
Computerised Muscle Modelling and Simulation for Interactive
Applications*
Martin Cervenka
1,∗ a
, Ondrej Havlicek
2 b
, Josef Kohout
1 c
and Libor V
´
a
ˇ
sa
1 d
1
The University of West Bohemia, Faculty of Applied Sciences, Department of Computer Science and Engineering,
Czech Republic
2
The University of West Bohemia, Faculty of Applied Sciences, NTIS - New Technologies for the Information Society,
Czech Republic
Keywords:
Muscle Modelling, Collision Detection, Collision Response, Position Based Dynamics, Discregrid, Scalar
Distance Field, as-Rigid-as-Possible, Radial Basis Functions.
Abstract:
The main challenges of collision detection and handling in muscle modelling are demonstrated. Then, a colli-
sion handling technique is tested, exploiting the issue of muscle penetrating the bone in some circumstances,
mainly when the movement is too rapid or the displacement of the bone is too high. Our approach also de-
tects the problem, using Discregrid to see the immediate direction change towards the penetrated bone. Some
alternatives to the described PBD (Position-Based dynamics) technique are presented: PBD with As-Rigid-
As-Possible modification and radial basis function approach.
1 INTRODUCTION
Osteoporosis (Wade et al., 2014), osteoarthritis
(Oatis, 2017), patellar dislocation (Barzan et al.,
2017), or hemiplegic diseases (Zhang et al., 2021) are
leading researchers to develop a satisfactory model of
the musculoskeletal system. Creating such a model is
a complex procedure with many issues. The problem
of collision detection (CD) between muscle and bone
models and its response (CR) is critical.
In this paper, we follow our previous work and
newly present the parameter upper bound, where the
simulation still works as expected. Another contribu-
tion of this paper is exploring novel modelling meth-
ods to overcome the current limitations. This paper
also briefly describes some techniques of CD and CR.
The paper is structured as follows. The next sec-
tion gives an idea of the whole muscle modelling
overview and the steps to obtain a usable comput-
erised muscle model. The state-of-the-art approaches
a
https://orcid.org/0000-0001-9625-1872
b
https://orcid.org/0000-0002-6944-7084
c
https://orcid.org/0000-0002-3231-2573
d
https://orcid.org/0000-0002-0213-3769
∗
This Work Was Supported by the Ministry of Educa-
tion, Youth and Sports of the Czech Republic, Project SGS-
2022-015.
∗∗
Corresponding author
to modelling and simulation of the muscles, based on
the position-based dynamics, are described in Sec-
tion 3. The description of existing CD and CR tech-
niques employed during these simulations follows.
Sections 5 and 6 present the current approach’s lim-
itations and propose several improvements to over-
come them. Discussion of future work and conclud-
ing remarks follow.
2 MUSCLE MODELLING
PIPELINE
The muscle modelling procedure involves many
steps, including acquiring relevant raw data and its
subsequent transformation into a useful form. The
last step is formulating the mathematical model,
where the main concern is (among others) the defi-
nition of the muscle-bone interaction.
An example of a complex pipeline (consisting of
data acquisition, model building and inverse kinemat-
ics) includes the following steps:
1. obtaining raw data of the patient at rest (such as
medical images) representing anatomical objects
(bones, muscles, muscle attachment areas, etc.),
and movement data,
2. extraction and transformation of the raw data into
214
Cervenka, M., Havlicek, O., Kohout, J. and Váša, L.
Computerised Muscle Modelling and Simulation for Interactive Applications.
DOI: 10.5220/0011688000003417
In Proceedings of the 18th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2023) - Volume 1: GRAPP, pages
214-221
ISBN: 978-989-758-634-7; ISSN: 2184-4321
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

a useful form, using:
(a) segmentation – separation of different types of
tissues present in medical images (if they are
distinguishable). Segmentation can be manual,
semi-automatic or automatic (depending on the
complexity of the segmentation),
(b) extraction – the conversion of segmented data
into geometrical models,
(c) registration – mapping of data and models from
different measurements and modalities into the
common reference space
(d) approximation and interpolation – reconstruc-
tion of missing parts or partially corrupted data.
3. acquiring some general apriori knowledge, deter-
mined by human anatomies, such as
(a) how the attachment areas will be determined
(whether based on apriori knowledge only or
the measured muscle attachment areas),
(b) defining how a bone is connected by a joint to
another bone or how a muscle is connected to a
set of bones (attachment areas), etc.,
(c) defining physiological parameters of studied
muscles, such as internal muscle architecture
(e.g. figure arrangement: parallel, pennate,
etc.), optimal (resting) length and others.
4. creating a mathematical model that requires:
(a) defining the space (discretised or continuous),
(b) defining the shape of the data (triangular sur-
face mesh, tetrahedral volumetric mesh, scat-
tered data... / surface defined by Fourier series,
implicit RBF,...)
(c) defining the interaction between muscle and
bone models and thus determining whether or
not a transformation of the measured data is
necessary.
5. transformation of the simulation output from its
model representation into the final form, e.g.,
from a triangular surface mesh into a set of in-
ternal fibres.
We recommend the following papers for a more
detailed view: the foremost step is well described
by (Fukuda et al., 2017) for the attachment area ac-
quisition strategy, (Lee et al., 2014) to determine the
pennate angle from the source data. There are also
data from invasive measurements, e.g. Visible Hu-
man Project (the National Library of Medicine) or
The Chinese Visible Human (Zhang et al., 2004). The
second step (registration) is also well described in
(Zhao et al., 2013). There is also an approach from
(Li et al., 2008), with promising results. The other
steps are highly dependent on the considered applica-
tion, whether the purpose is a plausible visualisation
of muscles in movement (see. e.g. (Romeo et al.,
2018)) or to calculate some physical phenomena of
the muscle (see. e.g. (Modenese and Kohout, 2020)),
and whether the user should be able to change the
modelling or simulation parameters interactively or
not.
We focus on interactive applications which do
not require specialised hardware. Consequently, any
model developed in the fourth step needs to trade off
some accuracy for the speed of simulation. In this pa-
per, we consider position-based dynamics (PBD) suit-
able for generating models of such a kind.
3 POSITION BASED DYNAMICS
Position-based dynamics (PBD) (M
¨
uller et al., 2007)
is a fast approach used mainly in the animation indus-
try to model elastic object (and cloth) deformations.
Nowadays, the PBD is making its way into physical
simulations as well. The original algorithm does not
consider the possibility of object anisotropy as far as
the algorithm has been developed for general objects.
The method accepts a manifold surface mesh and pro-
duces its deformed variant as the output.
The PBD also exists in the xPBD (eXtended ver-
sion of PBD, which respects the concept of elastic
potential energy) form. The xPBD incorporates elas-
tic potential energy and eliminates the necessity to
know the time step and iteration count (Macklin et al.,
2016).
Romeo et al.(Romeo et al., 2018) are the first who
proposed using the xPBD algorithm for muscle mod-
elling problems. Their fundamental idea is to build an
internal structure above the surface mesh to respect
the anisotropy of the muscle (the internal structure
respects the general direction of the muscle fibres).
They can form a volumetric model better suitable for
the PBD algorithm with an intelligent edge-creation
process. However, the paper needs to describe their
collision detection and handle thoroughly. According
to their video of the technique outcome, many col-
lisions occur, suggesting their approach did not ad-
dress the apparent requirement of avoiding muscle-
bone penetrations.
Angles et al. (Angles et al., 2019) developed a
PBD-based approach for muscle modelling in 2019.
Their approach virtually decomposes the muscle into
”rods” (which may approximate the muscle fibres).
These rods can adjust their diameter wherever they
want to preserve their volume. Their main con-
tribution is the ability to provide real-time simula-
tion, which Romeo’s approach cannot because ”its
≈ 40s/frame of processing time causes it unfitting
to interactive applications” (Angles et al., 2019).
Computerised Muscle Modelling and Simulation for Interactive Applications
215

They adopt ”Particle simulation using CUDA” from
(Green, 2010) for collision detection between rods
and response. Again, the problem of muscle-bone
penetration is not addressed, though in this case, the
extension is relatively straightforward.
The position-based dynamics for muscle mod-
elling is also described in the paper ”Fast and Real-
istic Approach to Virtual Muscle Deformation.” (Cer-
venka. and Kohout., 2020) where the PBD approach
has been proposed (finished the same year as Romeo’s
article (Romeo et al., 2018), working concurrently on
the same). The paper ”Muscle Deformation using
Position Based Dynamics” (Kohout and
ˇ
Cervenka,
2021) follows, which tests the approach and compares
the results to an existing FEM technique. The primary
benefit of our suggested system is that no interior is
needed. The anisotropy is computed on the surface
of the mesh only, utilising muscle fibres on the mesh
surface, representing the fibre direction. The voxeli-
sation technique has been used for collision detection
and response purposes.
4 COLLISION DETECTION AND
RESPONSE
Various approaches to detect and respond to an oc-
curring collision have been proposed. The most com-
mon algorithms exploit the D&C (divide & conquer)
paradigm. The bounding volume hierarchy is one of
them (Teschner et al., 2005), using a primitive (of-
ten an axis-aligned bounding box AABB or a sphere)
hierarchy to enclose the model and its parts. The spa-
tial hashing (Turk, 1990) is its generalisation over the
whole model space.
If there is a necessity to know not only if the col-
lision occurs but how far from the collision the model
is, the (signed) distance field approach is an excel-
lent way to go. Numerous techniques can be used
to construct such a field. The vast majority use vox-
elisation to obtain a cell array and then use an inter-
polation method to determine the value between the
cells. Some of these techniques are well described in
an older work by (Bærentzen and Aanæs, 2002).
In our research of the PBD approach, the first de-
cision was to use a simple voxelisation method to
simplify the collision detection problem. This sim-
ple idea, however, leads to some things that could be
improved. Luckily, some ideas have emerged to en-
tirely improve or even fix some problems, using more
complex collision detection algorithms. Havlicek
(Havlicek et al., 2022) changed the collision detec-
tion to Discregrid (using a signed distance field) and
FLC (using a binary search tree), beating the voxeli-
sation approach in terms of accuracy. However, there
is still some work because even those methods only
work correctly in extreme conditions, mainly if the
movement is rapid.
The collision response is a complicated task
as well. Assume that two bones move towards
each other and narrowly miss each other (like shear
blades). If a muscle is attached to both of the bones
and appears to be in between the bones, there is no
such room for the bones to move into. This problem
often happens on a smaller scale, near joints, espe-
cially where two bones move close. Our former so-
lution (Cervenka and Skala, 2020) was to assume, in
this particular case, only one of the bones and move
a muscle in the direction opposite of it, but it proved
insufficient. Havlicek (Havlicek et al., 2022) targets
this problem primarily, and he proposed a better ap-
proach of considering all adjacent bones and moving
opposite to the sum of all collision vectors. Even this
approach, however, does not always guarantee colli-
sion resolution.
Our current contribution, proposed in this paper,
follows our recent articles, mainly (Cervenka. and
Kohout., 2020; Kohout and
ˇ
Cervenka, 2021) and also
(Havlicek et al., 2022).In the first article, we devel-
oped a PBD-based approach for muscle modelling.
The issue with a muscle stuck inside a joint was
shown in that article. We believed that ”better col-
lision detection can fix the issue” (Cervenka. and Ko-
hout., 2020). In the second article, the voxelisation
collision detection approach was proven inaccurate in
some cases, mainly in the case of more ”complicated”
(e.g. concave) bone surfaces, which are located near
the joint areas more frequently. The last article ex-
plores two existing collision detection algorithms for
the PBD approach: Discregrid and Flexible Collision
Library. The Discregrid was shown to be more suit-
able for the problem.
4.1 Discregrid
Discregrid library can be considered a Signed Dis-
tance Field generator written in C++. The algorithm
computes for each point in 3D space the shortest dis-
tance and direction towards a given nearby surface
represented by a triangular mesh. Also, assuming
that the input mesh is at least watertight, the method
can make the inside/outside decision because the al-
gorithm provides the sign. A finite bounded subspace
is required for the approach to work.
The bounded surface is firstly divided (like in the
voxelisation method) into a rectangular grid with a
user-defined resolution, where each voxel is a 32-
node Serendipity type (Koschier and Bender, 2017).
GRAPP 2023 - 18th International Conference on Computer Graphics Theory and Applications
216
For each node, the distance and the direction to-
wards the nearest bounded surface are computed, see
(Bærentzen and Aanæs, 2002). The article describes
the problem of discontinuity of the mesh (where the
normal vectors have to be estimated differently).
The field creation process is time-consuming
(about half a minute for bone meshes consisting of
up to 45 000 vertices with the grid resolution of
64 × 64 × 64 on standard hardware) and unfeasible
for deformable objects like muscles, where recalcu-
lation is often needed. There is no such problem with
the bone models because their movement is only rigid
(allowing for Discregrid results to be transformed us-
ing a global transformation).
In the case of muscle modelling, a situation may
happen when the muscle collides with a bone or an-
other muscle, even at the start of the simulation. This
situation is caused by the different modalities used for
the data measurements (see section 2). To fix the is-
sue, the colliding surface vertices are pushed accord-
ing to the Discregrid result directional vectors so no
collision would occur at the start of the simulation.
5 MUSCLE SIMULATION
In this paper, we experimented with more types of
motion of the hip joint, not just flexion but also ro-
tation and adduction. We also tested a hip extension
scenario in the described PBD approach to find the
maximum amount of problematic cases possible. The
tests will be done on muscles and bones depicted in
Fig. 1.
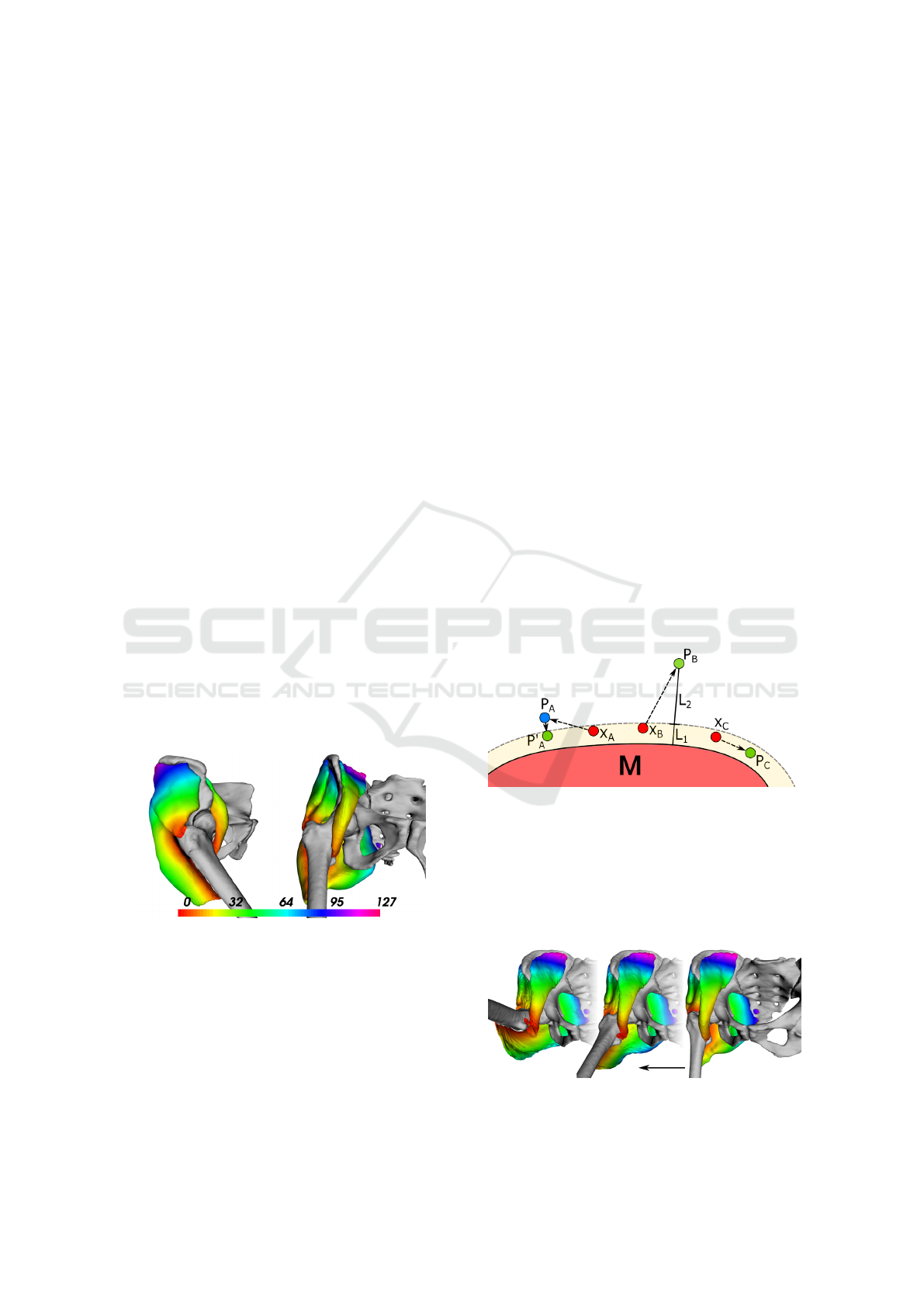
Figure 1: One view from the side and one from the front
of a muscle group surrounding the hip joint in rest pose.
The vertices of the muscles are coloured by their distance
to the femur bone (pointing down) in millimetres given by
Discregrid.
5.1 Test of Motion Types
For the test of different motion types, we tested the
original hip flexion (from 0
◦
to +90
◦
with the step of
2
◦
), hip rotation (from 0
◦
to +45
◦
and also to -45
◦
with the approximate degree of 1
◦
) and hip adduction
(from 0
◦
to +60
◦
with the approximate step of 2
◦
).
The results of hip flexion are shown in Fig. 3.
Near the joint area, the muscle nearly touches the fe-
mur bone; however, no collision occurs. Fig. 4 shows
the results for the rotational motion. As before, the
muscle nearly touches the femur’s upper extremity
without collisions. The adduction is demonstrated in
Fig. 5. In this case, the muscle is further from the hip
joint, lowering the possibility of collisions. The col-
lision detection and response approach solved all the
apriori collisions, so no collisions happened.
5.2 Test of Motion Speeds
To test for bone movement speed impact, we chose
the extension movement. The initial step of 2
◦
was
increased to 4
◦
, 5
◦
and finally 10
◦
. We also increased
the target angle to +80
◦
for rapid movement to have
time and space to show up fully.
In this case, when the angle finally reaches 72
◦
with the angle step of 4
◦
, first bone penetration occurs
(see Fig. 6). The muscle is not as fast as needed to
keep up with the bone movement, causing the distal
part of the muscle to enter the bone volume and go
through the whole cross-section. The muscle is also
being unnaturally pushed into itself by the bone.
Figure 2: Simplified 2D illustration of the sliding mecha-
nism. Points X
A
, X
B
and X
C
are being evaluated for the dis-
tance from the other muscle M. Consider the same thresh-
old distance for each of them (yellow margin). Point X
A
is
pushed to the position P
A
by previous PBD constraints but
returned towards the other muscle to the position P
′
A
. Point
X
B
is pushed away by the PBD so much it makes sense to
leave it to go ( L
2
> L
1
). Point X
C
is free to roam inside
the strip.
Figure 3: The result of the hip flexion progressing from
right to left.
Computerised Muscle Modelling and Simulation for Interactive Applications
217
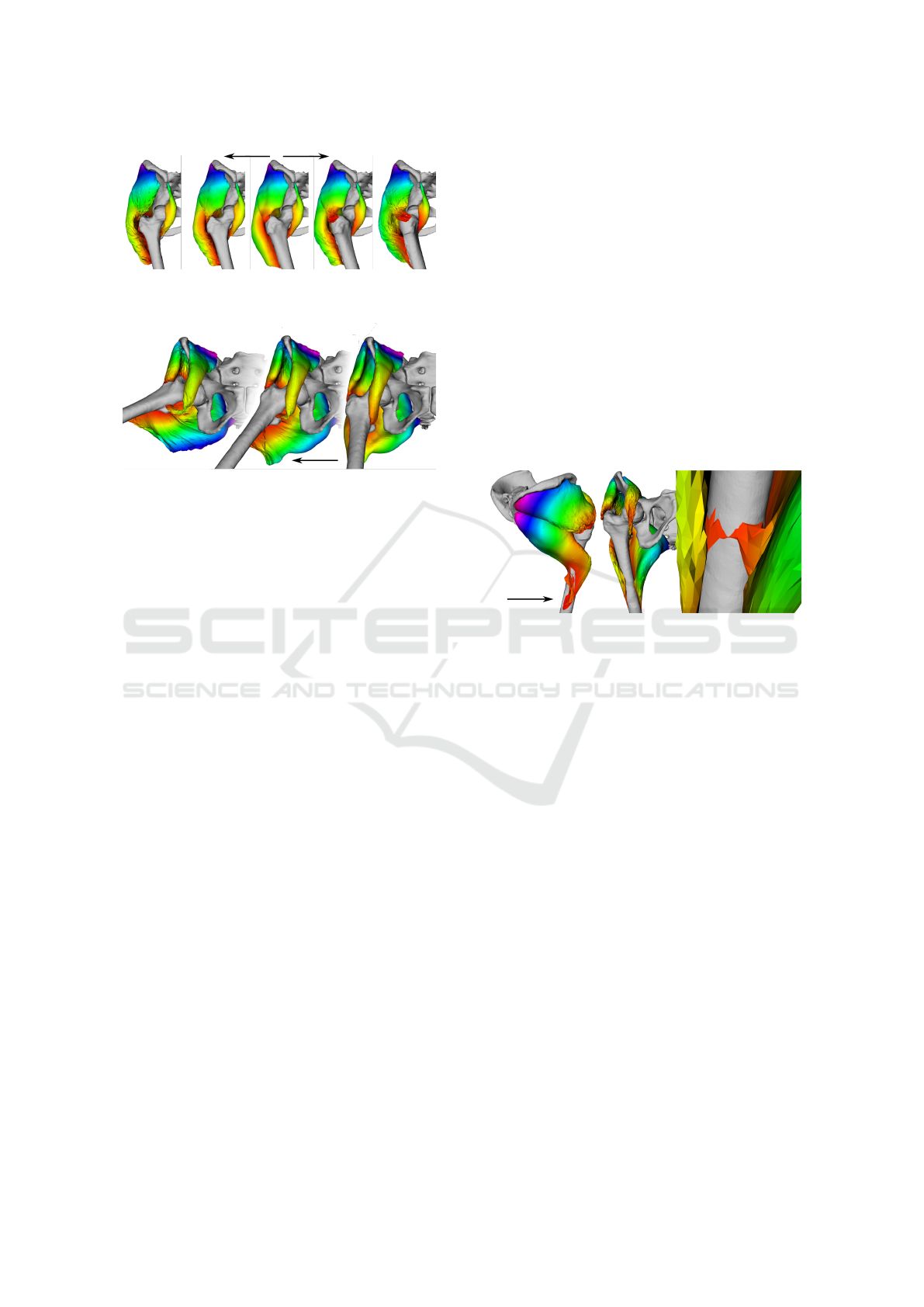
Figure 4: The result of the hip rotation progressing from the
centre to the sides.
Figure 5: The result of the hip adduction progressing right
to the left during the simulation.
5.3 Tunnelling Detection
The tunnelling problem (muscle ”jumps” in a single
iteration through the whole bone to the opposite side)
appears at places where the displacement of the bone
between two consecutive simulation frames exceeds
half the size of a muscle with which the bone collides.
According to (Havlicek et al., 2022), ”when [the fe-
mur] rotates about just 2
◦
(a typical step in simula-
tions), the displacement of the distal part of this bone
is nearly 3 cm”. In our experiments with the muscles
of the hip, i.e., no part of the muscle is near the dis-
tal portion of the bone, this problem arises when the
angle step is higher than 4
◦
.
This problem is relatively standard and does not
arise only with this particular approach. For example,
(Jan
´
ak, 2012) notes that ”if the movement of the ob-
ject is too fast in relation to the discrete time step, the
collision may not be detected”.
The scenario of the muscle movement is so rapid
that the whole muscle volume could go through the
entire bone model, which is possible (due to the al-
ready described displacement issue concerning the
change of the angle of the bone). Our solution would
be moving a muscle to the bone (rotate around the ex-
act centre of rotation and about the same angle), so
the muscle is closer to where it should lie. However,
before applying this correction, such an event must be
detected.
Generally, a continuous collision detection
method could be employed instead of the currently
used discrete one. However, such methods are
expensive. We, therefore, propose a simple (and fast)
test based on a comparison of the directional vectors
to the nearest bone surface, provided automatically
by the Discregrid.
If the direction of one muscle vertex suddenly
changes “too much”, we may expect that the penetra-
tion through the whole bone has happened. The “too
much” is defined as when the angle between the direc-
tional vector from the previous step and the new one
is greater than 135
◦
, as described in Equ. 1, where
d
i
is the direction to the bone in this computational
step and d
′
i
is the direction to the bone in the previous
computational step,
|
a
|
2
is the euclidean norm of the
vector a. A tunnelling case can be seen in Fig. 6.
arccos
d
i
· d
′
i
2
> 135
◦
(1)
d
i
· d
′
i
2
> −0.75
Figure 6: Due to the lack of contraction modelling, the mus-
cles are being dragged during the hip extension (left and
centre images). Finally, under the degree of 72
◦
, first tun-
nelling occurs (right) and is detected successfully by the
proposed algorithm.
5.4 Problem of Multiple Muscles
When multiple muscles are simulated in the scene,
they may also intersect each other. As noted in sec-
tion 4.1, building the Discregrid structure for the mus-
cle meshes is unusable for the interactive application
due to time requirements. An appropriate collision
handling system could be, e.g. a BVH structure (see
section 4), which would have to be updated each time
any muscle moves. As the nearby muscles often touch
each other - see Figure 1, this solution would proba-
bly be inefficient. Therefore, we propose the ”sliding”
technique using the so-called ”virtual edges” to keep
the muscles at a certain distance from each other to
prevent collisions and unrealistic detachments.
5.4.1 Sliding over Surfaces
The main idea is to allow the muscles to slide over
each other using the virtual edges between the mus-
cles. We may keep chosen vertices up to a certain
distance away from the other muscles, i.e. a thresh-
old. The vertices to keep close to the other muscles
GRAPP 2023 - 18th International Conference on Computer Graphics Theory and Applications
218

can be selected by their initial distance and kept in a
list. This list could also be updated if the PBD dis-
placement of a particular vertex away from the other
muscle would be greater than its initial distance, let-
ting the point go. Similarly, collecting more vertices
into this list could be achieved via checking, for ex-
ample, the neighbourhood vertices, which would be
the likely candidates.
In case the PBD forces are less in magnitude than
this attraction force but still point away from the mus-
cle, the vertex would be pushed in the closest direc-
tion to the other muscle surface, effectively making
the point slide along the other muscle surface and
hopefully preventing it from colliding or unrealisti-
cally spanning out, as illustrated in Fig. 2.
This functionality is implemented preliminarily,
using the Discregrid library to preserve the distances
between a bone and a muscle. The resulting vir-
tual edges can be seen in Figure 7. The reason for
this measure is that the original idea implementation
would be beyond the scope of this paper.
Figure 7: An example of virtual edges between a muscle
and a bone with the participating vertices in blue and red,
respectively. The points in blue are muscle vertices, which
are close enough to the bone at the start of the simulation.
For each of them, the closest bone vertex (red) is found. The
pairs of vertices are connected with a straight line segment,
symbolizing a virtual edge.
6 FUTURE WORK
The PBD approach on its own brings some problems
to muscle modelling. The shape is not well preserved
(see Fig. 5, on the middle image, the bottom central
part of the gluteus maximus muscle is unrealistically
deformed), and bone penetration happens. Any prob-
lems stem from low solver iteration count, essential
for real-time model interaction. The options to solve
these are to:
• increases the number of PBD iterations, ef-
fectively slowing down the simulation, which
would become no longer interactive (Kohout and
ˇ
Cervenka, 2021);
• uses the eXtended PBD, which converges more
consistently (the iteration count is not as signifi-
cant) but does not solve the penetration issues;
• uses a different muscle model.
6.1 RBF Representation
The radial basis function model opens a new possi-
bility to develop a new approach to the deformation
of this model, which would allow smooth and rapid
muscle simulation. Collision detection and response,
volume preservation, and muscle anisotropy are the
challenges for future work.
The critical decision for the suitable model is to
select the suitable radial basis function and shape pa-
rameters (if any). A comprehensive study of some
well-known RBFs has already been made (see, e.g.
(Majdisova and Skala, 2017)), and the shape param-
eters were explored (e.g. in (Skala and Cervenka,
2019) or (Afiatdoust and Esmaeilbeigi, 2015)).
6.2 ARAP & PBD
Because of the deformed and unrealistic shape of
the model during the simulation, the As-Rigid-As-
Possible (ARAP) approach from computer graphics is
proposed for merge with PBD to deform an object re-
specting its original shape (Sorkine and Alexa, 2007)
to obtain ”the best of both worlds”.
As the preliminary experiment, we tried to use a
single PBD iteration to preserve the muscle’s original
volume, followed by a single iteration from ARAP,
which should restore the initial shape of the muscle.
The problem is that these two restrictions force most
vertices to go in the opposite direction, resulting in a
rough surface. (see results in Fig. 8). The volume
preservation constraint is not solvable by introducing
a new condition into the system since the interleaving
approach does not work.
Dvorak et al. (Dvo
ˇ
r
´
ak et al., 2022) show how
to apply the ARAP approach to volume preservation.
However, their approach is not used directly for mus-
cle modelling problems. Our goal is to avoid the
introduction of an internal muscle structure to reach
lower computational complexity, meaning that their
approach would have to be altered drastically.
We see two options for fixing the mentioned is-
sues. The first is to start with PBD and replace the
shape preservation constraint with the shape preser-
vation constraint from the ARAP approach. A math-
Computerised Muscle Modelling and Simulation for Interactive Applications
219

Figure 8: ARAP approach with a simple volume preserva-
tion constraint. The initial model is in white, the user de-
formation is in yellow. The volume preserving model with
user defined constraint is in lime blue, the green model is
the simple interweaving approach of the ARAP and PBD.
ematical reformulation of the shape constraint and
finding a gradient expression would be required for
the ARAP shape preservation constraint. The other
option is to start with ARAP and replace the interleav-
ing approach with gradient descent from PBD. Then,
a volume constraint can be added. Either way, both
methods should end up with the same result.
7 CONCLUSION
All of the described techniques for muscle mod-
elling provide good outcomes; however, each has
some drawbacks. Some are inaccurate (Hill-type
model (Hill, 1938), incorrect according to (Burzy
´
nski
et al., 2021), Via-points too approximate according
to (Modenese and Kohout, 2020)), some are accu-
rate but difficult to set up or simply too slow to be
useful (Finite element methods used by, e.g. (Delp
and Blemker, 2005), proved difficult to set up by
(Romeo et al., 2018)). Other methods are ”com-
promise solutions” in terms of accuracy and com-
putational complexity (Mass-Spring system, (Georgii
and Westermann, 2005; Aubel and Thalmann, 2001;
Jan
´
ak, 2012), PBD, As-Rigid-As-Possible (Sorkine
and Alexa, 2007; Fasser et al., 2021; Wang et al.,
2021)). As the reader can probably imagine, many
open problems still exist.
ACKNOWLEDGEMENTS
The authors thank their colleagues and students at the
University of West Bohemia for their discussions and
suggestions. This work was supported by the Ministry
of Education, Youth and Sports of the Czech Repub-
lic, project SGS-2022-015 New Methods for Medical,
Spatial, and Communication Data.
REFERENCES
Afiatdoust, F. and Esmaeilbeigi, M. (2015). Optimal vari-
able shape parameters using genetic algorithm for ra-
dial basis function approximation. Ain Shams Engi-
neering Journal, 6(2):639–647.
Angles, B., Rebain, D., Macklin, M., Wyvill, B., Barthe,
L., Lewis, J., Von Der Pahlen, J., Izadi, S., Valentin,
J., Bouaziz, S., and Tagliasacchi, A. (2019). Viper:
Volume invariant position-based elastic rods. Proc.
ACM Comput. Graph. Interact. Tech., 2(2).
Aubel, A. and Thalmann, D. (2001). Interactive modeling
of the human musculature. In Proceedings Computer
Animation 2001. Fourteenth Conference on Computer
Animation (Cat. No.01TH8596), pages 167 – 255.
Barzan, M., Carty, C., Maine, S., Brito da Luz, S., Lloyd,
D., and Modenese, L. (2017). Subject-specific knee
kinematics during walking in children and adolescents
with recurrent patellar dislocation. In 23rd Australian
& New Zealand Orthopaedic Research Society.
Burzy
´
nski, S., Sabik, A., Witkowski, W., and Łuczkiewicz,
P. (2021). Influence of the femoral offset on the mus-
cles passive resistance in total hip arthroplasty. PLOS
ONE, 16(5):1–12.
Bærentzen, A. and Aanæs, H. (2002). Generating Signed
Distance Fields From Triangle Meshes. Informatics
and Mathematical Modelling.
Cervenka., M. and Kohout., J. (2020). Fast and realistic
approach to virtual muscle deformation. In Proceed-
ings of the 13th International Joint Conference on
Biomedical Engineering Systems and Technologies -
HEALTHINF,, pages 217–227. INSTICC, SciTePress.
Cervenka, M. and Skala, V. (2020). Behavioral study of
various radial basis functions for approximation and
interpolation purposes. In 2020 IEEE 18th World Sym-
posium on Applied Machine Intelligence and Infor-
matics (SAMI), pages 135–140.
Delp, S. and Blemker, S. (2005). Three-dimensional repre-
sentation of complex muscle architectures and geome-
tries. Annals of biomedical engineering, 33:661–73.
Dvo
ˇ
r
´
ak, J., K
´
a
ˇ
cerekov
´
a, Z., Van
ˇ
e
ˇ
cek, P., Hruda, L., and
V
´
a
ˇ
sa, L. (2022). As-rigid-as-possible volume track-
ing for time-varying surfaces. Computers & Graphics,
102:329–338.
GRAPP 2023 - 18th International Conference on Computer Graphics Theory and Applications
220

Fasser, M. ., Jokeit, M., Kalthoff, M., Gomez Romero,
D. A., Trache, T., Snedeker, J. G., Farshad, M.,
and Widmer, J. (2021). Subject-specific alignment
and mass distribution in musculoskeletal models of
the lumbar spine. Frontiers in Bioengineering and
Biotechnology, 9. Cited By :2.
Fukuda, N., Otake, Y., Takao, M., Yokota, F., Ogawa, T.,
Uemura, K., Nakaya, R., Tamura, K., Grupp, R., Far-
vardin, A., Sugano, N., and Sato, Y. (2017). Estima-
tion of attachment regions of hip muscles in ct im-
age using muscle attachment probabilistic atlas con-
structed from measurements in eight cadavers. Inter-
national Journal of Computer Assisted Radiology and
Surgery, 12.
Georgii, J. and Westermann, R. (2005). Mass-spring sys-
tems on the gpu. Simulation Modelling Practice and
Theory, 13:693–702.
Green, S. (2010). Particle simulation using cuda. In Particle
Simulation using CUDA.
Havlicek, O., Cervenka, M., and Kohout, J. (2022). Colli-
sion detection and response approaches for computer
muscle modelling. accepted for the IEEE 16th Inter-
national Scientific Conference on Informatics.
Hill, A. (1938). The heat of shortening and the dynamic
constants of muscle. Proc. R. Soc. Lond. B, 126:612–
745.
Jan
´
ak, T. (2012). Fast soft-body models for musculoskele-
tal modelling. Technical report, University of West
Bohemia, Faculty of Applied Sciences.
Kohout, J. and
ˇ
Cervenka, M. (2021). Muscle deformation
using position based dynamics. In Ye, X., Soares,
F., De Maria, E., G
´
omez Vilda, P., Cabitza, F., Fred,
A., and Gamboa, H., editors, Biomedical Engineer-
ing Systems and Technologies, pages 486–509, Cham.
Springer International Publishing.
Koschier, D. and Bender, J. (2017). Density maps for im-
proved sph boundary handling. In Proceedings of
the ACM SIGGRAPH / Eurographics Symposium on
Computer Animation, SCA ’17, New York, NY, USA.
Association for Computing Machinery.
Lee, D., Li, Z., Sohail, Q. Z., Jackson, K., Fiume, E., and
Agur, A. (2014). A three-dimensional approach to
pennation angle estimation for human skeletal mus-
cle. Computer methods in biomechanics and biomed-
ical engineering, 18:1–11.
Li, H., Sumner, R., and Pauly, M. (2008). Global correspon-
dence optimization for non-rigid registration of depth
scans. Computer Graphics Forum, 27.
Macklin, M., M
¨
uller, M., and Chentanez, N. (2016). Xpbd:
Position-based simulation of compliant constrained
dynamics. In Proceedings of the 9th International
Conference on Motion in Games, MIG ’16, page
49–54, New York, NY, USA. Association for Com-
puting Machinery.
Majdisova, Z. and Skala, V. (2017). Radial basis function
approximations: comparison and applications. Ap-
plied Mathematical Modelling, 51:728–743.
Modenese, L. and Kohout, J. (2020). Automated generation
of three-dimensional complex muscle geometries for
use in personalised musculoskeletal models. Annals
of Biomedical Engineering, 48.
M
¨
uller, M., Heidelberger, B., Hennix, M., and Ratcliff,
J. (2007). Position based dynamics. Journal of
Visual Communication and Image Representation,
18(2):109–118.
Oatis, C. A. (2017). Biomechanics of skeletal muscle. Lip-
pincott Williams & Wilkins.
Romeo, M., Monteagudo, C., and S
´
anchez-Quir
´
os, D.
(2018). Muscle Simulation with Extended Position
Based Dynamics. In Garc
´
ıa-Fern
´
andez, I. and Ure
˜
na,
C., editors, Spanish Computer Graphics Conference
(CEIG). The Eurographics Association.
Skala, V. and Cervenka, M. (2019). Novel rbf approxima-
tion method based on geometrical properties for sig-
nal processing with a new rbf function: Experimental
comparison. In 2019 IEEE 15th International Scien-
tific Conference on Informatics.
Sorkine, O. and Alexa, M. (2007). As-Rigid-As-Possible
Surface Modeling. In Belyaev, A. and Garland, M.,
editors, Geometry Processing. The Eurographics As-
sociation.
Teschner, M., Kimmerle, S., Heidelberger, B., Zachmann,
G., Raghupathi, L., Fuhrman, A., Cani, M.-P., Faure,
F., Magnenat-Thalmann, N., Strasser, W., and Volino,
P. (2005). Collision Detection for Deformable Ob-
jects. Computer Graphics Forum.
Turk, G. (1990). Interactive collision detection for molec-
ular graphics. Technical report, University of North
Carolina at Chapel Hill, USA.
Wade, S., Strader, C., Fitzpatrick, L., Anthony, M., and
O’Malley, C. (2014). Estimating prevalence of os-
teoporosis: Examples from industrialized countries.
Archives of osteoporosis, 9:182.
Wang, B., Matcuk, G., and Barbi
ˇ
c, J. (2021). Modeling
of personalized anatomy using plastic strains. ACM
Trans. Graph., 40(2).
Zhang, G., Wang, C., Liu, Q., Wei, J., Luo, C., Duan, L.,
Long, J., Zhang, X., and Wang, G. (2021). Develop-
ment of skeletal muscle model for bridge-style move-
ment rehabilitation. Journal of Physics: Conference
Series, 2026:012061.
Zhang, S.-X., Heng, P.-A., Liu, Z.-J., Tan, L.-W., Qiu, M.-
G., Li, Q.-Y., Liao, R.-X., Li, K., Cui, G.-Y., Guo,
Y.-L., Yang, X.-P., Liu, G.-J., Shan, J.-L., Liu, J.-J.,
Zhang, W.-G., Chen, X.-H., Chen, J.-H., Wang, J.,
Chen, W., Lu, M., You, J., Pang, X.-L., Xiao, H., Xie,
Y.-M., and Cheng, J. C.-Y. (2004). The chinese vis-
ible human (cvh) datasets incorporate technical and
imaging advances on earlier digital humans. Journal
of Anatomy, 204(3):165–173.
Zhao, Y., Clapworthy, G., Kohout, J., Dong, F., Tao, Y.,
Wei, S., and Mcfarlane, N. (2013). Laplacian muscu-
loskeletal deformation for patient-specific simulation
and visualisation. In 2013 17th International Confer-
ence on Information Visualisation, pages 505–510.
Computerised Muscle Modelling and Simulation for Interactive Applications
221
