
Ontology-Powered Boosting for Improved Recognition of Ontology
Concepts from Biological Literature
Pratik Devkota
1 a
, Somya D. Mohanty
1 b
and Prashanti Manda
2 c
1
Department of Computer Science, University of North Carolina, Greensboro, NC, U.S.A.
2
Informatics and Analytics, University of North Carolina, Greensboro, NC, U.S.A.
Keywords:
Named Entity Recognition, Automated Ontology Curation, Deep Learning, Biological Ontology.
Abstract:
Automated ontology curation involves developing machine learning models that can learn patterns from sci-
entific literature to predict ontology concepts for pieces of text. Deep learning has been used in this area with
promising results. However, these models often ignore the semantically rich information that’s embedded in
the ontologies and treat ontology concepts as independent entities. Here, we present a novel approach called
Ontology Boosting for improving prediction accuracy of automated curation techniques powered by deep
learning. We evaluate the performance of our models using Jaccard semantic similarity – a metric designed
to assess similarity between ontology concepts. Semantic similarity metrics have the capability to estimate
partial similarity between ontology concepts thereby making them ideal for evaluating the performance of
annotation systems such as deep learning where the goal is to get as close as possible to human performance.
We use the CRAFT gold standard corpus for training our architectures and show that the Ontology Boosting
approach results in substantial improvements in the performance of these architectures.
1 INTRODUCTION
Biological ontologies are critical for consistent
knowledge representation that can be accessed by
humans and computers alike. Since the introduc-
tion of the Gene Ontology, over 900 bio-ontologies
have been created for representing knowledge in sub-
domains such as anatomy, human disease, chemicals
and drugs, etc. Scientists and curators now use these
ontology concepts to describe various aspects of bi-
ological objects thereby creating knowledge bases of
ontology annotations. These annotations are critical
for transferring knowledge in free text such as publi-
cations into a computationally amenable format that
can power large-scale comparative analyses. Ontol-
ogy annotation is still largely accomplished via hu-
man curation - a process where scientists manually
read text and select ontology concepts that accurately
represent the information in the text. While there has
been a rapid growth in the number of ontologies as
well as the number of ontology-powered annotations,
ontology curation has not experienced the same level
a
https://orcid.org/0000-0001-5161-0798
b
https://orcid.org/0000-0002-4253-5201
c
https://orcid.org/0000-0002-7162-7770
of advances.
Automated curation methods that can scale to the
pace of publishing and show efficiency and accuracy
are direly needed. The goal of these methods would
be to process scientific literature and mark words or
phrases in text with one or more ontology concepts
thereby conducting automated curation. Automated
curation tools can be used as stand-alone approaches
that perform annotation without supervision or pre-
liminary annotators that can make suggestions for hu-
man curators to accept or reject. In either case, it is
important for the models to replicate the process of
a human curator as closely as possible. One of the
ways in which curators select appropriate ontology
concepts is by looking at the concepts in the context
of the ontology including the hierarchy and relation-
ships in the ontology and not as concepts as individual
constructs. This indicates that for automated methods
to be successful at curation, they need to be cognizant
of the ontology structure as well as relationships be-
tween different concepts.
The ultimate goal of automated ontology concept
recognition is to develop intelligent systems that can
understand the ontology hierarchy and make predic-
tions that are cognizant of those relationships. For
example, if a phrase in text corresponds to ontology
80
Devkota, P., Mohanty, S. and Manda, P.
Ontology-Powered Boosting for Improved Recognition of Ontology Concepts from Biological Literature.
DOI: 10.5220/0011683200003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 3: BIOINFORMATICS, pages 80-90
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

concept X in the gold standard, the model should ide-
ally recognize that X corresponds to the phrase. How-
ever, automated models are not perfect and sometimes
make mistakes. In this case, the desired outcome
would be for the model to recognize that semantically
similar concepts to X (such as X’s parent) exist in the
ontology and that those concepts should be associated
with the text. Ontology sentient models take the on-
tology structure or the ontology graph as input while
training and make predictions accordingly. However,
developing accurate ontology sentient models can be
a challenge due to the size and complexities of the on-
tology graph that often results in models that are too
large or require an inordinate time for training.
The automated annotation models previously de-
veloped by this team (Manda et al., 2018; Manda
et al., 2020; Devkota et al., 2022b; Devkota et al.,
2022a) have shown good accuracy in recognizing on-
tology concepts from text. In most cases, these sys-
tems are able to predict the same ontology concept as
the ground truth in the gold standard data achieving
perfect accuracy. However, ontology-based predic-
tion systems can also achieve partial accuracy. This
happens when a model might not predict the exact
ontology concept as the gold standard but a related
concept (sub-class or super-class), thereby achieving
partial accuracy. In cases where our models were not
able to predict the ground truth exactly, they failed
to achieve reasonable partial accuracy and predicted
concepts that were highly unrelated to the ground
truth. Thus, our models’ accuracy could be improved
by focusing on improving the partial accuracy for in-
stances when the model fails to make an exact predic-
tion.
With the above motivation, here, we present an al-
ternative approach called Ontology-powered Boost-
ing (OB) to improve the prediction performance
of automated curation models by using informa-
tion about the ontology hierarchy to post-process the
model’s predictions after training has completed. The
goal of OB is to combine the model’s preliminary
predictions with knowledge of the ontology hierarchy
to selectively increase the confidence of certain pre-
dictions to improve overall prediction accuracy. The
method relies on a computationally inexpensive cal-
culation and avoids bloated machine learning models
that cannot be trained or deployed without requiring
enormous resources.
Note that the contribution of this work is not in
presenting novel architectures for recognizing ontol-
ogy concepts but rather in presenting the Ontology
Boosting approach for further improving prediction
accuracy of our previously published deep learning
architectures. Hence, we will present architecture de-
tails briefly and will refer the reader to our prior work
for complete details.
2 BACKGROUND
Automated methods of recognizing ontology con-
cepts in literature have been developed in the last
decade and the approaches range from lexical anal-
ysis to traditional machine learning to deep learning
in more recent times.
Text mining tools that use traditional machine
learning based methods employ supervised learning
techniques using gold standard corpora (Beasley and
Manda, 2018). In 2018, we conducted a survey of
ontology-based Named Entity Recognition and con-
ducted a formal comparison of methods and tools
for recognizing ontology concepts from scientific lit-
erature. Three concept recognition tools (MetaMap
(Aronson, 2001), NCBO Annotator (Jonquet et al.,
2009), Textpresso (M
¨
uller et al., 2004) were com-
pared (Beasley and Manda, 2018).These methods can
form generalizable associations between text and on-
tology concepts leading to improved accuracy.
The rise of deep learning in the areas of image
and speech recognition has translated into text-based
problems as well. Preliminary research has shown
that deep learning methods result in greater accu-
racy for text-based tasks including identifying ontol-
ogy concepts in text (Lample et al., 2016; Habibi
et al., 2017; Lyu et al., 2017; Wang et al., 2018;
Manda et al., 2020). Deep learning methods use vec-
tor representations that enable them to capture depen-
dencies and relationships between words using en-
riched representations of character and word embed-
dings from training data (Casteleiro et al., 2018). We
evaluated the feasibility of using deep learning for the
task of recognizing ontology concepts in a 2018 study
(Manda et al., 2018). We compared Gated Recurrent
Units (GRUs), Long Short Term Memory (LSTM),
Recurrent Neural Networks (RNNs), and Multi Layer
Perceptrons (MLPs) and evaluated their performance
on the CRAFT gold standard dataset. We also intro-
duced a new deep learning model/architecture based
on combining multiple GRUs with a character+word
based input. We used data from five ontologies in
the CRAFT corpus as a Gold Standard to evaluate
our model’s performance. Results showed that our
GRU-based model outperformed prior models across
all five ontologies. These findings indicated that deep
learning algorithms are a promising avenue to be ex-
plored for automated ontology-based curation of data.
This study also served as a formal comparison and
guideline for building and selecting deep learning
Ontology-Powered Boosting for Improved Recognition of Ontology Concepts from Biological Literature
81

models and architectures for ontology-based curation.
In 2020, we presented new architectures based
on GRUs and LSTM combined with different in-
put encoding formats for automated Named Entity
Recognition (NER) of ontology concepts from text.
We found that GRU-based models outperform LSTM
models across all evaluation metrics. We also created
multi level deep learning models designed to incorpo-
rate ontology hierarchy into the prediction. Surpris-
ingly, inclusion of ontology semantics via subsump-
tion reasoning yielded modest performance improve-
ment (Manda et al., 2020). This result indicated that
more sophisticated approaches to take advantage of
the ontology hierarchy are needed.
Continuing this work, a 2022 study (Devkota
et al., 2022b) presented state of the art deep learning
architectures based on GRUs for annotating text with
ontology concepts. We augmented the models with
additional information sources including NCBI’s Bio-
Thesauraus and Unified Medical Language System
(UMLS) to augment information from CRAFT for in-
creasing prediction accuracy. We demonstrated that
augmenting the model with additional input pipelines
can substantially enhance prediction performance.
Considering that our previous attempt at creating
intelligent prediction systems that use the ontology
hierarchy was not successful, we developed a differ-
ent approach to providing the ontology as input to
the deep learning model (Manda et al., 2020). In
2022, we presented an intelligent annotation system
(Devkota et al., 2022a) that uses the ontology hierar-
chy for training and predicting ontology concepts for
pieces of text. Here, we used a vector of semantic
similarity scores to the ground truth and all ancestors
in the ontology to train the model. This representation
allowed the model to identify the target GO term fol-
lowed by “similar” GO terms that are partially accu-
rate predictions. This output label representation also
helped the model optimize the weights to target more
than one prediction label. We showed that our ontol-
ogy aware models can result in 2% - 10% (depend-
ing upon choice of embedding) improvements over a
baseline model that doesn’t use ontology hierarchies.
3 METHODS
3.1 Ontology Boosting
A key component of our approach is to combine the
prediction of the deep learning architectures with the
graph structure of ontological concepts. Here we
“boost” the predictions of the deep learning model by
supporting them with similar predictions while tak-
ing model uncertainty into account. Here we take a
two step approach, 1) identify candidates for boost-
ing, and 2) boost the predictions with semantically
similar concepts.
In the first step, we identify the candidate pre-
dictions where the deep learning models had low
confidence. We calculate the uncertainty in the
predictions based on the last layer softmax out-
put. The layer outputs a probability vector (ν
i
=<
p(x
0
), p(x
1
),··· p(x
m
) >, where i is the i’th input to-
ken and p(x
j
) probability of tag x
j
), corresponding to
all possible tags (0···m), for each token that is pro-
vided as input to the model. In general, we calcu-
late the argmax(ν
i
) to select the tag with the highest
probability as the model output. Here we leverage the
top k probabilities from ν
i
vector to calculate the un-
certainty in the model’s predictions by evaluating the
entropy H(ν
i
k
) using Shannon’s information entropy
where ν
i
k
= p(argmax
k
(ν
i
)).
The highest predicted probability of the ν
i
prob-
ability vector and the entropy (H(ν
i
)) value are used
to determine the threshold for the predictions where
boosting needs to be applied. The intuition behind
this is to only boost specific predictions where the
model has low confidence. We choose the thresh-
olds for the two parameters by analyzing the pre-
dictions graphs (Figures 3, 3a), i.e. visualizing the
thresholds of the parameters where the model makes
the most errors. We also select the top k predictions
(argmax
k
(x)) from the ν
i
vector to boost. Boosting
all of the possible tag predictions does not benefit the
model’s performance and has a detrimental effect on
computational overhead.
The second step boosts the predicted probabili-
ties of the identified candidates by combining them
with the model predictions of the candidate’s ances-
tors/subsumers. Specifically, for each token i, we
boost the probabilities of top k tags (p(argmax
k
(ν
i
)))
with the probabilities of their subsumers using the fol-
lowing computation:
I (x
j
) = −log(f
x
/C)
˜p(x
j
) = β ∗ p(x
j
) ∗I (x
j
) +
d
∑
n=1
α ∗ p(x
n
j
) ∗I (x
n
j
)
n
where, we first calculate the information content
(I (x
j
)) of tag x
j
(0 ≤ j ≤ m, where m is the num-
ber of tags) as the negative log of concept frequency
( f
x
) over the total number of available concepts (C).
I (x
j
) is then utilized to calculate the boosted proba-
bility, ˜p(x
j
), which consists of two components, the
modulated original probability (β ∗ p(x
j
) ∗ I (x
j
)) and
supportive parent boosting (
∑
d
n=1
α∗p(x
n
j
)∗I (x
n
j
)
n
).
The modulated original probability combines the
original probability with a weighting factor β and the
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
82

information content of the concept I (x
j
). The second
part of the computation evaluates all of the subsumer
probabilities of x
j
by individually calculating the
modulated probabilities of parents (α∗ p(x
n
j
)∗ I (x
n
j
)),
where x
j
has d ancestors, while controlling the influ-
ence by normalizing with the depth factor n. Here α is
a weighting parameter used to control the influence of
the ancestor probabilities to the boosting. The calcu-
lated parent probabilities are then summed and added
to the modulated probability of x
j
.
Using the aforementioned approach, ˜p(x
j
) com-
bines the predicted probabilities of the tags with their
ancestor’s predictions from a single model. This is
done only for the GO annotations, where a specific
annotation probability might be boosted to make it the
top prediction if it had supporting parent predictions
from the model. The information content parameter
modulates the effect on the boosted probability by
taking frequency of occurrence of a concept and its
hierarchy into account. α and β parameters can fur-
ther control the emphasis we put on the parental con-
tribution vs original probability, where a β value of
0 nullifies the parental contribution and of 1 includes
the parental support completely. As we go higher in
the ancestor path, the depth factor n enforces lower
contributions coming from higher subsumers.
We utilize Bayesian optimization to evaluate the
different values of k (top k predictions), entropy
threshold (H(ν
i
)), α and β to maximize the model
prediction accuracy. Specifically, we utilize Tree-
structured Parzen Estimators (TPE) (Bergstra et al.,
2013) approach to derive the optimal values for each
of parameters for maximizing our objective function.
The objective function in the experiment is defined to
maximize the mean semantic similarity. α and β are
evaluated with continuous values between 0.0 to 1.0,
while k was evaluated for values between 1 - 10. The
range of entropy was defined to be continuous values
ranging from 0.0 to the highest value of entropy of the
top k predictions for each token.
We demonstrate the efficacy of the Ontology
Boosting approach on two deep learning architectures
from our previous work (Devkota et al., 2022b; De-
vkota et al., 2022a).
3.2 Training Dataset
This study uses GO annotations from version v4.0.1
(https://github.com/UCDenver-ccp/CRAFT/releases/
tag/v4.0.1) of The Colorado Richly Annotated Full
Text Corpus (CRAFT) (Bada et al., 2012), a manually
annotated corpus containing 97 articles each of which
is annotated to 10 ontologies.
3.3 Data Preprocessing
The following preprocessing steps are performed to
translate annotations from the CRAFT corpus to the
desired input formats for the deep learning models.
Please refer to previous work (Devkota et al., 2022b)
for full details.
3.3.1 Sentence Segmentation and Tokenization
Annotations for each CRAFT article are recorded
in the corresponding xml annotations file via char-
acter index spans. In order to obtain annotations
per word, we utilize a sentence segmentation library
called SpaCy (https://spacy.io/). First, the segmenter
splits the text into sentences by accounting for sen-
tence end marks (such as periods, exclamation, ques-
tion marks, etc.) and then uses a tokenizer to split
the sentences into individual words (or tokens) by ac-
counting for word boundaries (such as space, hyphen,
tab, etc.).
3.3.2 IOB Tagging
Each extracted word/token is mapped to a GO term or
an out-of-concept annotation. Each token is mapped
to one of three tags: 1) GO to indicate an annotation,
‘O’ for a non-annotation, and ‘EOS’ to indicate the
end of sentence.
3.3.3 POS Tagging and Token Encoding
POS tagging looks at the contextual information of
the word based on the words surrounding it in a sen-
tence or a phrase. Here we used the SpaCy POS tag-
ger to evaluate and tag the tokens of sentences with
15 parts of speech tags — adjective, adposition (such
as - in, to, during), adverb, auxiliary (such as - is, has
done, will do, should do), conjunction, coordinating
conjunction, determiner, interjection, noun, numeral,
particle, pronoun, proper noun, punctuation, subor-
dinating conjunction, symbol, verb, other (not anno-
tated to any of the others), and space.
We represent character level aspects of a token
using character encodings. These encodings repre-
sent upper-case and lower-case characters with ‘C’ or
‘c’ respectively. Numbers are represented using an
‘N’ and punctuation (such as commas, periods, and
dashes) are retained in the encoding.
3.3.4 BioThesaurus Encoding
One of the architectures tested in this study uses exter-
nal information from existing large scale knowledge
bases. The first data source we use is BioThesaurus
(Liu et al., 2006), which is a database of protein and
Ontology-Powered Boosting for Improved Recognition of Ontology Concepts from Biological Literature
83
gene names mapped to the UniProt Knowledgebase.
If a token is present in the database, we map if it iden-
tifies as a protein name, biomedical terms, chemical
terms, and/or macromolecule.
3.3.5 Unified Medical Language System
(UMLS) Encoding
Next, we query the UMLS (Lindberg et al., 1993)
database for tokens extracted from the articles.
Words/tokens associated with a UMLS term are en-
coded as 1 or 0 otherwise. If a phrase (sequence of
tokens) is found in UMLS, all tokens from the phrase
are encoded as 1.
3.4 Deep Learning Architecture
We evaluate our boosting approach on two deep learn-
ing architectures from our prior work (Devkota et al.,
2022b; Devkota et al., 2022a). We describe the two
architectures briefly here and refer the reader to the
articles for comprehensive details.
3.4.1 Architecture 1 - Externally Augmented
Predictor (A
1
)
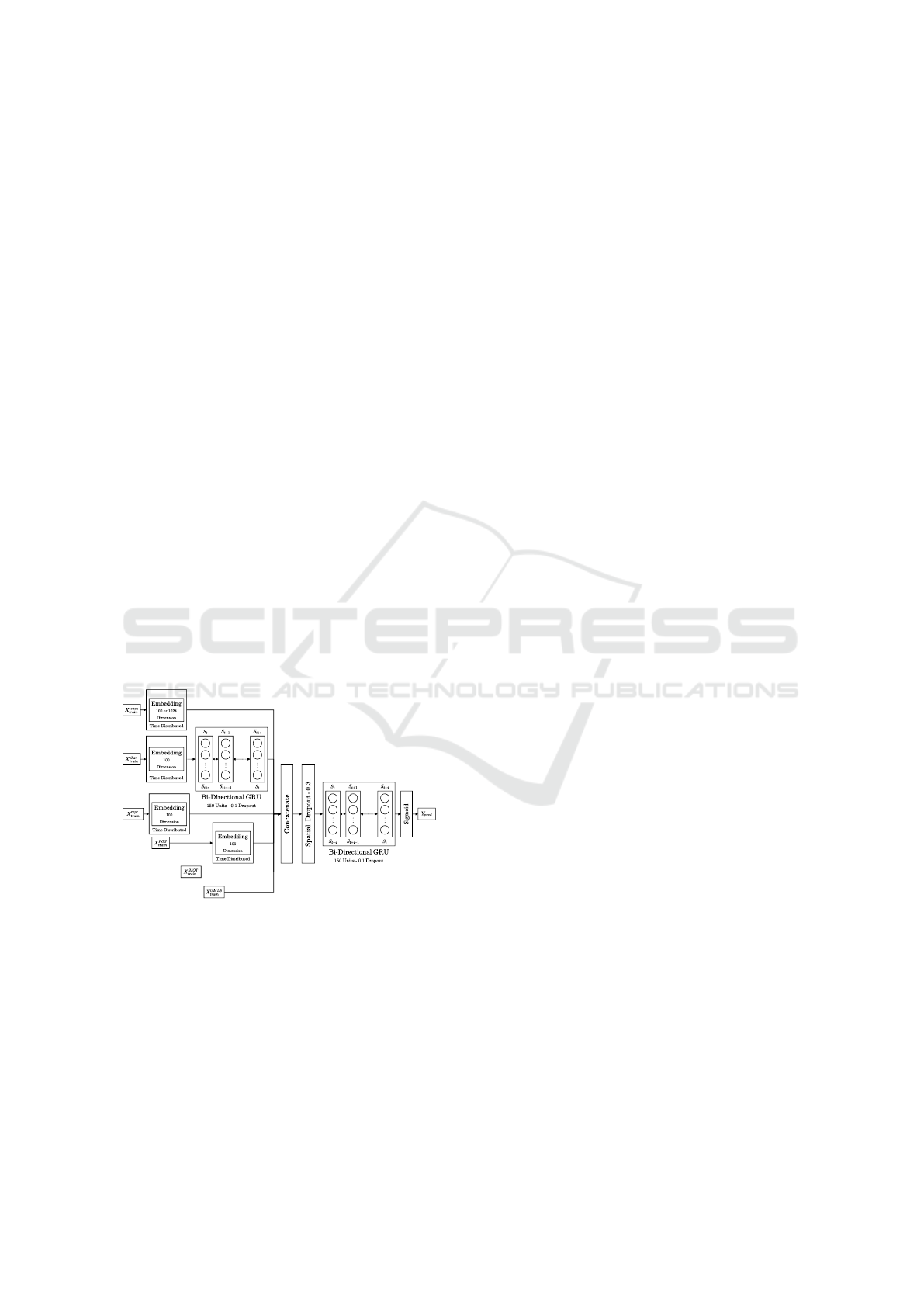
Figure 1 shows the three key components of A
1
—
1) Input Pipelines; 2) Embedding/Latent Represen-
tations; and 3) Sequence Modeler. This architecture
was originally published in (Devkota et al., 2022b).
Figure 1: Architecture of a GRU based model for ontology
concept recognition. Figure originally published in (De-
vkota et al., 2022b).
Input Pipelines. Each sentence and each token are
provided six different components as input — 1) to-
ken (X
token
tr ain
), 2) character sequence (X
char
tr ain
), 3) token-
character representation (X
repr
tr ain
), 4) parts-of-speech
(X
POS
tr ain
), 5) BioThesaurus (X
BIOT
tr ain
), and 6) UMLS
(X
UMLS
tr ain
).
The token (X
token
tr ain
) input, is a sequential tensor
where each token is represented with a high di-
mensional one hot encoded vector. Similarly, the
character sequence (X
char
tr ain
) is also a sequential ten-
sor consisting of character sequences present in a
word/token.
Character representations (X
repr
tr ain
) and POS tags
(X
POS
tr ain
) are based on words/tokens in sentences. Bio-
thesaurus encodings (X
BIOT
tr ain
) contain a four dimen-
sional vector sequence where each token is one hot
encoded for its association with protein, biomedical,
chemical and macromolecule categories. UMLS en-
codings (X
UMLS
tr ain
) are also provided as an one hot en-
coded vector sequence, where 1 indicates a token’s
presence and 0 indicates absence in UMLS.
Embedding/Latent Representations. The super-
vised embedding is a bottleneck layer which learns
to map the one hot encoded input into a smaller di-
mensional representation. We used ELMo (Peters
et al., 2018) pretrained embeddings for the X
token
tr ain
in-
put. Embeddings in ELMo are learned via a bidi-
rectional language model where the sequence of the
words are also taken into account. We use the pre-
trained model on 1 Billion Word Benchmark, which
consists of approximately 800M tokens of news crawl
data and has an embedding of 1024 dimensional out-
put embedding vectors.
Sequence Modeler. We utilize Bi-GRUs in two lo-
cations in the architecture, first to model the sequence
of characters present in each token and a second
main Bi-GRU model to concatenate input pipelines
together. After the embedding of the characters, they
are passed via the first Bi-GRU (consisting of 150
units) resulting in a sequence representation of the
characters in a sentence. 10% dropout is used in this
pipeline to regularize the output to prevent overfitting.
The character sequence representation is then con-
catenated with the ELMo embeddings, character rep-
resentation, and parts of speech, and input tensors
from Bio-Thesaurus and UMLS. This concatenated
feature map representing each sentence is then passed
to a spatial dropout, which removes 30% of the 1-D
sequence features from the input to the main Bi-GRU.
The main Bi-GRU processes the feature maps (with
10% dropout), and outputs to a single time-distributed
dense layer of 1774 nodes (representing each of the
output tags).
Architecture hyper-parameters, which include —
supervised embedding shape ({20, 50, 100, 150,
200}), dropout ({01, .2, .3, .5, .7}), number of epochs
({50, 100, 200, 300}), and class weighting, were eval-
uated using a grid search approach. We used Adam
(Kingma and Ba, 2017) as our optimiser for all of the
experiments with a default learning rate of 0.0001.
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
84
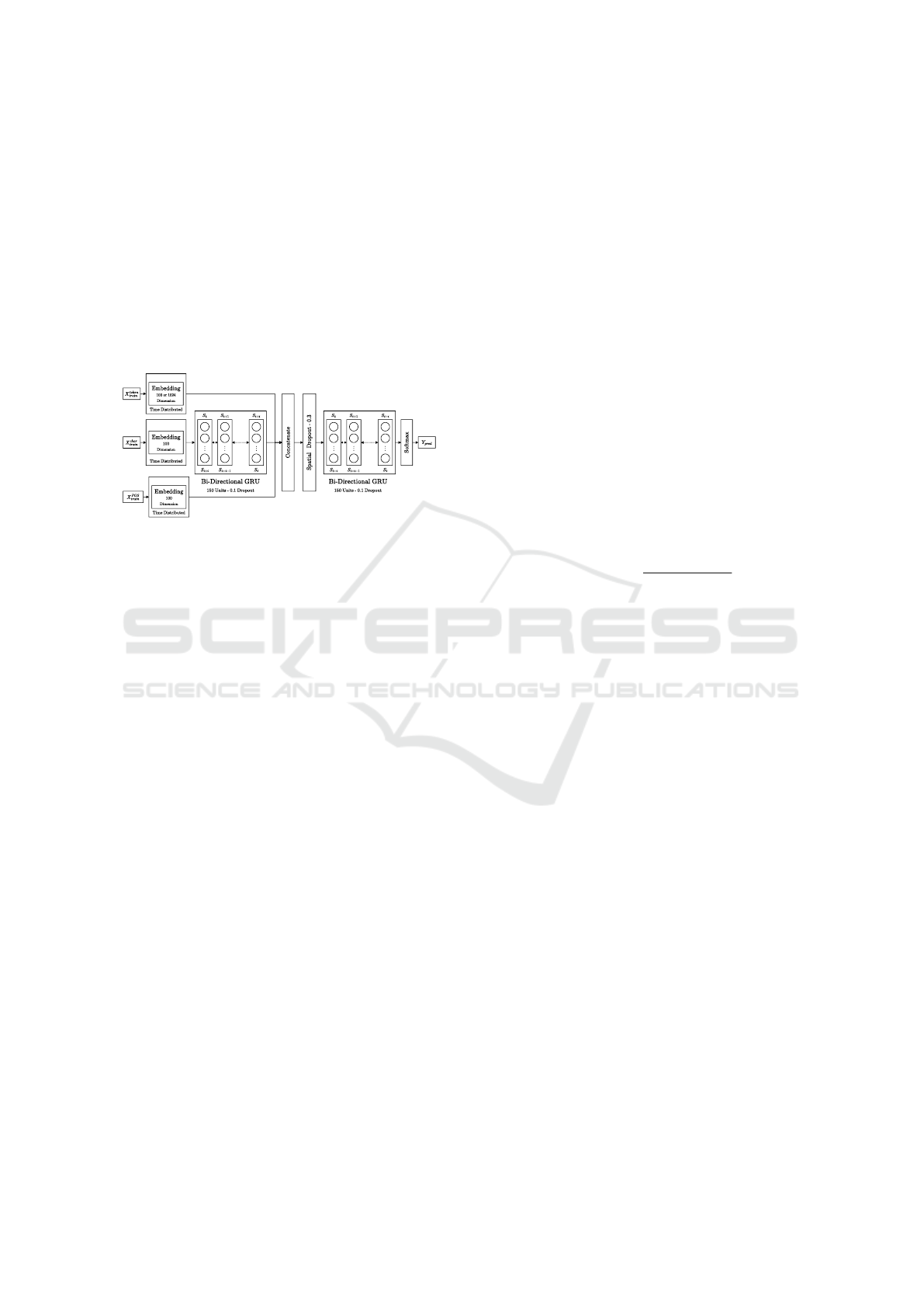
3.4.2 Architecture 2 - Intelligent Predictor (A
2
)
A
2
, as shown in figure 2 uses an intelligent predic-
tion system by using the ontology hierarchy structure
as opposed to A
1
(Devkota et al., 2022a). This ar-
chitecture was originally published in (Devkota et al.,
2022a). The overall structure of the A
2
is similar to
A
1
in terms of the Sequence Modeler and Embed-
ding/Latent Representation components. This archi-
tecture varies in the Input Pipelines provided as well
as how the target vector is represented for training the
model.
Figure 2: Architecture of an intelligent ontology prediction
system based on GRUs. Figure originally published in (De-
vkota et al., 2022a).
Input Pipelines. We provide three inputs for each
word in a sentence - 1) token (X
token
tr ain
), 2) character
sequence (X
char
tr ain
), 3) parts-of-speech (X
POS
tr ain
).
Target Vector Representation. Target labels to be
predicted are typically provided as a one-hot encoded
vector where the size of the vector equals the number
of output labels (as in the case of A
1
). In our stud-
ies, the output labels correspond to the set of all GO
terms. Typically, the value of the GO term to be pre-
dicted is set to a 1 and the value of all other GO terms
is set to 0. This approach of representing the target
labels, however, does not allow the model to learn the
ontology hierarchy nor does it allow for semantically
similar partial predictions.
In A
2
, we use Jaccard semantic similarity scores
as values in the label vector. The value of the GO
term to be predicted is set to 1 and the value of all
other GO terms in the vector is set to the Jaccard sim-
ilarity score between that term and the GO term to
be predicted. This representation allows the model to
identify the target GO term followed by “similar” GO
terms that are partially accurate predictions. This out-
put label representation also helps the model optimize
the weights to target more than one prediction label.
So for each output tag, the representation is as fol-
lows:
Y =
(
l = [1], if T ==
b
T
l = [β ∗ J
sim
(T ,
b
T )], if T ̸=
b
T & T ̸= O
(1)
where, Y is the final target vector, l is the label
for the word, T is the ground truth tag,
b
T is the pre-
dicted tag, β is the Jaccard weight, and J
sim
is the Jac-
card similarity between T and
b
T . The target vector
Y is computed by comparing the true tag (T ) with the
possible tags (
b
T ), where if the T ==
b
T == O (no an-
notation) OR a GO annotation then the value is set
to 1. Else, if the ground truth tag (T ) is a GO term,
then we calculate its Jaccard similarity to all possible
GO terms and create the target vector by weighting
it with β. We evaluate β values between {0, .25, .5,
1}, where a β value 0 indicates the traditional one-hot
vectorization and a β value 1 indicates the full Jaccard
score taken into account.
The Jaccard similarity (J
sim
) of the ground truth
concept T and a predicted concept
b
T (Pesquita et al.,
2009) is calculated as:
J
sim
(T ,
b
T ) =
|S(T ) ∩ S(
b
T )|
|S(T ) ∪ S(
b
T )|
(2)
where, S(T ) is the set of ontology subsumers of
T . Specifically, J
sim
of two concepts (A, B) in an on-
tology is defined as the ratio of the number of con-
cepts in the intersection of their subsumers over the
number of concepts in their union of their subsumers
(Pesquita et al., 2009).
Sequence Modeler. The sequence modeler for A
2
is similar to the sequence modeler in A
1
. The differ-
ences are in the optimizer, the loss function and the
activation function in the final output layer. A soft-
max activation is used in the final layer which normal-
izes the output of the model to a probability distribu-
tion over the output tags. We use the Adam algorithm
with weight decay (Loshchilov and Hutter, 2017) as
our optimiser with an initial learning rate of 0.001.
The learning rate is reduced by a factor of 0.1 after
the first 10000 training steps, and reduced further by
a factor of 0.1 after the next 15000 steps. The weight
decays by a factor of 0.0001 after the first 10000 steps
and further by another factor of 0.0001 after the next
15000 steps. We use sigmoid focal cross entropy
(Lin et al., 2017) as the loss function. Sigmoid focal
cross entropy is particularly useful for cases where we
have highly imbalanced classes. It reduces the relative
loss for easy to classify, higher frequency examples,
putting more focus on harder to classify, misclassified
examples. This loss function uses α, also called bal-
Ontology-Powered Boosting for Improved Recognition of Ontology Concepts from Biological Literature
85

ancing factor and β or modulating factor, which are
set to 0.25 and 2.0 respectively.
3.5 Performance Evaluation Metrics
Our primary evaluation metric in this study is seman-
tic similarity. Metrics such as F1 are designed for tra-
ditional information retrieval systems that either re-
trieve a piece of information or fail to do so (a binary
evaluation). However, this is not a true indication of
the performance of ontology-based retrieval or pre-
diction systems where the notion of partial accuracy
applies. A model might not predict the exact concept
as a gold standard but might predict the parent or an
ancestor of the ground truth as indicated by the ontol-
ogy. The parent will have a higher degree of similarity
to the ground truth thereby resulting in high semantic
similarity (but would count as a failure for F1 compu-
tation). Semantic similarity metrics (Pesquita et al.,
2009) designed to measure different degrees of sim-
ilarity between ontology concepts can be leveraged
to measure the similarity between the predicted con-
cept and the actual annotation to quantify the partial
prediction accuracy. Here, we use Jaccard similarity
(Pesquita et al., 2009) that measures the ontological
distance between two concepts to assess partial simi-
larity.
We also provide a modified F1 score for our ar-
chitectures. The model is tasked with predicting non-
annotations (indicated by an ‘O’ tag) or annotations
(indicated by a ‘GO’ tag). Since the majority of tags
in the training corpus are non-annotations, the model
predicts them with great accuracy. In order to avoid
biasing the F1 score, we omit accurate predictions of
‘O’ tags from the calculation to report a relatively
conservative F1 score.
4 RESULTS AND DISCUSSION
Table 1 presents a summary of how boosting affected
the results of the two architectures. First, we see that
the majority of tokens are selected for boosting via
the Bayesian selection process (Row 2). However, the
majority of tokens that are boosted remain unchanged
indicating that when the model makes correct predic-
tions, the boosting process largely retains the correct
prediction (Row 3). Reassuringly, boosting does not
change any of the GO predictions to an ‘O’ tag (Row
4). The majority of incorrect predictions made by the
models happen when the ground truth is a GO con-
cept but the model incorrectly predicts an ‘O’ (non-
annotation). Boosting makes a substantial difference
in this case by changing these instances from an ‘O’ to
a GO concept (Row 5). Of these instances, 37% (A
1
)
- 41% (A
2
) are corrected from an ‘O’ prediction to
an exactly matching GO concept as the ground truth
(Row 6). When boosting corrects an ‘O’ prediction to
a GO term (exact or partial match to the ground truth),
the average semantic similarity of these instances lies
between 53 % - 60%.
We examine the impact of our boosting approach
on improving prediction accuracy of the two architec-
tures presented above (Table 2). The base scores refer
to the output of the architecture before boosting was
applied and the boosted scores reflect performance af-
ter boosting is applied. We see that boosting improves
Jaccard semantic similarity scores by 7% for A
1
and
5.8% for the A
2
. The F1 scores experience a modest
improvement of 2.5% for A
1
and no improvement for
A
2
.
Ontology Boosting corrected 201 incorrect pre-
dictions (A
1
) while changing 6 correct predictions to
a semantically similar concept to the ground truth.
Similarly, boosting corrected 174 incorrect predic-
tions (A
2
) while changing 113 correct predictions to
a different GO concept (semantically similar). The
net effect of these two contributions appears to re-
sult in modest improvements or keeps the F1 score
unchanged. However, the real contribution of boost-
ing is reflected in the semantic similarity scores which
show an improvement.
Figures 3a and 3b show the probability of the
highest prediction and entropy (of the top 5 predic-
tions) across all tokens in the dataset. Correct predic-
tions are shown in blue and incorrect ones are shown
in red. These graphs indicate the probability and en-
tropy zones where the architectures make incorrect
predictions that can be corrected using boosting. We
use the Bayesian method described above to select in-
correct predictions on this graph to be boosted.
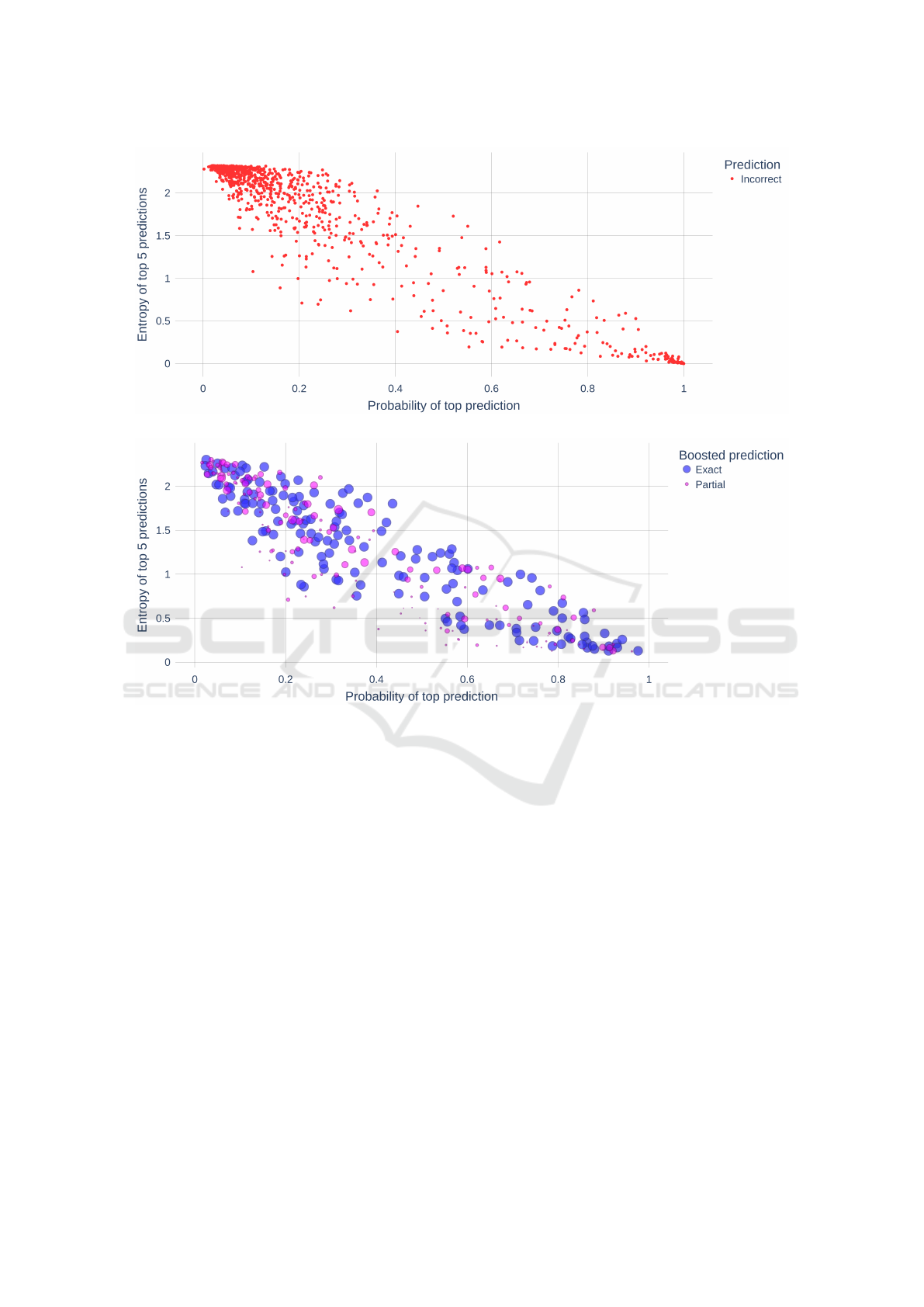
Figure 4a shows the incorrect predictions by A
1
.
Like above, the majority of these instances were cases
where the ground truth is a GO concept and the
prediction is an ‘O’ term (non-annotation). Figure
4b shows instances incorrectly predicted as ‘O’ that
were corrected by boosting. In this figure, blue in-
stances represent cases where the boosted prediction
was an exact match to the ground truth (100% ac-
curacy) whereas the purple instances represent cases
where the boosted prediction was a partial match to
the ground truth. The size of the purple instances re-
flect the degree of partial relatedness to the ground
truth - larger indicates higher semantic similarity to
the ground truth. We see that boosting has a substan-
tial effect on correcting inaccurate predictions.
Figure 5a shows the incorrect predictions by A
2
.
The majority of these instances were cases where the
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
86
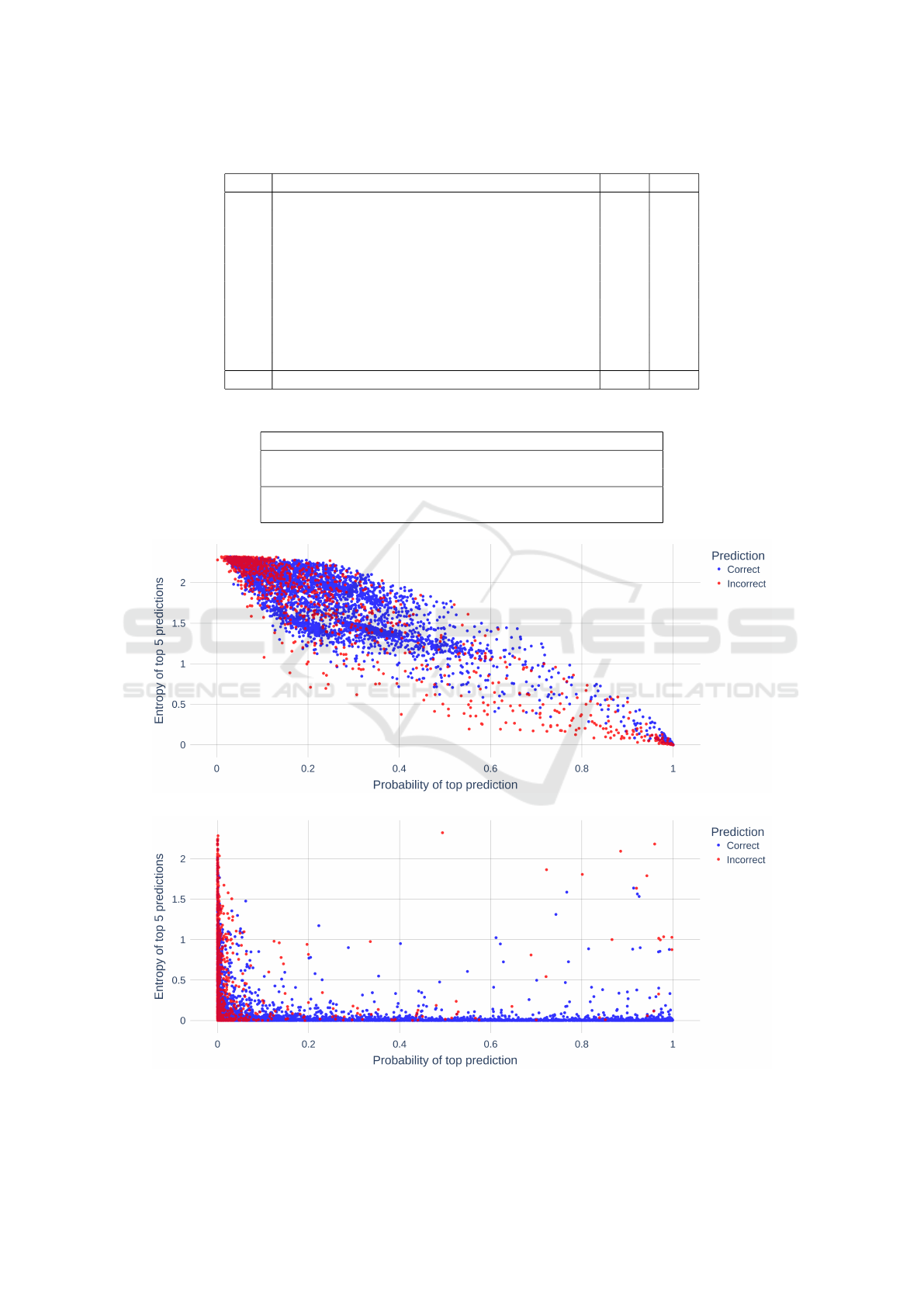
Table 1: Effect of ontology boosting on the two architectures.
Row Description A
1
A
2
1 Total number of tokens 5439 5495
2 Number of tokens boosted 4972 5197
3 Number of tokens boosted but unchanged 4411 4587
4 Number of tokens boosted from GO to O 0 0
5 Number of tokens boosted from O to GO 534 366
6 Number of tokens boosted from O to an exact GO 197 152
7 Average Semantic Similarity for O to GO 0.53 0.60
Table 2: Impact of boosting on the two architectures.
Architecture Semantic Similarity Modified F1
A
1
Base 0.84 0.83
Boosted 0.90 0.85
A
2
Base 0.85 0.81
Boosted 0.90 0.81
(a) Architecture A
1
.
(b) Architecture A
2
.
Figure 3: Distribution of correct and incorrect predictions with respect to probability and entropy of predictions.
Ontology-Powered Boosting for Improved Recognition of Ontology Concepts from Biological Literature
87
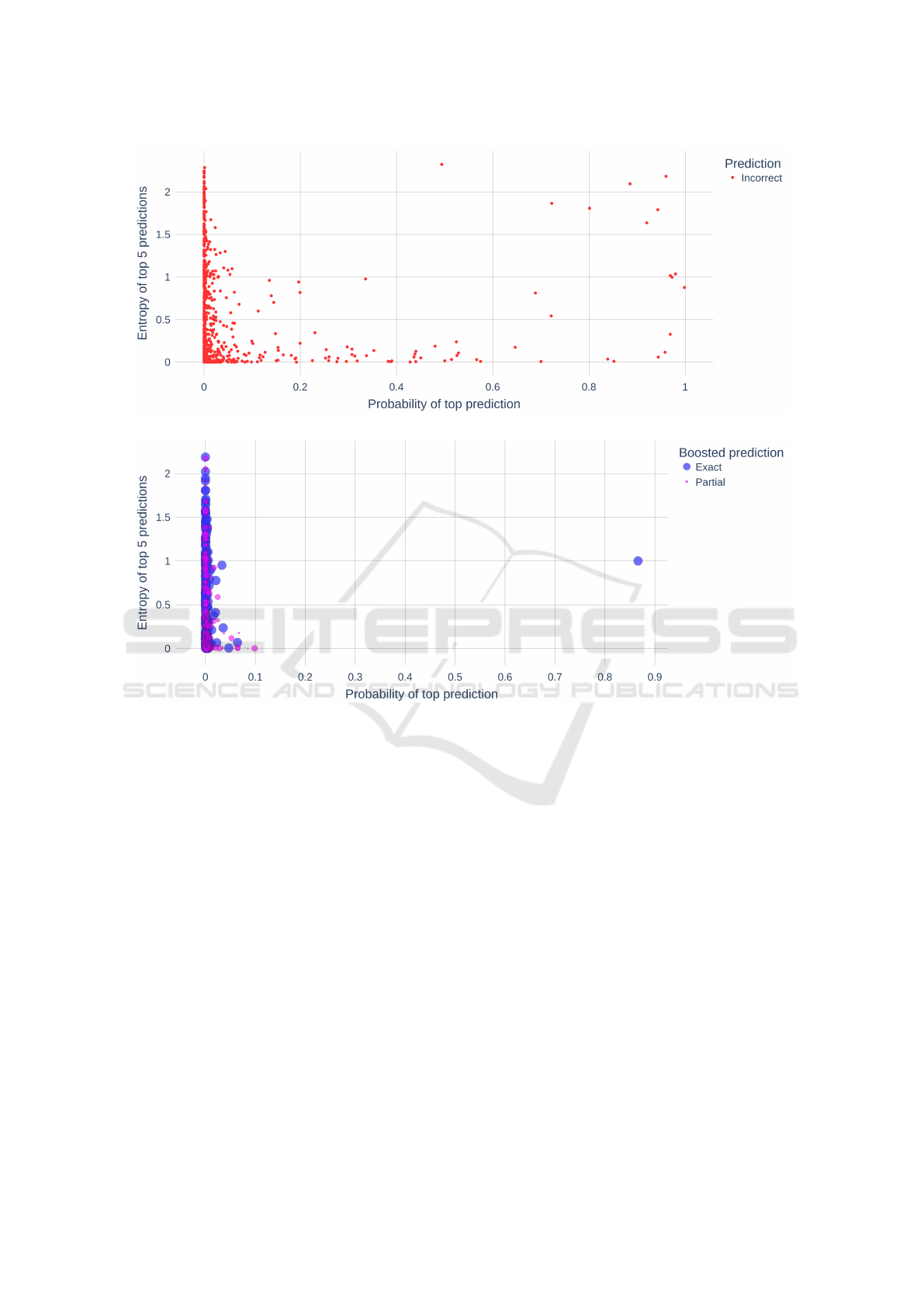
(a) Incorrect predictions.
(b) Corrected predictions after boosting.
Figure 4: A
1
predictions corrected via Ontology Boosting.
ground truth is a GO concept and the prediction is an
‘O’ term (non-annotation). Figure 5b shows instances
incorrectly predicted as ‘O’ that were corrected by
boosting.
5 CONCLUSIONS
Automated methods for ontology-based annotation
of scientific literature are important for represent-
ing knowledge in a consistent machine-readable for-
mat. Deep learning models have shown promising
improvements and performance at this task. How-
ever, the majority of these architectures do not ac-
count for or take advantage of the ontology hierarchy
thereby losing valuable information. Encoding and
representing complex ontologies can lead to massive
models that are computationally and financially infea-
sible to train. Here, we presented a novel approach
called Ontology Boosting that allows post processing
of ontology predictions by deep learning architectures
to selectively improve the confidence of certain pre-
dictions by using information from the ontology such
as subsumers, information content, depth of a con-
cept in the ontology, etc. We show that this computa-
tionally inexpensive step can result in substantial im-
provements to our key performance metric - semantic
similarity. Our results clearly show that the predic-
tions made by the deep learning model are closer to
the human ground truth after applying the Boosting
process as compared to before.
ACKNOWLEDGEMENTS
This work is funded by a CAREER grant from the
Division of Biological Infrastructure at the National
Science Foundation of United States of America
(#1942727).
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
88
(a) Incorrect predictions.
(b) Corrected predictions after boosting.
Figure 5: A
2
predictions corrected via Ontology Boosting.
REFERENCES
Aronson, A. R. (2001). Effective mapping of biomedical
text to the umls metathesaurus: the metamap program.
In Proceedings of the AMIA Symposium, page 17.
American Medical Informatics Association.
Bada, M., Eckert, M., Evans, D., Garcia, K., Shipley, K.,
Sitnikov, D., Baumgartner, W. A., Cohen, K. B., Ver-
spoor, K., Blake, J. A., and Hunter, L. E. (2012). Con-
cept annotation in the craft corpus. BMC Bioinformat-
ics, 13(1):161.
Beasley, L. and Manda, P. (2018). Comparison of natu-
ral language processing tools for automatic gene on-
tology annotation of scientific literature. Proceedings
of the International Conference on Biomedical Ontol-
ogy.
Bergstra, J., Yamins, D., and Cox, D. (2013). Making a
science of model search: Hyperparameter optimiza-
tion in hundreds of dimensions for vision architec-
tures. In Dasgupta, S. and McAllester, D., editors,
Proceedings of the 30th International Conference on
Machine Learning, volume 28 of Proceedings of Ma-
chine Learning Research, pages 115–123, Atlanta,
Georgia, USA. PMLR.
Casteleiro, M. A., Demetriou, G., Read, W., Prieto, M. J. F.,
Maroto, N., Fernandez, D. M., Nenadic, G., Klein,
J., Keane, J., and Stevens, R. (2018). Deep learning
meets ontologies: experiments to anchor the cardio-
vascular disease ontology in the biomedical literature.
Journal of biomedical semantics, 9(1):13.
Devkota, P., Mohanty, S., and Manda, P. (2022a). Knowl-
edge of the ancestors: Intelligent ontology-aware an-
notation of biological literature using semantic sim-
ilarity. Proceedings of the International Conference
on Biomedical Ontology.
Devkota, P., Mohanty, S. D., and Manda, P. (2022b). A
gated recurrent unit based architecture for recognizing
ontology concepts from biological literature. BioData
Mining, 15(1):1–23.
Habibi, M., Weber, L., Neves, M., Wiegandt, D. L., and
Leser, U. (2017). Deep learning with word embed-
dings improves biomedical named entity recognition.
Bioinformatics, 33(14):i37–i48.
Jonquet, C., Shah, N., H Youn, C., Musen, M., Callendar,
Ontology-Powered Boosting for Improved Recognition of Ontology Concepts from Biological Literature
89

C., and Storey, M.-A. (2009). Ncbo annotator: Se-
mantic annotation of biomedical data.
Kingma, D. P. and Ba, J. (2017). Adam: A method for
stochastic optimization.
Lample, G., Ballesteros, M., Subramanian, S., Kawakami,
K., and Dyer, C. (2016). Neural architectures
for named entity recognition. arXiv preprint
arXiv:1603.01360.
Lin, T.-Y., Goyal, P., Girshick, R., He, K., and Doll
´
ar, P.
(2017). Focal loss for dense object detection.
Lindberg, D. A., Humphreys, B. L., and McCray, A. T.
(1993). The unified medical language system. Year-
book of Medical Informatics, 2(01):41–51.
Liu, H., Hu, Z.-Z., Zhang, J., and Wu, C. (2006). Bio-
thesaurus: a web-based thesaurus of protein and gene
names. Bioinformatics, 22(1):103–105.
Loshchilov, I. and Hutter, F. (2017). Decoupled weight de-
cay regularization.
Lyu, C., Chen, B., Ren, Y., and Ji, D. (2017). Long short-
term memory rnn for biomedical named entity recog-
nition. BMC bioinformatics, 18(1):462.
Manda, P., Beasley, L., and Mohanty, S. (2018). Taking
a dive: Experiments in deep learning for automatic
ontology-based annotation of scientific literature. Pro-
ceedings of the International Conference on Biomedi-
cal Ontology.
Manda, P., SayedAhmed, S., and Mohanty, S. D. (2020).
Automated ontology-based annotation of scientific lit-
erature using deep learning. In Proceedings of The
International Workshop on Semantic Big Data, pages
1–6.
M
¨
uller, H.-M., Kenny, E. E., and Sternberg, P. W. (2004).
Textpresso: An ontology-based information retrieval
and extraction system for biological literature. PLOS
Biology, 2(11).
Pesquita, C., Faria, D., Falcao, A. O., Lord, P., and Couto,
F. M. (2009). Semantic similarity in biomedical on-
tologies. PLoS computational biology, 5(7).
Peters, M. E., Neumann, M., Iyyer, M., Gardner, M.,
Clark, C., Lee, K., and Zettlemoyer, L. (2018).
Deep contextualized word representations. CoRR,
abs/1802.05365.
Wang, X., Zhang, Y., Ren, X., Zhang, Y., Zitnik, M.,
Shang, J., Langlotz, C., and Han, J. (2018). Cross-
type biomedical named entity recognition with deep
multi-task learning. arXiv preprint arXiv:1801.09851.
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
90
