
Skin Lesion Segmentation Using Attention-Based DenseUNet
Anwar Jimi
1
, Hind Abouche
1
, Nabila Zrira
2
and Ibtissam Benmiloud
1
1
MECAtronique Team, CPS2E Laboratory National Superior School of Mines Rabat, Morocco
2
ADOS Team, LISTD Laboratory National Superior School of Mines Rabat, Morocco
Keywords:
Skin Lesion Segmentation, DenseNet, Deep Learning, DenseUNet, Attention.
Abstract:
Skin lesion segmentation in dermoscopic images is still a challenging problem due to the blurry borders and
low contrast of the lesions. Deep learning networks, like U-Net, have been successfully used to segment med-
ical images over the past few years, and their performance has improved in terms of time and accuracy. This
paper proposes an automated method for segmenting lesion boundaries that combines two architectures (i.e.,
the U-Net and the DenseNet as backbone) as well as the attention mechanism. Moreover, we also used adap-
tive gamma correction to enhance the contrast of the image, which considerably enhanced the segmentation
results. Furthermore, we trained our model on the ISIC 2016, the ISIC 2017, and the ISIC 2018 datasets. Fi-
nally, the qualitative and quantitative experimental results of the skin lesion segmentation are very promising.
1 INTRODUCTION
Skin cancer is the most prevalent type of cancer
worldwide. As ozone levels decrease, the atmosphere
increasingly loses its protective filtering function, and
the surface of the Earth receives more solar ultraviolet
(UV) radiation. According to the World Health Orga-
nization (WHO), every 10% reduction in the ozone
layer would lead to 4,500 melanoma cases and more
than 300,000 non-melanoma instances of skin can-
cer (Organization et al., 2017). The prevalence of
melanoma is increasing globally, but UV radiation is
the principal cause of melanoma growth. Melanoma
causes more than 20,000 deaths in Europe each year.
Currently, 132,000 cases of melanoma and 2 to 3 mil-
lion cases of non-melanoma skin cancer are reported
annually worldwide. According to the Skin Cancer
Foundation (SCF), skin cancer accounts for one in
three cancer diagnoses and one in five lifetime cases
of cancer in the United States (US).
Basal cell carcinoma and squamous cell carci-
noma are two types of non-melanoma skin malignan-
cies. Although they are rarely fatal, surgical treat-
ments are often disfiguring and traumatic. It is chal-
lenging to identify historical trends in the occurrence
of non-melanoma skin cancers since trustworthy reg-
istries for these malignancies have not yet been estab-
lished. Nevertheless, particular research in Australia,
Canada, and the US shows that the prevalence of non-
melanoma skin cancers more than tripled between the
1960s and 1980s.
The most common type of skin cancer that re-
sults in mortality is malignant melanoma, which is
also the one that is reported and diagnosed more fre-
quently than non-melanoma skin cancer. The preva-
lence of malignant melanoma has considerably in-
creased since the early 1970s, by an average of 4%
per year in the US. Several studies have shown that the
risk of malignant melanoma is related to genetic and
personal characteristics, as well as a person’s UV ra-
diation behavior. Malignant melanoma is more com-
mon in white people with blue eyes and red or blond
hair. Australia has the highest incidence, where the
annual incidence is more than 10 and 20 times higher
than in European women and men, respectively.
Automatic skin lesion segmentation is a critical
step in Computer-Aided Diagnosis (CAD). However,
because skin lesions vary significantly in shape, size,
and color, this task remains difficult. Furthermore,
the borders of certain lesions are uneven and hazy.
Thus, today, computer vision and image processing
approaches are being used to improve dermoscopy in
order to develop tools that are capable of correctly di-
agnosing lesions, with the goal of improving access
to reliable data to assist doctors. This enhancement
can be implemented in a number of ways, including
the detection of lesions, their borders, and colors, as
well as the segmentation of different types of lesions.
Deep learning, which is based on Convolutional
Neural Networks (CNNs), has recently gained promi-
Jimi, A., Abouche, H., Zrira, N. and Benmiloud, I.
Skin Lesion Segmentation Using Attention-Based DenseUNet.
DOI: 10.5220/0011686400003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 3: BIOINFORMATICS, pages 91-100
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
91
nence in machine learning and computer vision, par-
ticularly in the semantic image segmentation area
(Litjens et al., 2017). In this paper, we propose a new
automatic approach for the segmentation of skin le-
sions using attention-based DenseUNet. In addition,
we used adaptive gamma correction to enhance the
contrast of the image and hence improve the segmen-
tation result.
The following is a summary of this paper. An
overview of skin cancer is presented in Section 2. The
state-of-the-art of skin lesion segmentation is briefly
introduced in Section 3. Section 4 describes the used
datasets and our proposed approach. Section 5 illus-
trates implementation details, segmentation metrics
and experimental results. Section 6 is about discus-
sion and future work. Finally, we conclude this paper
in Section 7.
2 SKIN CANCER
The most dangerous kind of skin cancer is melanoma
(Capdehourat et al., 2011). It spreads easily to
any organ and expands swiftly. Skin cells called
melanocytes are the source of melanoma. These cells
create the dark pigment known as melanin, which
gives skin its color. Though it only accounts for
around 1% of all skin malignancies, melanoma is
the most common death from skin cancer. Early
melanomas are often recoverable, so it’s crucial to be
able to identify them. Melanoma can present as raised
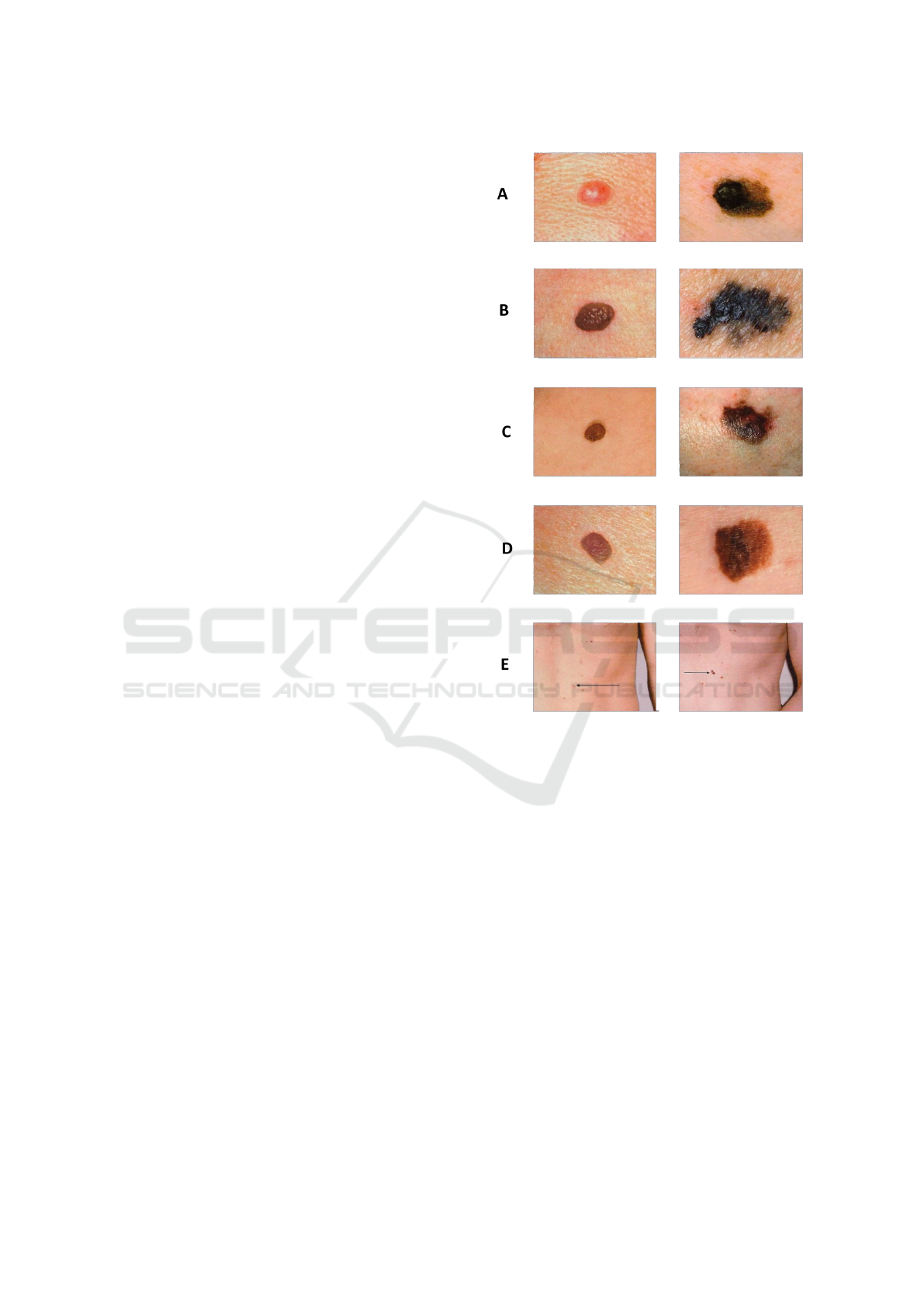
bumps, scaly patches, open sores, or moles. Table
1 illustrates the indicators which are offered by the
”ABCDE” memory aid from the American Academy
of Dermatology (Nachbar et al., 1994) to determine if
a lesion on the skin can be melanoma:
Asymmetry: The two halves are not identical;
Border: There are rough edges;
Color: With varying tones of brown, black; gray, red,
or white, the color is mottled and irregular;
Diameter: The spot is larger than the eraser’s tip (6.0
mm);
Evolving: The spot is either brand-new or is altering
in size, shape, or color.
Moreover, Figure 1 shows the comparison be-
tween malenoma and non melanoma skin lesion based
on the ABCDE rule.
3 RELATED WORK
In this section, we describe and present relevant work
performed on the issue of skin lesion segmentation. It
Figure 1: Right: normal lesion. Left: melanoma lesion.
focuses on recent approaches that have incorporated
deep learning methods for lesion segmentation.
In 2015, Wang et al. presented the U-Net (Ron-
neberger et al., 2015) network for segmenting medical
images. A neural network called U-Net uses symmet-
ric encoders and decoders, a structure that has demon-
strated exceptional productivity in the area of medical
imaging. Additionally, a variety of enhanced models
built on the U-Net framework have been put forth to
further increase the accuracy of computer-aided med-
ical imaging diagnostic activities.
Inspired by the U-Net, Sulaiman et al. (Vesal
et al., 2018) suggested the SkinNet model based on
the CNN. The CNN architecture that has been sug-
gested represents a redesign of the U-Net. SkinNet
uses dilated convolutions specifically in the lowest
layer of an encoder branch in the U-Net, to provide a
more global context to the extracted features from the
image. Furthermore, the authors swap out usual con-
volution layers in both the U-Net encoder and decoder
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
92

Table 1: Comparison between melanoma and normal lesion.
Indicator Melanoma Normal
Asymetry (A) Asymmetrical Symmetrical
Border (B) Uneven Even
Color (C) Multiple colors One color
Diameter (D) Larger than
1
4
inch Smaller than
1
4
inch
Evolving (E) Changing in size, color, shape Ordinary mole
parts, using dense convolution blocks, to more effec-
tively combine multi-scale visual information. The
ISIC 2017 dataset was used to assess the SkinNet
model, which received an IOU score of 76.7% and
a dice coefficient of 85.10%. Galdran et al. (Galdran
et al., 2017) utilized the U-Net architecture as well as
color constancy techniques to maintain the estimated
illumination information while normalizing the color
over the whole dataset. This makes it possible for
normalized images to fluctuate in color and lighting
at random while being trained. On the ISIC 2017
dataset, they attained a dice coefficient of 84.60%.
Berseth et al. (Berseth, 2017) created a U-Net archi-
tecture for segmenting skin lesions based on the prob-
ability map of the image dimension, then trained the
model using ten-fold cross-validation.
Currently, in deep learning algorithms, certain
models are frequently employed as pre-trained en-
coders. Many pre-trained algorithms like ResNet, Ef-
ficientNet, and MobileNet can train the U-Net model
with greater accuracy. Kashan et al. (Zafar et al.,
2020) presented a system for automatically segment-
ing lesion borders, that created a new architecture
known as Res-Unet by combining the U-Net and
ResNet architectures. Additionally, they employed
image inpainting to remove the hair, which dramat-
ically enhanced the segmentation outcomes. The
model was assessed using the ISIC 2017 and PH2
datasets. On the ISIC 2017 test set, the approach
achieved a Jaccard Index of 0.772. Whereas, on the
PH2 dataset it achieved a Jaccard Index of 0.854. Ba-
heti et al. (Baheti et al., 2020) introduced a novel
architecture called Eff-UNet that integrated the effi-
ciency of compound-scaled EfficientNet as the en-
coder for feature extraction with the U-Net decoder
for recreating the fine-grained segmentation map. Wi-
bowo et al. (Wibowo et al., 2021) suggested a
lightweight encoder-decoder built on U-Net and Mo-
bileNetV3 to enhance the network architecture’s per-
formance. Also, they employed some methods like
the filling-in-the-hole post-processing method and
stochastic weight averaging learning schema, to en-
hance the segmentation map during testing. To pre-
vent overfitting, the authors utilized random augmen-
tation by increasing image variety in the training
dataset. Zahangir et al. (Alom et al., 2018) proposed
a Recurrent Residual Convolutional Neural Network
(RRCNN) and a Recurrent Convolutional Neural Net-
work (RCNN) based on the U-Net. Proposed models
make use of the capabilities of RCNN, Residual Net-
works, and U-Net. RCNN and RRCNN both facilitate
quick network training and provide excellent feature
representation for segmentation tasks.
The Google Deep Mind team made the initial sug-
gestion for the attention mechanism in an image clas-
sification challenge, triggering a wave of attention
mechanism research (Mnih et al., 2014). Wang et al.
(Wang et al., 2018) introduced a non-local block to
obtain the reliance of the global information on the
pixel-level relationship. Chaitanya et al. (Kaul et al.,
2019) suggested a novel technique to incorporate at-
tention within CNN using feature maps produced by
a different convolutional auto-encoder. Hu et al. (Hu
et al., 2018) affirmed that by explicitly describing the
interdependencies between channels, SE-Net adap-
tively recalibrates channeled feature responses. Woo
et al. (Woo et al., 2018) developed Convolutional
Block Attention Module (CBAM), a straightforward
yet efficient attention module for feed-forward con-
volutional neural networks, using a feature map in be-
tween.
4 MATERIALS AND METHODS
In this section, we firstly introduce the used dermo-
scopic images of melanocytic lesions. Secondly, we
present all the techniques used in the preprocessing
step . Thirdly, we describe in detail the model archi-
tecture that is used in the context of lesion segmenta-
tion. As shown in Figure 2, the approach is divided
into three major steps.
4.1 Used Datasets
We evaluated the proposed network on three dermo-
scopic image datasets, including the ISIC-2016 chal-
lenge dataset (Gutman et al., 2016), the ISIC-2017
challenge dataset (Codella et al., 2018) and the ISIC-
2018 challenge dataset (Codella et al., 2019; Tschandl
et al., 2018). The International Skin Imaging Collab-
Skin Lesion Segmentation Using Attention-Based DenseUNet
93
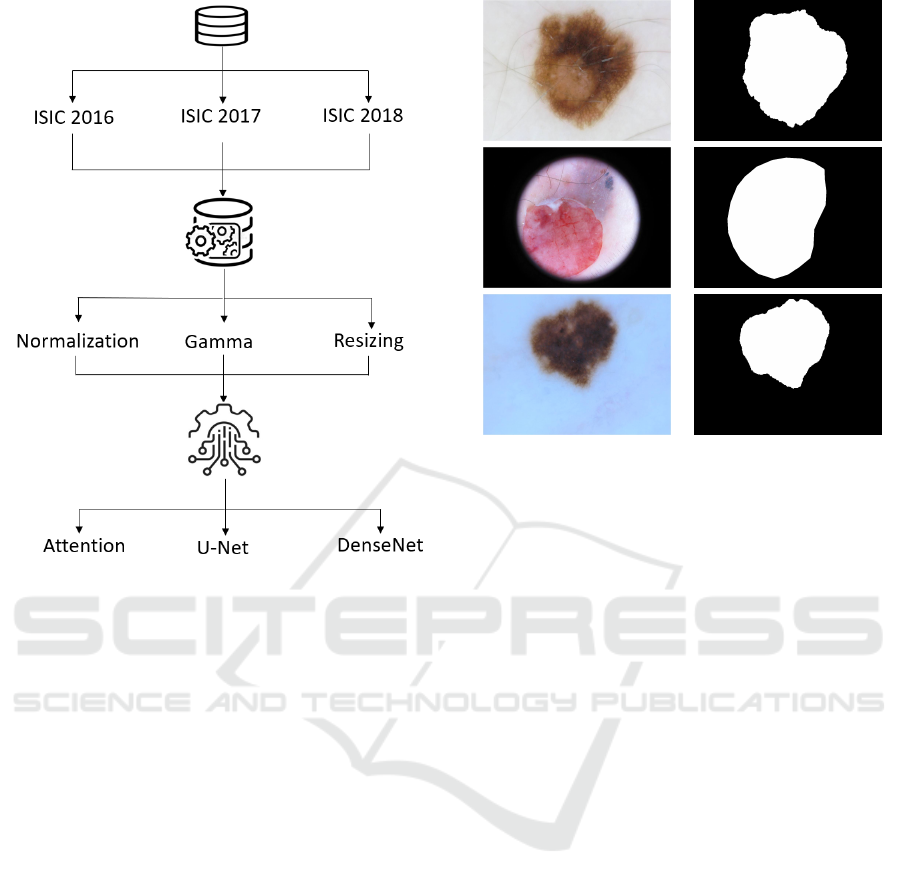
Figure 2: Diagram of the proposed model.
orative (ISIC) offers expertly annotated digital skin
lesion image datasets from all over the world to sup-
port the computer-aided diagnosis of melanoma and
other skin lesions. These images will help to provide
an automated and effective computer diagnosis. An
overview of the ISIC 2016, ISIC 2017, and ISIC 2018
datasets is shown in Table 2.
There are 900 training images and 379 test images
in the ISIC 2016 challenge dataset. The ISIC 2017
skin lesion challenge dataset included 2,000, 150,
and 600 images for training, validation, and testing,
respectively. The dimensions of the images ranged
from 556 × 679 to 4499 × 6748 pixels. The ISIC
2018 skin lesion dataset challenge included 2,594 im-
ages for training. This dataset was divided into train-
ing (1,815), validation (259), and test sets consecu-
tively (not randomly). The image sizes ranged from
556×679 pixels to 4, 499×6, 748 pixels. The sample
images from the datasets are displayed in Figure 3.
4.2 Image Preprocessing
Deep learning architectures can successfully learn
from unprocessed image data. However, on prop-
erly preprocessed images, they usually perform bet-
ter. The preprocessing used in this work is described
as follows.
RGB images Ground truths
Figure 3: Examples of skin lesion images from ISIC
datasets.
4.2.1 Image Resizing
The images and associated ground truths were scaled
to 256 ×256 pixels (height ×width) to adjust for vari-
ances in image size within the datasets.
4.2.2 Image Normalization
Each pixel in the images and ground truth masks has
8 bits in size and can have a value between 0 and 255.
The input image was divided by 255 to normalize
the images, changing each pixel’s normal value range
from 0 to 1. When the ground truth mask is rounded
up or set to the ceiling, it is converted to a binary rep-
resentation (0 for background and 1 for foreground).
4.2.3 Contrast Enhancement
Contrast enhancement plays an important role in im-
proving visual quality in computer vision, pattern
recognition, and image processing.
In this paper, we use adaptive gamma correction
with weighting distribution (Huang et al., 2012) to
improve the image quality for better segmentation.
Three main steps make up the method. The flowchart
of the approach is shown in Figure 4.
First, based on probability and statistical infer-
ence, the histogram analysis provides the spatial in-
formation of a single image. The weighting distribu-
tion is employed in the second stage to smooth the
fluctuant phenomenon and prevent the creation of un-
wanted artifacts. Gamma correction can automati-
cally improve the image contrast in the third and final
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
94
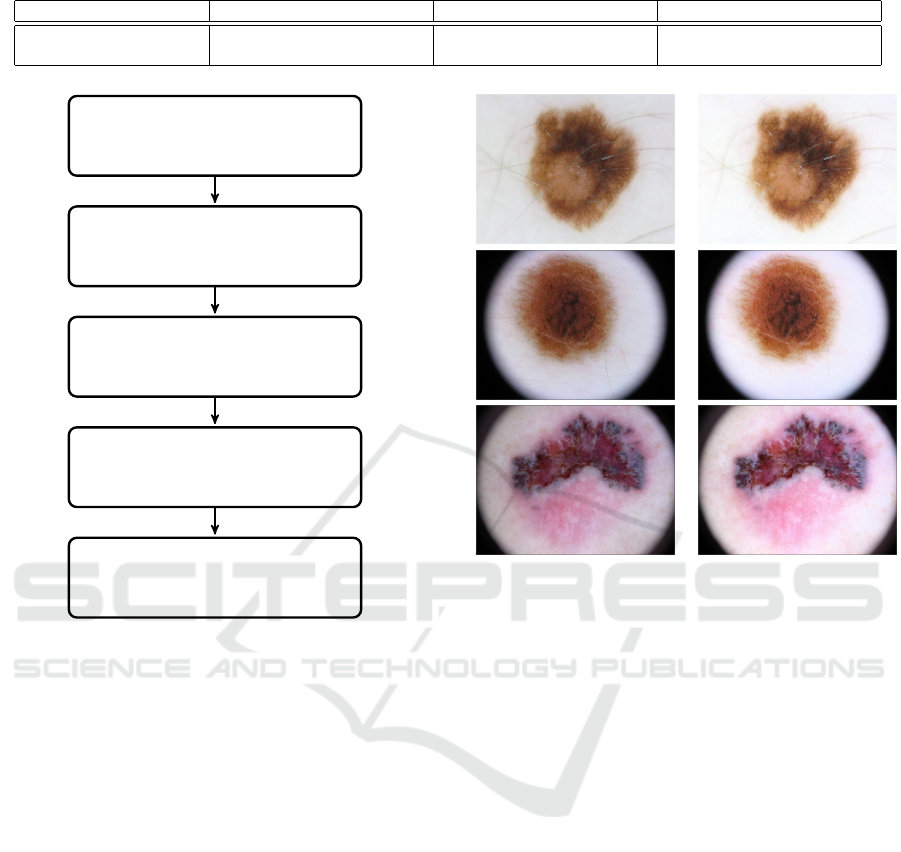
Table 2: Description of the three datasets.
Dataset ISIC 2016 ISIC 2017 ISIC 2018
Total number of images 1,279 2,750 2,594
Image size (pixel) 576 × 768 to 2, 848× 4, 288 556 × 679 to 4, 499 × 6, 748 556 × 679 to 4, 499 × 6, 748
Input image
Histogram analysis
Weighting distribution
Gamma correction
Enhanced image
Figure 4: Flowchart of Adaptive Gamma Correction With
Weighting Distribution.
step by using a smoothing curve. The results of the
image enhancement are shown in Figure 5.
4.3 Model Architecture
Deep learning models are currently being utilized to
solve object detection and visual recognition prob-
lems. For semantic segmentation, CNN models
have demonstrated a significant advantage over semi-
automated techniques. The U-Net architecture based
on the encoder-decoder approach has achieved great
results in the segmentation of medical images.
Common layer combinations make up CNN mod-
els (i.e., convolutional layer, max-pooling, batch nor-
malization, and activation layer). In the area of med-
ical diagnostics, CNN architectures have been widely
applied.
For this purpose, ISIC datasets are used to train
a CNN architecture. The network architecture takes
insight from both U-Net and DenseNet as well as the
mechanism of the attention gate as shown in Figure 6.
The convolutional side (i.e., contracting path) is
based on the DenseNet architecture. The idea of
Before gamma After gamma
Figure 5: Gamma correction.
DenseNet was first suggested by Huang et al. (Huang
et al., 2017) and leads to significant advancements
in state-of-the-art scores compared to earlier models
like ResNet (He et al., 2016) and ResNeXt (Xie et al.,
2017) in image classification tasks like ImageNet.
DenseNet is made up of a dense block and a tran-
sition block as its two main construction blocks. A
dense block consists of several normalized 3 × 3 con-
volution layers, where the outputs of each layer are
concatenated with each of the feature maps entering
the succeeding layers to encourage feature reuse. A
dense block has n layers and n! skip connections.
Each layer produces a feature map with a constant
depth of k, causing n × k channels to leave the dense
block. The transition block is made up of a normal-
ized 1 × 1 convolution to decrease the depth of the
feature maps and a 2 × 2 average pool with stride 2 to
halve the resolution.
In the U-Net architecture, Oktay et al. (Oktay
et al., 2018) first suggested the Attention Gate (AG).
The AG attention module automatically and adap-
tively learns to concentrate on the various sizes and
shapes of the target structures in medical images. The
model under the AG strain implicitly learns to em-
phasize important features useful for a particular task
while removing unnecessary regions from an input
Skin Lesion Segmentation Using Attention-Based DenseUNet
95
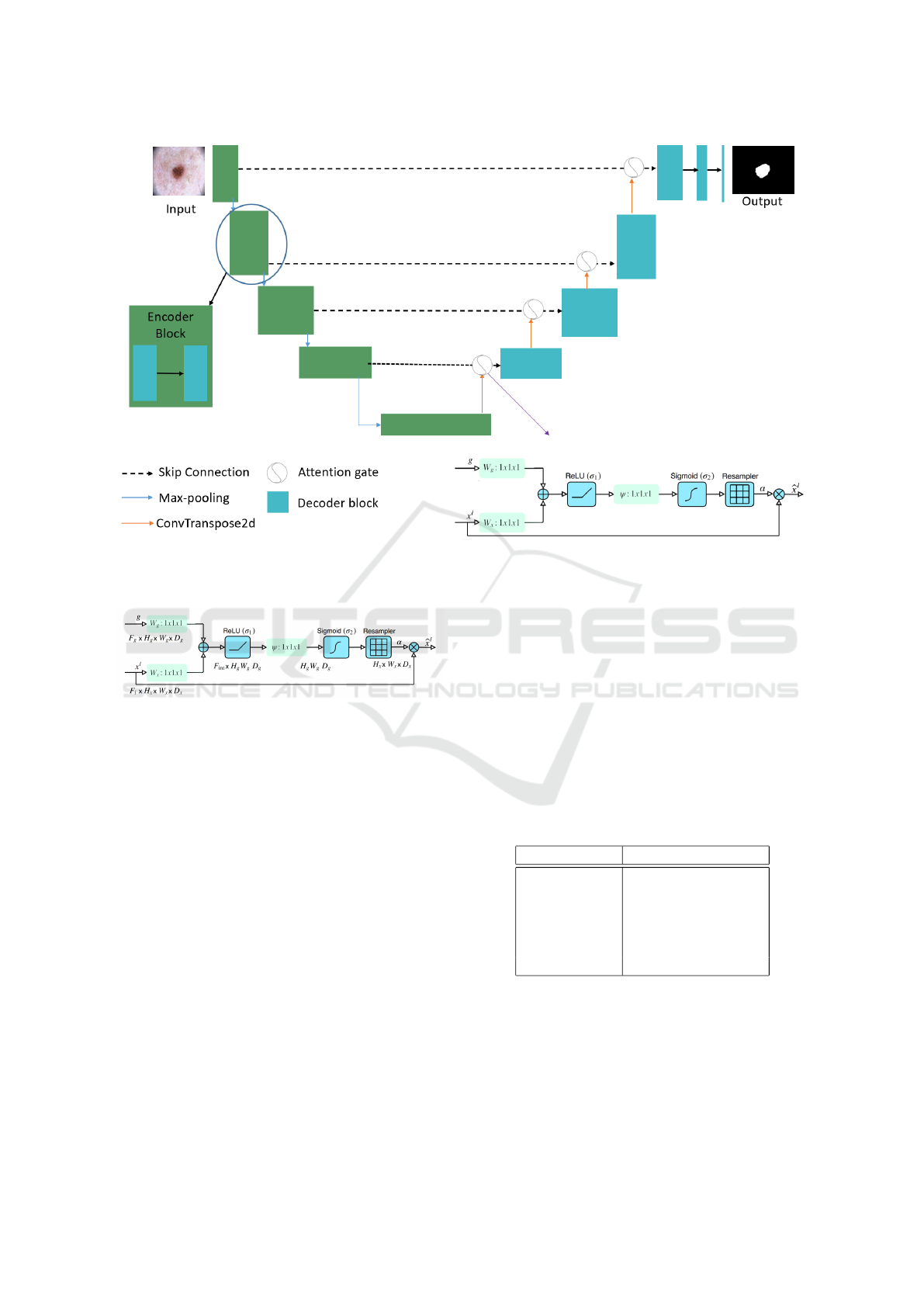
Figure 6: Diagram of the proposed model.
image. Figure 7 shows a schematic of the AG.
Figure 7: Attention gate (Oktay et al., 2018).
As following is how the attention mechanism
functions:
• There are two inputs to the attention gate, vectors
x and g.
• g, gating signal comes from the next lowest layer
of the network.
• x, comes from skip connections.
• The two vectors are added element by element.
This process results in aligned weights getting
larger while unaligned weights getting relatively
smaller.
• The resulting vector passes through a Rectified
Linear Unit (ReLU) activation layer and a 1 × 1
convolution that reduces the dimensions.
• This vector passes through a sigmoid layer that
scales it between [0, 1], generating the attention
coefficients (weights), where coefficients nearer 1
indicate more pertinent features.
• The attention coefficients are upsampled to the
original dimensions of the x vector using trilinear
interpolation. The attention coefficients are mul-
tiplied element-wise to the original x vector, this
scales the vector based on relevance.
4.4 Network Training
Our model was trained over 100 epochs with early
stopping to avoid overfitting. The learning rate is de-
creased if, after 10 epochs, the model’s loss is not re-
duced. After nearly 40 epochs, our model came to an
end. The hyperparameters utilized to train our model
are listed in the Table 3.
Table 3: Hyperparameters maintained during training.
Name Value
Input Size 256 × 56 × 3
Batch Size 32
Learning Rate 1 × 10
−4
Optimizer Adam
Epoch 100
Loss Function Binary Crossentropy
5 EXPERIMENTAL RESULTS
In this section, we first explain the implementation de-
tails of our approach, and then we present the results
of our model compared to the other state-of-the-art al-
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
96

gorithms that utilize the same datasets using segmen-
tation metrics.
5.1 Details of Implementation
We implemented our network using TensorFlow on a
GPU T4 and P100 in Google Colab. All training and
testing phases were performed in the same environ-
ment using Python 3.5 as the programming language
and the TensorFlow 2.5.0 framework for deep learn-
ing.
5.2 Segmentation Metrics
In order to evaluate semantic segmentation techniques
in the literature the following measures have been em-
ployed (Pereira et al., 2016):
• Accuracy (AC) is a review of how well the lesion
image was segmented overall.
AC =
T P + T N
T P + T N + FP + FN
(1)
• Jaccard index (JS) is a union over intersection of
segmented lesions and ground truth masks (Pow-
ers, 2020).
JS =
T P
T P + FN + FP
(2)
• Dice Coefficient (DC) is the similarity between
the predicted results and the annotated ground
truths.
DC =
2 × T P
2 × (T P + FN + FP)
(3)
• Sensitivity (SE) shows the percentage of correctly
identified skin lesion pixels.
SE =
T P
T P + FN
(4)
• Specificity (SP) represents the percentage of pix-
els segmented as non skin lesions.
SP =
T N
T N + FP
(5)
5.3 Comparative Experiments
5.3.1 Comparison on the ISIC 2016 Dataset
We trained and evaluated the suggested model using
the ISIC 2016 dataset. The comparison of the sug-
gested method with the state-of-the-art on the ISIC
2016 dataset is summarized in the Table 4. Different
techniques have been used for segmentation. Yuan
et al. (Yuan and Lo, 2017) achieved an AC value of
0.957 and a DC of 0.921. Also, Bi et al. (Bi et al.,
2017) obtained an AC value of 0.953 and a DC of
0.921. Our method obtained promising results. We
achieved an AC value of 0.9803 and a DC of 0.9433.
Figure 8 provides a visual representation of our
suggested segmentation method of skin lesions. The
experimental renderings can also be used to see how
well the method works.
Figure 8: Segmentation results of our model on ISIC 2016
dataset.
5.3.2 Comparison on the ISIC 2017 Dataset
On the ISIC 2017 dataset, we further trained and
tested the suggested network in this section. A com-
parison of the segmentation performance of the pro-
posed network and other approaches is shown in Ta-
ble 5. The metrics scores from the other models
on this dataset are hardly sufficient because there
are more images in this dataset that are difficult
to segment precisely. Our suggested network still
achieves satisfactory evaluation metrics. Attention-
based DenseUNet showed that the segmentation of
skin lesions was sufficiently successful to produce
good results.
Figure 9 displays the results of the suggested
model on this dataset of partially segmented skin le-
sion images. The outcomes also demonstrated how
well our suggested network performed.
5.3.3 Comparison on the ISIC 2018 Dataset
We further evaluated the architecture using the ISIC
2018 dataset and compared our segmentations with
the current state-of-the-art to determine how robust
our suggested model was. The results are shown in
Skin Lesion Segmentation Using Attention-Based DenseUNet
97

Table 4: Model performance on the ISIC 2016.
Approaches AC JS DC SE SP
U-Net (Ronneberger et al., 2015) 0.936 0.782 0.868 0.930 0.935
FCN (Long et al., 2015) 0.941 0.813 0.886 0.917 0.949
Bi et al. (Bi et al., 2017) 0.953 0.859 0.921 0.962 0.945
Yuan et al. (Yuan and Lo, 2017) 0.957 0.849 0.913 0.924 0.965
Ours 0.9803 0.8564 0.9433 0.9680 0.9855
Table 5: Model performance on the ISIC 2017.
Approaches AC JS DC SE SP
U-Net (Ronneberger et al., 2015) 0.913 0.687 0.781 0.825 0.976
SkinNet (Vesal et al., 2018) 0.932 0.767 0.851 0.930 0.905
MobileNetV3-UNet (Wibowo et al., 2021) 0.938 0.805 0.877 0.8624 0.963
Galdran et al. (Galdran et al., 2017) 0.948 0.767 0.846 0.865 0.980
Ours 0.9619 0.7160 0.8661 0.8490 0.9892
Figure 9: Segmentation results of our model in ISIC 2017
dataset.
Table 6 below. Our approach produced encouraging
outcomes. We achieved an AC value of 0.9788 and
a DC of 0.9228. Whereas, MobileNetV3-UNet (Wi-
bowo et al., 2021) reached an AC of 0.9479 and DC
of 0.9098. Another architecture, Unet++ (Zhou et al.,
2019) obtained an AC of 0.952 and a DC of 0.872.
According to the results, our model performed better
than the current methods employed in the associative
research area.
A visual representation of our suggested method
for segmentation of skin lesions is shown in Fig-
ure 10. Experimental renderings can also be used to
see how effective the algorithm is.
Figure 10: Segmentation results of our model in ISIC 2018
dataset.
6 DISCUSSION AND
PERSPECTIVES
There have been deep learning techniques based on
DenseNet and U-Net used in medical images. The
biggest challenges are the noise of images and the
low contrast. Since U-Net has the ability to pre-
cise pixel-level localization, we suggested a model
named DenseUNet based on DenseNet and U-Net. In
the meantime, the attention mechanism (Arora et al.,
2021) has been used in our module. The attention
mechanism can enhance the precision of feature ex-
traction by preventing missing pixel-level informa-
tion. However, we improved the image contrast by
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
98
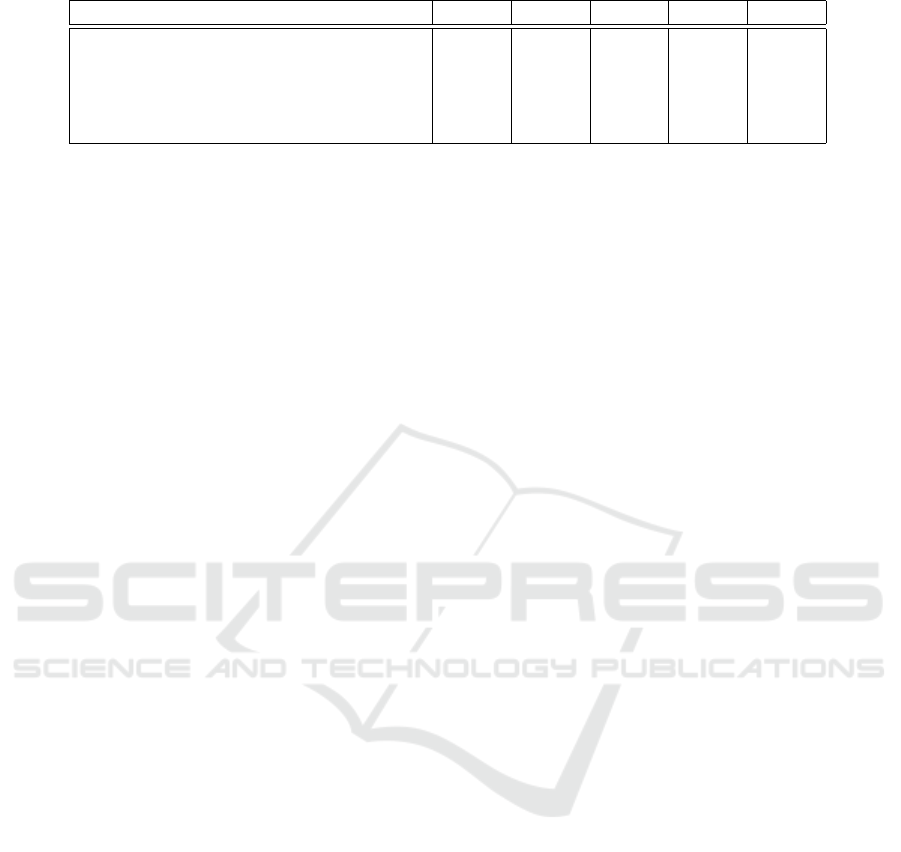
Table 6: Model performance on the ISIC 2018.
Approaches AC JS DC SE SP
U-Net (Ronneberger et al., 2015) 0.890 0.549 0.647 0.708 0.964
R2U-Net (Alom et al., 2019) 0.880 0.581 0.679 0.792 0.928
Unet++ (Zhou et al., 2019) 0.952 0.796 0.872 0.89 0.970
MobileNetV3-UNet (Wibowo et al., 2021) 0.9479 0.8344 0.9098 0.9089 0.9638
Ours 0.9788 0.7990 0.9228 0.9385 0.9897
applying the adaptive gamma correction with weight-
ing distribution.
Experimental results show that our model
achieves state-of-the-art performance on three pub-
licly available datasets due to the robustness of our
model.
For future research, we will use Vision Transform-
ers (ViT) for lesion segmentation. Also, we will cre-
ate a software application to help the dermatologist
segment the skin lesion for further diagnosis.
7 CONCLUSION
One of the hardest and most prevalent issues in im-
age processing is image segmentation. Even human
vision may not be accurate enough for this task, and
in some situations, it may make a wrong or inaccu-
rate diagnosis. Consequently, image segmentation is
a challenging process. However, with the develop-
ment of new approaches in recent years, consider-
able advancements in this field have been realized. In
this paper, we successfully created a skin lesion seg-
mentation method by combining CNN with a power-
ful algorithm that efficiently increases the contrast of
the dermoscopic images. The combination of U-Net,
DenseNet, and attention gate in our proposed method
provides excellent results when compared to state-of-
the-art.
REFERENCES
Alom, M. Z., Hasan, M., Yakopcic, C., Taha, T. M.,
and Asari, V. K. (2018). Recurrent residual con-
volutional neural network based on u-net (r2u-net)
for medical image segmentation. arXiv preprint
arXiv:1802.06955.
Alom, M. Z., Yakopcic, C., Hasan, M., Taha, T. M., and
Asari, V. K. (2019). Recurrent residual u-net for med-
ical image segmentation. Journal of Medical Imaging,
6(1):014006.
Arora, R., Raman, B., Nayyar, K., and Awasthi, R. (2021).
Automated skin lesion segmentation using attention-
based deep convolutional neural network. Biomedical
Signal Processing and Control, 65:102358.
Baheti, B., Innani, S., Gajre, S., and Talbar, S. (2020). Eff-
unet: A novel architecture for semantic segmentation
in unstructured environment. In Proceedings of the
IEEE/CVF Conference on Computer Vision and Pat-
tern Recognition Workshops, pages 358–359.
Berseth, M. (2017). Isic 2017-skin lesion analy-
sis towards melanoma detection. arXiv preprint
arXiv:1703.00523.
Bi, L., Kim, J., Ahn, E., Kumar, A., Fulham, M., and Feng,
D. (2017). Dermoscopic image segmentation via mul-
tistage fully convolutional networks. IEEE Transac-
tions on Biomedical Engineering, 64(9):2065–2074.
Capdehourat, G., Corez, A., Bazzano, A., Alonso, R., and
Mus
´
e, P. (2011). Toward a combined tool to assist der-
matologists in melanoma detection from dermoscopic
images of pigmented skin lesions. Pattern Recogni-
tion Letters, 32(16):2187–2196.
Codella, N., Rotemberg, V., Tschandl, P., Celebi, M. E.,
Dusza, S., Gutman, D., Helba, B., Kalloo, A., Liopy-
ris, K., Marchetti, M., et al. (2019). Skin lesion anal-
ysis toward melanoma detection 2018: A challenge
hosted by the international skin imaging collaboration
(isic). arXiv preprint arXiv:1902.03368.
Codella, N. C., Gutman, D., Celebi, M. E., Helba, B.,
Marchetti, M. A., Dusza, S. W., Kalloo, A., Liopy-
ris, K., Mishra, N., Kittler, H., et al. (2018). Skin
lesion analysis toward melanoma detection: A chal-
lenge at the 2017 international symposium on biomed-
ical imaging (isbi), hosted by the international skin
imaging collaboration (isic). In 2018 IEEE 15th in-
ternational symposium on biomedical imaging (ISBI
2018), pages 168–172. IEEE.
Galdran, A., Alvarez-Gila, A., Meyer, M. I., Saratxaga,
C. L., Ara
´
ujo, T., Garrote, E., Aresta, G., Costa, P.,
Mendonc¸a, A. M., and Campilho, A. (2017). Data-
driven color augmentation techniques for deep skin
image analysis. arXiv preprint arXiv:1703.03702.
Gutman, D., Codella, N. C., Celebi, E., Helba, B.,
Marchetti, M., Mishra, N., and Halpern, A. (2016).
Skin lesion analysis toward melanoma detection: A
challenge at the international symposium on biomed-
ical imaging (isbi) 2016, hosted by the international
skin imaging collaboration (isic). arXiv preprint
arXiv:1605.01397.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resid-
ual learning for image recognition. In Proceedings of
the IEEE conference on computer vision and pattern
recognition, pages 770–778.
Hu, J., Shen, L., and Sun, G. (2018). Squeeze-and-
excitation networks. In Proceedings of the IEEE con-
Skin Lesion Segmentation Using Attention-Based DenseUNet
99

ference on computer vision and pattern recognition,
pages 7132–7141.
Huang, G., Liu, Z., Van Der Maaten, L., and Weinberger,
K. Q. (2017). Densely connected convolutional net-
works. In Proceedings of the IEEE conference on
computer vision and pattern recognition, pages 4700–
4708.
Huang, S.-C., Cheng, F.-C., and Chiu, Y.-S. (2012). Ef-
ficient contrast enhancement using adaptive gamma
correction with weighting distribution. IEEE trans-
actions on image processing, 22(3):1032–1041.
Kaul, C., Manandhar, S., and Pears, N. (2019). Focus-
net: An attention-based fully convolutional network
for medical image segmentation. In 2019 IEEE 16th
international symposium on biomedical imaging (ISBI
2019), pages 455–458. IEEE.
Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A.,
Ciompi, F., Ghafoorian, M., Van Der Laak, J. A.,
Van Ginneken, B., and S
´
anchez, C. I. (2017). A survey
on deep learning in medical image analysis. Medical
image analysis, 42:60–88.
Long, J., Shelhamer, E., and Darrell, T. (2015). Fully con-
volutional networks for semantic segmentation. In
Proceedings of the IEEE conference on computer vi-
sion and pattern recognition, pages 3431–3440.
Mnih, V., Heess, N., Graves, A., et al. (2014). Recurrent
models of visual attention. Advances in neural infor-
mation processing systems, 27.
Nachbar, F., Stolz, W., Merkle, T., Cognetta, A. B., Vogt,
T., Landthaler, M., Bilek, P., Braun-Falco, O., and
Plewig, G. (1994). The abcd rule of dermatoscopy:
high prospective value in the diagnosis of doubtful
melanocytic skin lesions. Journal of the American
Academy of Dermatology, 30(4):551–559.
Oktay, O., Schlemper, J., Folgoc, L. L., Lee, M., Heinrich,
M., Misawa, K., Mori, K., McDonagh, S., Hammerla,
N. Y., Kainz, B., et al. (2018). Attention u-net: Learn-
ing where to look for the pancreas. arXiv preprint
arXiv:1804.03999.
Organization, W. H. et al. (2017). Radiation: Ultraviolet
(uv) radiation and skin cancer. Published October, 16.
Pereira, S., Pinto, A., Alves, V., and Silva, C. A. (2016).
Brain tumor segmentation using convolutional neural
networks in mri images. IEEE transactions on medi-
cal imaging, 35(5):1240–1251.
Powers, D. M. (2020). Evaluation: from precision, recall
and f-measure to roc, informedness, markedness and
correlation. arXiv preprint arXiv:2010.16061.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-net:
Convolutional networks for biomedical image seg-
mentation. In International Conference on Medical
image computing and computer-assisted intervention,
pages 234–241. Springer.
Tschandl, P., Rosendahl, C., and Kittler, H. (2018). The
ham10000 dataset, a large collection of multi-source
dermatoscopic images of common pigmented skin le-
sions. Scientific data, 5(1):1–9.
Vesal, S., Ravikumar, N., and Maier, A. (2018). Skin-
net: A deep learning framework for skin lesion seg-
mentation. In 2018 IEEE Nuclear Science Sympo-
sium and Medical Imaging Conference Proceedings
(NSS/MIC), pages 1–3. IEEE.
Wang, X., Girshick, R., Gupta, A., and He, K. (2018). Non-
local neural networks. In Proceedings of the IEEE
conference on computer vision and pattern recogni-
tion, pages 7794–7803.
Wibowo, A., Purnama, S. R., Wirawan, P. W., and Rasyidi,
H. (2021). Lightweight encoder-decoder model for
automatic skin lesion segmentation. Informatics in
Medicine Unlocked, 25:100640.
Woo, S., Park, J., Lee, J.-Y., and Kweon, I. S. (2018). Cbam:
Convolutional block attention module. In Proceed-
ings of the European conference on computer vision
(ECCV), pages 3–19.
Xie, S., Girshick, R., Doll
´
ar, P., Tu, Z., and He, K. (2017).
Aggregated residual transformations for deep neural
networks. In Proceedings of the IEEE conference on
computer vision and pattern recognition, pages 1492–
1500.
Yuan, Y. and Lo, Y.-C. (2017). Improving dermoscopic
image segmentation with enhanced convolutional-
deconvolutional networks. IEEE journal of biomed-
ical and health informatics, 23(2):519–526.
Zafar, K., Gilani, S. O., Waris, A., Ahmed, A., Jamil, M.,
Khan, M. N., and Sohail Kashif, A. (2020). Skin
lesion segmentation from dermoscopic images using
convolutional neural network. Sensors, 20(6):1601.
Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N., and Liang, J.
(2019). Unet++: Redesigning skip connections to ex-
ploit multiscale features in image segmentation. IEEE
transactions on medical imaging, 39(6):1856–1867.
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
100
