cancer Inform, 4:259-274.
Pietrowska, M., Jelonek, K., Michalak, M., Roś, M.,
Rodziewicz, P., Chmielewska, K., Polański, K.,
Polańska, J., Gdowicz- Kłosok, A., Giglok, M.,
Suwiński, R., Tarnawski, R., Dziadziuszko, R.,
Rzyman, R., and Widłak, P. (2014). Identification of
serum proteome components associated with
progression of non-small cell lung cancer, Acta
Biochem. Pol. 61 (2), 325–331.
Popper, H.H. (2016). Progression and metastasis of lung
cancer. Cancer Metastasis Rev. Mar1;35(1):75–91.
Saidel, G.M., Liotta, L.A., Kleinerman, J. (1976). System
dynamics of a metastatic process from an implanted
tumor. J Theor Biol 56(2):417-434.
Smieja, J., Psiuk-Maksymowicz, K., Swierniak, A. (2022).
A Framework for Modeling and Efficacy Evaluation of
Treatment of Cancer with Metastasis. In:
Biocybernetics and Biomedical Engineering – Current
Trends and Challenges. Pijanowska, D.G., Zieliński,
K., Liebert, A., Kacprzyk, J. (eds). Lecture Notes in
Networks and Systems, vol 293. Springer, Cham.
Suwinski, R., Klusek, A., Tyszkiewicz, T., Kowalska, M.,
Szczesniak-Klusek, B., Gawkowska-Suwinska, M.,
Tukiendorf, A., Kozielski, J., Jarzab, M. (2012). Gene
Expression from Bronchoscopy Obtained Tumour
Samples as a Predictor of Outcome in Advanced
Inoperable Lung Cancer, Plos One 7 ( 7): e41379.
Suwinski, R., Giglok, M., Galwas-Kliber, K., Idasiak, A.,
Jochymek, B., Deja, R., Maslyk, B., Mrochem-Kwar-
ciak, J., Butkiewicz, D. (2019). Blood serum proteins as
biomarkers for prediction of survival, locoregional
control and distant metastasis rate in radiotherapy and
radio-chemotherapy for non-small cell lung cancer,
BMC Cancer, 19:427.
Swierniak A., Krześlak M. (2013). Application of
evolutionary games to modeling carcinogenesis, Math
Biosci Eng, 10(3), 873-911.
Swierniak A, Kimmel M, Smieja J, Puszynski K, Psiuk-
Maksymowicz K. (2016). System Engineering
Approach to Planing Anticancer Therapies, Springer,
New York, Heidelberg Dordrecht London.
Swierniak, A., Krzeslak, M., Borys, D., and Kimmel M.
(2018). The role of interventions in the cancer
evolution-an evolutionary games approach. Math
Biosci. Eng.16(10); 265-291.
Swierniak, A., Krzeslak, M. (2016). Cancer heterogeneity
and multilayer spatial evolutionary games. Biology
Direct, 11(1):53-61.
Tomlinson I.P.M., Bodmer W.F. (1997). Modeling the
consequences of interactions between tumour cells
.
British Journal of Cancer, 75, 1997, 157-180.
System Modeling and Machine Learning in Prediction of Metastases in Lung Cancer
227
