
Hybrid Genetic U-Net Algorithm for Medical Segmentation
Jon-Olav Holland
1
, Youcef Djenouri
2
, Roufaida Laidi
3
and Anis Yazidi
1
1
OsloMet, Oslo, Norway
2
NORCE Norwegian Research Centre, Oslo, Norway
3
CERIST, Algiers, Algeria
Keywords:
Hyperparameter Optimization, Genetic Algorithm, U-Net, Medical Applications.
Abstract:
U-Net based architecture has become the de-facto standard approach for medical image segmentation in recent
years. Many researchers have used the original U-Net as a skeleton for suggesting more advanced models such
as UNet++ and UNet 3+. This paper seeks to boost the performance of the original U-Net via optimizing its
hyperparameters. Rather than changing the architecture itself, we optimize hyperparameters which does not
affect the architecture, but affects the performance of the model. For this purpose, we use genetic algorithms.
Intensive experiments on medical dataset have been carried out which document a performance gain at a low
computation cost. In addition, preliminary results reveal the benefit of the proposed framework for medical
image segmentation.
1 INTRODUCTION
Image segmentation models have been gaining trac-
tion over the last years (Kheradmandi and Mehranfar,
2022; Iqbal et al., 2022; Lin et al., 2022b). Segmen-
tation models are used in a variety of important fields
not only in experimental settings but also in produc-
tion environments as some of these models are now
being applied to real-world applications such as au-
tonomous driving (Wang et al., 2022), and remote
sensing (Wu et al., 2022). One of the most notable
fields is medical imaging (You et al., 2022; Kherad-
mandi and Mehranfar, 2022). In fact, these networks
have become so useful that Bergen hospital in Nor-
way has begun using them for tumor detection (E-
Helse, 2019). The models are used as an assistance
tool for doctors, yielding the probability of the patient
having a tumor. Our goal is to optimize the U-Net
model, which is a wildly used segmentation model.
We seek to optimize the hyperparameters of the model
using genetic algorithms, further increasing the per-
formance of the model. The work reported in (Ron-
neberger et al., 2015) introduced U-Net in 2015, and
since then, the model has been applied to several do-
mains within deep learning computer vision. U-Net is
a successful segmentation model with many succes-
sors in medical applications (Lin et al., 2022a). The
successors use U-Net as a skeleton, but seek to fur-
ther improve the model by making minor changes to
the architecture (Fang et al., 2022; Wu et al., 2019).
However, none of those successors seeks to improve
hyperparameters of the U-Net model itself. Deciding
the value of the U-Net hyperparameters may seem ar-
bitrary, as it is extremely difficult to assess the optimal
value. In this research work, we propose hyperparam-
eter optimization to enhance and improve the U-Net
model by assessing the optimal hyperparameter val-
ues. It is an end-to-end framework which uses the ge-
netic algorithm for guiding the training of the U-Net
architecture. The main contributions of this research
work can be given as follows:
1. We propose a new genetic algorithm which allows
exploring the possible combination of the U-Net
architecture.
2. We develop new crossover, and mutation op-
erators which intelligently explore the solutions
space of the different combinations of the hyper-
parameter optimization of the U-Net.
3. We test the proposed framework on large data for
medical image segmentation. The initial results of
the proposed framework are very promising.
The remaining of the paper is presented as fol-
lows. Section 2 presents the related work. Section 3
describes the image segmentation problem. Section 4
explains the main components of the proposed frame-
work. Section 5 gives the experimental analysis part,
while Section 6 concludes the paper.
558
Holland, J., Djenouri, Y., Laidi, R. and Yazidi, A.
Hybrid Genetic U-Net Algorithm for Medical Segmentation.
DOI: 10.5220/0011703700003393
In Proceedings of the 15th International Conference on Agents and Artificial Intelligence (ICAART 2023) - Volume 3, pages 558-564
ISBN: 978-989-758-623-1; ISSN: 2184-433X
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

2 RELATED WORK
Ultrasound image segmentation has the goal to iden-
tify the different labels in a given ultrasound image. In
the context of deep learning, the aim is to design effi-
cient models in order to learn the segmentation func-
tion. The input of the model is an ultrasound image,
and the output will be the label of each pixel in that
image. Huang et al. (Huang et al., 2020) proposed
a machine learning method for breast ultrasound im-
age segmentation in order to identify tumors. The ul-
trasound images are first cropped and pre-processed
using bilateral filtering, histogram equalization, and
pyramid mean shift filtering to remove noise. Simple
linear iterative clustering is then performed for group-
ing the pixel of images into super-pixels. Features are
extracted for each super-pixel, where two labels are
created, the tumor label if the super-pixel contains a
tumor, the normal label, otherwise. The kNN clas-
sifier is then performed to classify the pixels located
to the super-pixels into tumor or normal. Adjacent
tumor super-pixels are finally merged to segment the
tumor of the new image. Amiri et al. (Amiri et al.,
2020) proposed a two-stage ultrasound image seg-
mentation approach for breast lesion detection. The
first use of the U-Net model aims to detect the lesions.
The second use of the U-Net model aims to segment
the detected lesions. Lee et al. (Lee et al., 2020)
introduced the use of channel attention mechanisms
to improve CNN performance for breast cancer seg-
mentation in an ultrasound image. Interdependencies
of the channels of the image are trained by injecting
the statistical feature of each channel features (mean
of pixel values) on fully connected layers-based net-
work. The output of this network with the input
images are injected into CNN for the segmentation.
Wu et al. (Wu et al., 2020) proposed the encoder-
decoder deep learning model for thyroid nodule seg-
mentation on ultrasound image data. It contains: i)
dense block structure, where any two layers are con-
nected. Batch Normalization is used in order to train
this dense block. ii) Atrous spatial pyramid pooling
is used for creating contextual multiscale information
of input feature map. iii) Model size optimization for
reducing the number of parameters learned, where a
further 1 × 1 convolution operation is computed be-
fore each convolution layer. The semantic features
are obtained from the contextual information, and in-
jected into each layer of the decoder module. The
hierarchical feature fusion is also performed to merge
the feature maps of the blocks of the decoder mod-
ule. Zeng et al. (Zeng et al., 2021) proposed a hybrid
deep learning architecture for fetal ultrasound image
segmentation. A combination of V-Net with attention
mechanism is carried out in order to reach the bet-
ter accuracy of the segmentation results. To deal with
large range of batch size, global normalization is used
instead of batch normalization. A mixed loss func-
tion based on the dice similarity coefficient is devel-
oped in order to minimize the error ratio. Note that the
dice similarity coefficient is determined by the inter-
section over the union of the ground truth, and the out-
put of the network. Xue et al. (Xue et al., 2021) ad-
dressed three issues related to breast lesion ultrasound
image segmentation, which are: in homogeneous in-
tensity distributions inside the breast lesion region,
ambiguous boundary due to similar appearance be-
tween lesion and non-lesion regions, and irregular
breast lesion shapes. CNN is first used for multiscale
feature maps generation. Each CNN layer is con-
nected to a 1 × 1 convolutional layer with maxpool-
ing operation for detecting the breast lesion bound-
aries. The features of all CNN layers are concate-
nated and combined with spatial-wise, channel-wise
blocks for learning the correlation among the gener-
ated feature maps, and predict the output image. Liu
et al. (Liu et al., 2021) proposed a hybrid deep learn-
ing algorithm for detecting prostate cancer using ul-
trasound images. Feature extraction is performed us-
ing the Sobel filter, the features are injected into a
RCNN (Regional Convolution Neural Network) for
ultrasound image segmentation. Ouahabi et al. (Oua-
habi and Taleb-Ahmed, 2021) developed an encode-
decoder deep learning model for thyroid segmenta-
tion. It adds a new layer which integrates the mer-
its of dense connectivity, dilated convolutions for ex-
tracting relevant features, and dealing with varied-size
regions, respectively.
The image segmentation models in particular U-
Net based architecture require high number of hyper-
parameters to be tuned. This research work develops
an end-to-end framework based on genetic algorithm,
and U-Net for improving the image segmentation pro-
cess.
3 BACKGROUND ON IMAGE
SEGMENTATION
An image is “segmented” or “partitioned” into vari-
ous groups throughout the image segmentation pro-
cess. For instance, image segmentation is used to
distinguish the speaker from the background in the
Zoom call functionality that lets you alter your back-
ground. This is but one use case for image segmen-
tation in the real world. Face identification, video
surveillance, object detection, medical imaging, and
other fields can all benefit from image segmentation.
Hybrid Genetic U-Net Algorithm for Medical Segmentation
559
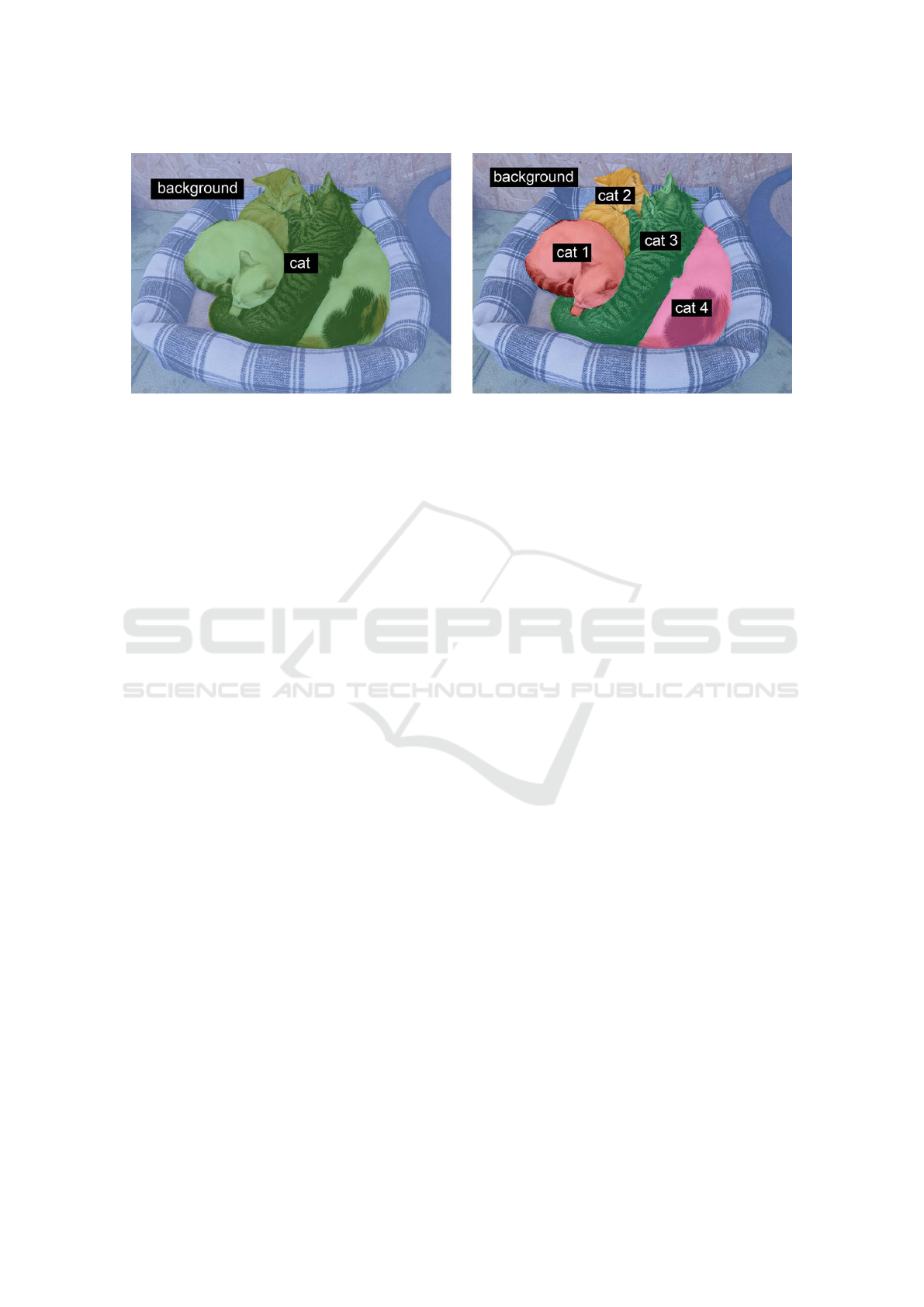
Figure 1: Semantic segmentation vs. instance segmentation (Chollet, 2021).
These applications use two-dimensional data in some
cases and three-dimensional data in others.
There are two types of image segmentation: se-
mantic segmentation, and instance segmentation.
• Semantic segmentation, where the objective is to
assign a category to each pixel in an image. Se-
mantic segmentation aims to classify each tree in
a forest image into the appropriate category.
• Instance segmentation, This accomplishes the
same thing as semantic segmentation but goes a
step farther. An instance segmentation of the for-
est image would then separate the trees into tree 1,
tree 2, and so on. Instance segmentation aims to
separate items of the same category into a series.
Figure 1 illustrates the distinction between in-
stance and semantic segmentation visually. The terms
“semantic segmentation” and “image segmentation”
will be used interchangeably. Object segmentation
and detection share similarities. Finding the various
object classes in a given image is the aim of object de-
tection. The object is marked by object detection with
a square frame represented by a bounding box. Ob-
ject detection just displays the location of an object; it
does not identify its shape. Object detection does not
meet the criterion for several tasks. For instance, the
form of the malignant cell is important when estimat-
ing the extent of the disease while trying to identify
it.
4 PROPOSED FRAMEWORK
As illustrated in Figure 2, the goal is to find the op-
timal learning rate, number of epochs, and batch size
for the U-Net architecture. All these hyperparameters
have a direct impact on the performance of the model
without changing the architecture. To optimize the
hyperparameters, we use genetic algorithms. Genetic
algorithms (GA) consist of a set of population of in-
dividuals, each of which consists of genes. For our
task, the genes are the hyperparameters; learning rate,
number of epochs, and batch size. The individuals are
then tested in the environment. Their genes, or hyper-
parameters, are applied to the model and the model
produces a loss. The individuals are then granted fit-
ness based on the loss, higher loss yields lower fitness,
and vice versa. After each individual in the population
is tested and has received their fitness, this generation
is complete. The last step is for the current popula-
tion of the next generation. To do so, parents are se-
lected based on their fitness to reproduce, higher fit-
ness yields a higher chance of becoming a parent; sur-
vival of the fittest. The next generation’s population is
then repopulated by the set of parents, and the process
begins anew. Optimally, the GA will converge toward
a global optimum where most individuals consist of
the optimal hyperparameter values for the model.
1. Defining the Search Space: The learning rate,
the number of epochs, and the batch size have
been specified as the components of the GA’s
search space. However, the need to specify and
explain the scope of the hyperparameters is still
existing. Depending on the restrictions placed by
the hyperparameter itself, the range may be quite
arbitrary. For instance, the learning rate could
only be set to between 0 and 1. It is somewhat
wasteful to add such a high learning rate even
when it is never utilized.
2. Selection: Additionally, we need to choose the in-
dividual who will make up the future generation.
We applied the tournament-based strategy, which
randomly chooses a group of individual from the
population. The winner of the “tournament” is
chosen to be a parent after the set competes in
it. The winner of the competition will be the one
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
560
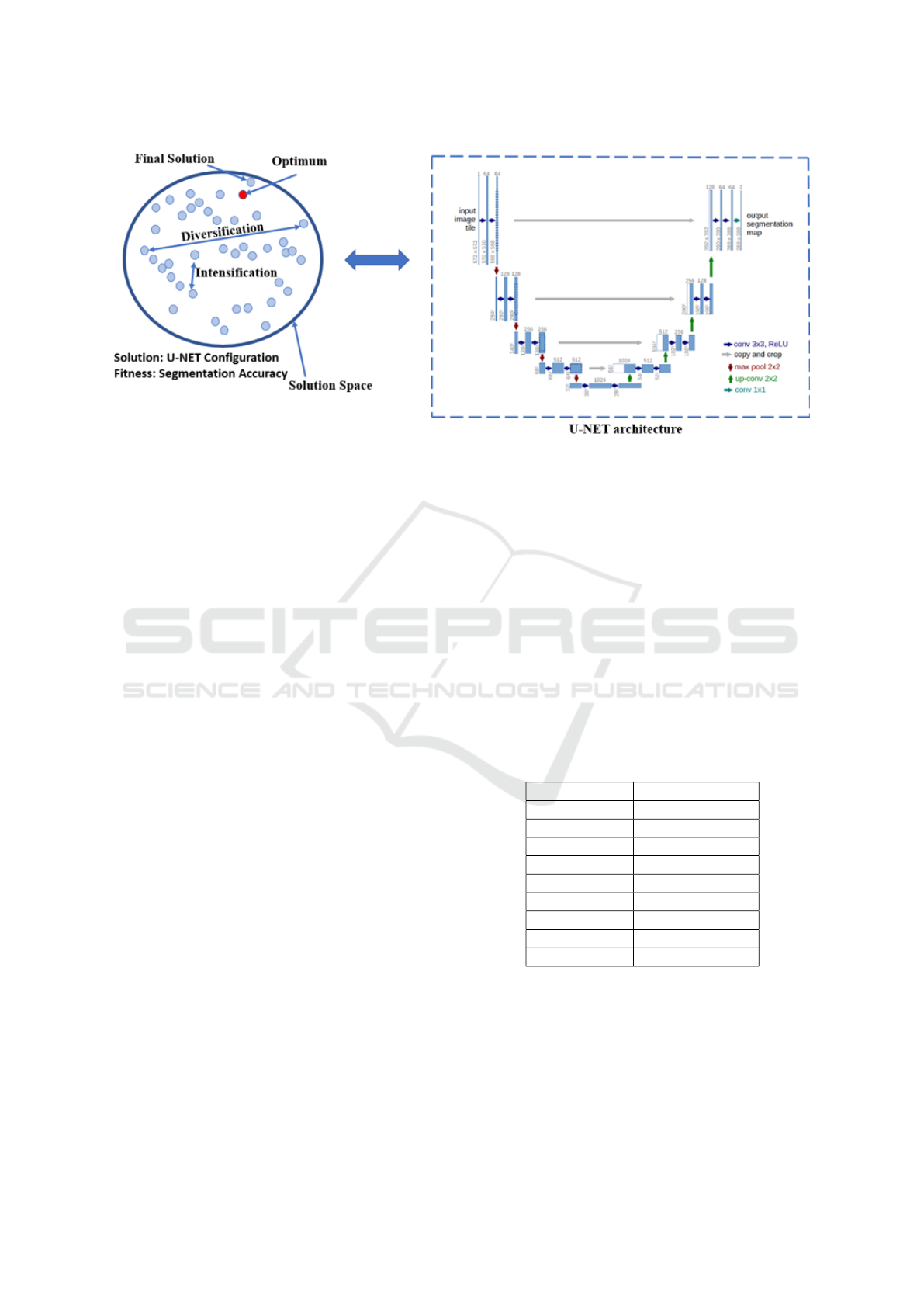
Figure 2: The proposed framework that combines both the genetic algorithm, and the U-Net architecture to improve the
medical segmentation process (U-NET diagram is retrieved from (Ronneberger et al., 2015)).
with the highest level of fitness. Therefore, the
outcome is somewhat predetermined. The com-
petitors in the competition are not subjected to
any new tests. The selection process for tourna-
ments includes some built-in exclusions for the
very lowest performers. For example, in a pop-
ulation of 100 individuals, where five individuals
are competing, the four less performing individ-
uals can never be chosen as parents, given these
values. There is no tournament set in the populace
where these four individuals can prevail. Figure 3
illustrates a selection tournament example that is
applied to the suggested framework.
3. Crossover: The chosen parents must reproduce
and give creation to new individuals after submit-
ting an application for tournament selection and
locating the set of parents. The new individual
is made up of a combination of the DNA from
the parents. We can transform the values to bi-
nary representations or use the genes as their val-
ues while executing crossover. Successful parents
receive the crossover process, but it has the po-
tential to alter the order of the many genes. Ev-
ery possible gene combination is included in the
search space, and the crossover process aids in
the systematic exploration of the various combi-
nations. Utilizing a binary format allows for even
more profound investigation of the crossover pro-
cess because it gives each gene’s value the oppor-
tunity to be changed.
4. Mutation: There is a possibility that a newly pro-
duced individual will become mutated. The in-
dividual’s genes are changed via mutation. By
changing the gene’s value, mutation might occur
to one or many genes. Within the limitations of
the gene, alternation causes the gene to be ran-
domized. New genes are added to the population
through mutation. The modified individual bear-
ing the new genes will be quickly wiped out of
the population if these genes are bad, that is, if
they score a poor fitness. If the genes are sound,
however, the GA will continue to use the newly
discovered genes and procreate the population.
5 PERFORMANCE EVALUATION
Table 1: The table contains the best individual after 50 gen-
erations, given the different mutation rates.
Mutation Rate IoU Performance
5% 0.7274
10% 0.7273
15% 0.7465
20% 0.7278
25% 0.7289
30% 0.7366
35% 0.7274
40% 0.7276
45% 0.7272
We apply our solution to the ultrasound nerve dataset.
As with most medical imaging datasets, the data is
imbalanced. The dataset contains 5600 images where
all the images are labeled. We use 90% of images to
train a new model with each individual hyperparame-
ters, and we use 10% of images for testing the model.
The evaluation of the proposed framework is calcu-
lated using the Intersection-over-Union (IoU), (equa-
Hybrid Genetic U-Net Algorithm for Medical Segmentation
561
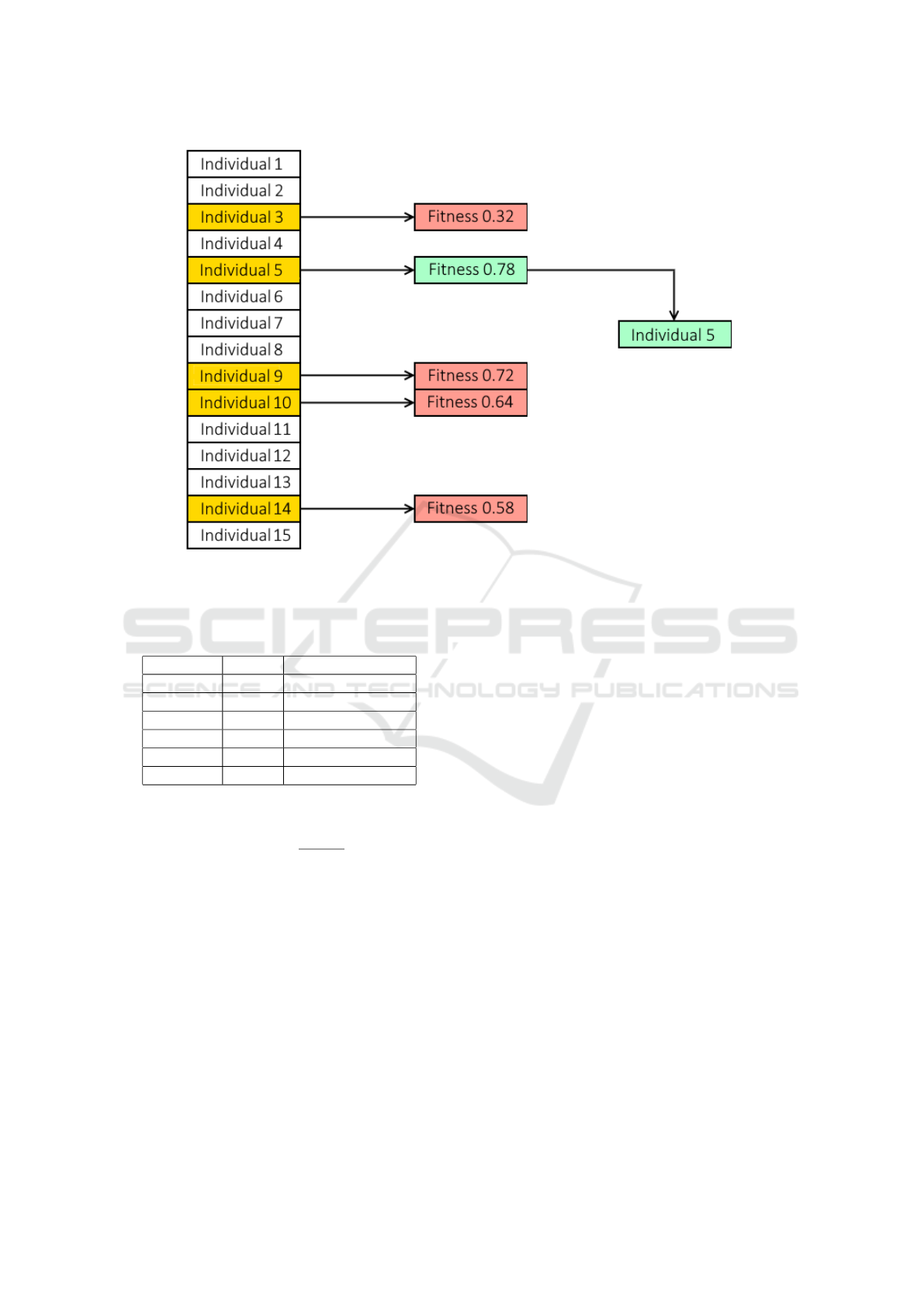
Figure 3: A population containing 15 individuals where tournament selection is applied. At random, 5 individuals are ran-
domly chosen to compete in the tournament. The winner of the tournament is based on the predetermined fitness of the
individual. In this case, individual 5 wins the tournament and is selected to be a parent.
Table 2: Comparison of the proposed solution with UNet
algorithm.
% Images UNet Proposed Solution
10% 0.7104 0.7239
20% 0.7329 0.7692
30% 0.7567 0.7985
50% 0.7859 0.8003
80% 0.7971 0.8120
100% 0.8001 0.8431
tion 1).
IoU(U, V ) =
|U ∩V |
|U ∪V |
(1)
IoU is a method of measuring the overlapping la-
bels. It measures the overlapping true and false labels,
then divides it by the union of the labels. As expected,
this heavily punishes the model by falsely predicting
a nerve in the wrong spot. But in addition, it also pun-
ishes the model if it were to only predict false labels
(no nerve) for every input. Encouraging the model to
actually find the nerves and not stall the learning. We
run a set of short tests to evaluate the optimal muta-
tion rate. These tests consist of a training set of 100
images from the nerve dataset, and the epoch range
is set between 0 and 1. Due to the low amount of
data used, and the low number of epochs, we evaluate
from the training loss, and not the validation loss. We
run these tests to find the mutation rate we want to
use and to see if there are any discrepancies between
the different mutation rates. Table 1 shows the differ-
ent range of mutation rates and the performance. The
performance is calculated by the dice loss functions.
Regardless of the mutation rate, there is no large dif-
ferent in the performance. The 15% mutation rate is
slightly above the rest, this may be random. Regard-
less, we will use 15% mutation rate for our initial ex-
periment. In the following experiments, we will use
the hyperparameters of the top individual found by
the genetic algorithm. Table 2 compares the results
of the proposed solution with the UNet algorithm. By
varying the number of images from 10% to 100%, the
proposed solution outperforms the UNet algorithm in
terms of IoU. For instance, when training 100% of im-
ages, the IoU of UNet is only 0.8001%, where the IoU
of the proposed solution is 0.8431%. These results
are achieved thanks to the hyperparameter optimiza-
tion technique used to find the best parameters of the
UNet model. The last experiment aims to visualize
some results of the developed model. Figure 4 shows
three images; the input, the hand annotated nerve cell,
and the predicted segmentation. The results indicate
a low gap between the ground truth and the predicted
segmentation of the proposed model. These results
are achieved thanks to the strategy used in the seg-
mentation process, where an efficient hyperparameter
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
562

Figure 4: Ultrasound images of the neck as input on the left side, center shows the hand annotated nerve, right side shows the
prediction by the model. These are some of the best results.
optimization is running to retrieve the optimal param-
eters of the designed model. These results demon-
strate the applicability of the developed model in real
settings to help the practitioners and doctors for med-
ical decision-making.
6 CONCLUSION
Although U-Net model shows a great behavior for
solving segmentation problem in medical applica-
tions, some limitations remain unsolved. In this pa-
per, we solved the hyperparameter optimization is-
sue by developing an end-to-end intelligent frame-
work which combines the genetic algorithm with the
U-Net architecture to achieve the optimal accuracy in
training complex medical data. We used genetic algo-
rithms to optimize the hyperparameters of the UNet
architecture for medical segmentation. The results re-
veal the superiority of the developed model compared
to the UNet model. As future perspective, we aim to
explore other evolutionary algorithms such as particle
swarm optimization, Ant Colony, and mimetic algo-
rithm in order to speed the convergence to the opti-
mum. Exploring evolving learning, and in particular
the NEAT optimization, is also in our future agenda.
Hybrid Genetic U-Net Algorithm for Medical Segmentation
563

REFERENCES
Amiri, M., Brooks, R., Behboodi, B., and Rivaz, H. (2020).
Two-stage ultrasound image segmentation using u-net
and test time augmentation. International journal of
computer assisted radiology and surgery, 15(6):981–
988.
Chollet, F. (2021). Deep Learning with Python, Second Edi-
tion.
E-Helse (2019). Utredning om bruk av kunstig intelligens i
helsesektoren.
Fang, Y., Huang, H., Yang, W., Xu, X., Jiang, W., and
Lai, X. (2022). Nonlocal convolutional block atten-
tion module vnet for gliomas automatic segmentation.
International Journal of Imaging Systems and Tech-
nology, 32(2):528–543.
Huang, Q., Huang, Y., Luo, Y., Yuan, F., and Li, X. (2020).
Segmentation of breast ultrasound image with seman-
tic classification of superpixels. Medical image anal-
ysis, 61:101657.
Iqbal, A., Sharif, M., Yasmin, M., Raza, M., and Aftab, S.
(2022). Generative adversarial networks and its appli-
cations in the biomedical image segmentation: a com-
prehensive survey. International Journal of Multime-
dia Information Retrieval, pages 1–36.
Kheradmandi, N. and Mehranfar, V. (2022). A critical re-
view and comparative study on image segmentation-
based techniques for pavement crack detection. Con-
struction and Building Materials, 321:126162.
Lee, H., Park, J., and Hwang, J. Y. (2020). Channel at-
tention module with multiscale grid average pooling
for breast cancer segmentation in an ultrasound im-
age. IEEE transactions on ultrasonics, ferroelectrics,
and frequency control, 67(7):1344–1353.
Lin, M., Cai, Q., and Zhou, J. (2022a). 3d md-unet: A novel
model of multi-dataset collaboration for medical im-
age segmentation. Neurocomputing, 492:530–544.
Lin, Z., Zhang, Z., Han, L.-H., and Lu, S.-P. (2022b). Multi-
mode interactive image segmentation. In Proceedings
of the 30th ACM International Conference on Multi-
media, pages 905–914.
Liu, Z., Yang, C., Huang, J., Liu, S., Zhuo, Y., and Lu, X.
(2021). Deep learning framework based on integra-
tion of s-mask r-cnn and inception-v3 for ultrasound
image-aided diagnosis of prostate cancer. Future Gen-
eration Computer Systems, 114:358–367.
Ouahabi, A. and Taleb-Ahmed, A. (2021). Deep learn-
ing for real-time semantic segmentation: Application
in ultrasound imaging. Pattern Recognition Letters,
144:27–34.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-net:
Convolutional networks for biomedical image seg-
mentation. volume 9351.
Wang, H., Chen, Y., Cai, Y., Chen, L., Li, Y., Sotelo, M. A.,
and Li, Z. (2022). Sfnet-n: An improved sfnet al-
gorithm for semantic segmentation of low-light au-
tonomous driving road scenes. IEEE Transactions on
Intelligent Transportation Systems.
Wu, D., Zhuang, Z., Xiang, C., Zou, W., and Li, X. (2019).
6d-vnet: End-to-end 6-dof vehicle pose estimation
from monocular rgb images. In Proceedings of the
IEEE/CVF Conference on Computer Vision and Pat-
tern Recognition Workshops, pages 0–0.
Wu, L., Lu, M., and Fang, L. (2022). Deep covariance
alignment for domain adaptive remote sensing image
segmentation. IEEE Transactions on Geoscience and
Remote Sensing, 60:1–11.
Wu, Y., Shen, X., Bu, F., and Tian, J. (2020). Ultrasound
image segmentation method for thyroid nodules using
aspp fusion features. IEEE Access, 8:172457–172466.
Xue, C., Zhu, L., Fu, H., Hu, X., Li, X., Zhang, H., and
Heng, P.-A. (2021). Global guidance network for
breast lesion segmentation in ultrasound images. Med-
ical Image Analysis, page 101989.
You, C., Zhou, Y., Zhao, R., Staib, L., and Duncan, J. S.
(2022). Simcvd: Simple contrastive voxel-wise repre-
sentation distillation for semi-supervised medical im-
age segmentation. IEEE Transactions on Medical
Imaging.
Zeng, Y., Tsui, P.-H., Wu, W., Zhou, Z., and Wu, S. (2021).
Fetal ultrasound image segmentation for automatic
head circumference biometry using deeply supervised
attention-gated v-net. Journal of Digital Imaging,
34(1):134–148.
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
564
