
Cardiac Arrhythmia Classification in Electrocardiogram Signals with
Convolutional Neural Networks
Igor Lopes Souza
a
and Daniel Oliveira Dantas
b
Departamento de Computac¸
˜
ao, Universidade Federal de Sergipe, S
˜
ao Crist
´
ov
˜
ao, SE, Brazil
Keywords:
Electrocardiography, ECG, Atrial Fibrillation.
Abstract:
Electrocardiography is a frequently used examination technique for heart disease diagnosis. Electrocardiogra-
phy is essential in the clinical evaluation of patients who have heart disease. Through the electrocardiogram
(ECG), medical doctors can identify whether the cardiac muscle dysfunctions presented by the patient have
an inflammatory origin and early diagnosis of serious diseases that primarily affect the blood vessels and the
brain. The basis of arrhythmia diagnosis is the identification of normal and abnormal heartbeats and their
classification into different diagnoses based on ECG morphology. Heartbeats can be divided into five cate-
gories: non-ectopic, supraventricular ectopic, ventricular ectopic, fusion, and unknown beats. It is difficult to
distinguish these heartbeats apart on the ECG as these signals are typically corrupted by outside noise. The
objective of this study is to develop a classifier capable of classifying a patient’s ECG signals for the detec-
tion of arrhythmia in clinical patients. We developed a convolutional neural network (CNN) to identify five
categories of heartbeats in ECG signals. Our experiment was conducted with ECG signals obtained from a
publicly available MIT-BIH database. The number of instances was even out to five classes of heartbeats. The
proposed model achieved an accuracy of 99.33% and an F1-score of 99.44% in the classification of ventricular
ectopic beats (VEB).
1 INTRODUCTION
According to the World Health Organization, cardio-
vascular diseases (CVDs) are the leading cause of
death in the world (McAloon et al., 2016). Arrhyth-
mia, a heart rhythm disorder, is considered one of the
most common disorders of the heart. Arrhythmia is
a problem with the rate or rhythm of the heartbeat.
During an arrhythmia, the heart may beat too fast, too
slow, or with an irregular rhythm. Atrial fibrillation
(AF) is the most prevalent case of arrhythmia. AF
causes irregular heartbeats. In AF, the electrical ac-
tivity of the atria (the heart’s upper chambers) is ir-
regular, inconsistent, and not synchronized with ven-
tricles (Hagiwara et al., 2018).
AF is diagnosed by interpreting the ECG. Au-
tomatic diagnosis is useful in home settings, where
an ECG interpretation specialist is not available to
diagnose AF (Mant et al., 2007). Classification of
ECG signals is necessary for the automatic diagno-
sis of arrhythmia. To improve AF detection, machine
learning methods were used by various authors (Lown
et al., 2020; Pollock et al., 2020; Shoemaker et al.,
a
https://orcid.org/0000-0002-2499-4607
b
https://orcid.org/0000-0002-0142-891X
2020). Recently, Sanchez successfully experimented
with the latest and most innovative convolutional neu-
ral networks (CNN) (S
´
anchez and Cervera, 2019).
Deep convolutional neural networks have the capa-
bility of hierarchical feature learning, which allows
the neural network to differentiate and generalize
ECG signal patterns with higher accuracy than an ex-
pert (Chen et al., 2022; Kiranyaz et al., 2021). CNNs
have been used to diagnose arrhythmias, and coro-
nary artery diseases, and classify strokes (Zhiqiang
and Jun, 2017).
Many approaches to arrhythmia heartbeat classi-
fication with CNN have been proposed. Han (Han
and Shi, 2020) presents a method to detect and local-
ize myocardial infarction by combining a multiple-
lead residual neural network (ML-ResNet) frame-
work with three residual blocks and feature fusion us-
ing 12-lead ECG recordings.
Qiyang Xie (Xie et al., 2021) used ResNet34 to
train a model with the morphological characteristics
of ECG signals and obtain meaningful information
from ECG signals.
Xiong (Xiong et al., 2017) proposed a purely data-
driven, deep learning pipeline, a 16-layer CNN, for
the automatic classification of ECG signals from the
356
Souza, I. and Dantas, D.
Cardiac Arrhythmia Classification in Electrocardiogram Signals with Convolutional Neural Networks.
DOI: 10.5220/0011682800003411
In Proceedings of the 12th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2023), pages 356-362
ISBN: 978-989-758-626-2; ISSN: 2184-4313
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

Computing in Cardiology (CinC) Challenge 2017 into
four categories, including AF. The large dataset of
ECG data recorded from patients and labeled by ex-
perts provided a framework for developing and vali-
dating their approach to ECG diagnosis.
Zhi Li (Li et al., 2020) developed a deep learning
method for cardiac arrhythmia classification based on
ResNet. The design consists of a 1D 31-layer con-
volutional residual network. The algorithm includes
four residual blocks, each of which consists of three
layers of 1D convolutions, three layers of batch nor-
malization (BN), three layers of the rectified linear
unit (ReLU) activation function, and a structure of
identity shortcut connections. The 2-lead ECG sig-
nals were used in combination with deep learning
techniques to automatically identify the normal, left
group, right group, premature atrial, and premature
ventricular contraction heartbeats.
Zhu (Zhu et al., 2020) developed a deep learn-
ing approach for the automated diagnosis of multiple
cardiac rhythm labels or conduction abnormalities by
real-time ECG signal analysis. The dataset used was
obtained from ECG data with 10s in length and 12-
channel format. The data is from adult patients, with
21 distinct rhythm classes for the diagnosis of simul-
taneous cardiac arrhythmias, i.e., patients with multi-
ple heart diseases.
Kiranyaz (Kiranyaz et al., 2017) proposed a per-
sonalized health monitoring system that can detect
early occurrences of arrhythmias from a patient’s
ECG signal by modeling common causes of arrhyth-
mias in the signal domain as degradation from nor-
mal ECG beats to abnormal beats. Using the degra-
dation models, abnormal beats were created from the
patient’s average normal beat. A simple 1D convolu-
tional neural network was trained using real normal
beats and synthesized abnormal beats.
Han (Han and Shi, 2020) presented a method to
detect and localize myocardial infarction by com-
bining an ML-ResNet framework with three residual
blocks and feature fusion using 12-lead ECG record-
ings. The single-lead feature branching network is
trained to automatically learn local features of dif-
ferent levels between different layers, which can be
used to characterize the spatial representation of the
ECG. The main features are merged as global fea-
tures. For the generalization and evaluation of the
proposed method in clinics, intra-patient and inter-
patient schemes were used.
Acharya (Acharya et al., 2017) developed a nine-
layer deep convolutional neural network to identify
five different categories of heartbeats in ECG signals:
non-ectopic beat, supraventricular ectopic beat, ven-
tricular ectopic beat, fusion beat, and unknown beat.
The experiment was conducted on noise-attenuated
and non-attenuated data sets from a public database,
MIT-BIH. This set was artificially augmented to equal
the number of instances of the five heartbeat classes
and filtered to remove high-frequency noise.
Xiang (Xiang et al., 2018) proposed an accu-
rate method for patient-specific ECG beat classifica-
tion, which adopts morphological features and tim-
ing information. As to the morphological features
of a heartbeat, attention-based two-level 1-D CNN
is incorporated in the proposed method to extract
different-grained features automatically by focusing
on various parts of a heartbeat. The timing informa-
tion, the difference between previous and post RR in-
tervals, is computed as a dynamic feature. Both the
extracted morphological features and the interval dif-
ference are used by multi-layer perceptron (MLP) for
classifying ECG signals.
Ali Sellami (Sellami and Hwang, 2019) proposed
a new type of deep convolutional neural network for
heartbeat classification. A batch-weighted loss func-
tion was created to quantify the loss and decrease
the imbalance between classes. The loss weights
change dynamically as the distribution of classes in
each batch changes.
Schwab (Schwab et al., 2017) proposed a machine
learning approach based on recurrent neural networks
(RNN) to analyze different cardiac arrhythmias with
only a single lead and short ECG recordings, below
10s. To facilitate training dependencies on the tem-
poral dimension, a new task formulation was intro-
duced that takes advantage of the natural beat-based
segmentation of ECG signals.
Rahhal (Al Rahhal et al., 2019) proposed a novel
end-to-end architecture based on a dense convolu-
tional network (DCN) for ECG signal classification.
The architecture is based on two main modules: the
first is a generative module and the second is a dis-
criminative module. The generative module con-
verts the one-dimensional ECG signal into an image
through fully connected up-sampling layers and con-
volutional layers. The discriminative module receives
the image from the generative module and performs
feature learning and classification.
Zhai (Zhai and Tin, 2018) developed a high-
performance ECG-based arrhythmic beat classifica-
tion system. The classifier was designed based on a
CNN. The single-channel ECG signal was segmented
into heartbeats according to the change in beats. Zhai
provided accurate ECG classification tools.
We believe that it is possible to further improve
the accuracy, sensitivity, specificity, precision, and
F1-score of CNN heartbeat classifiers. Our study
aims to improve the classification metrics by increas-
Cardiac Arrhythmia Classification in Electrocardiogram Signals with Convolutional Neural Networks
357
ing the number of convolution layers in the couple-
convolution implementation and using a new archi-
tecture with triple-convolution (Uchida et al., 2018)
for the classifier in conjunction with fine-tuning for
further optimization. The classification of arrhyth-
mia signals in public ECG datasets will generate more
precise and accurate results. The improved classifica-
tion of ECG signals will generate more accurate re-
sponses in the detection of cardiac arrhythmias, fa-
cilitating the health care of patients. Our neural net-
work architecture was inspired by Xiong architecture
design (Xiong et al., 2017), but with a faster architec-
ture, fewer loops, more convolutions for feature ex-
traction, and a higher resulting F1-Score.
2 METHODOLOGY
In this study, we created a classifier capable of distin-
guishing the different types of heartbeats and detect-
ing cardiac arrhythmia. This architecture was fine-
tuned so that the models achieved the highest valida-
tion accuracy and F1-score possible. Our model was
evaluated in the test set. The ECG heartbeat classifier
is composed of two main steps: preprocessing and
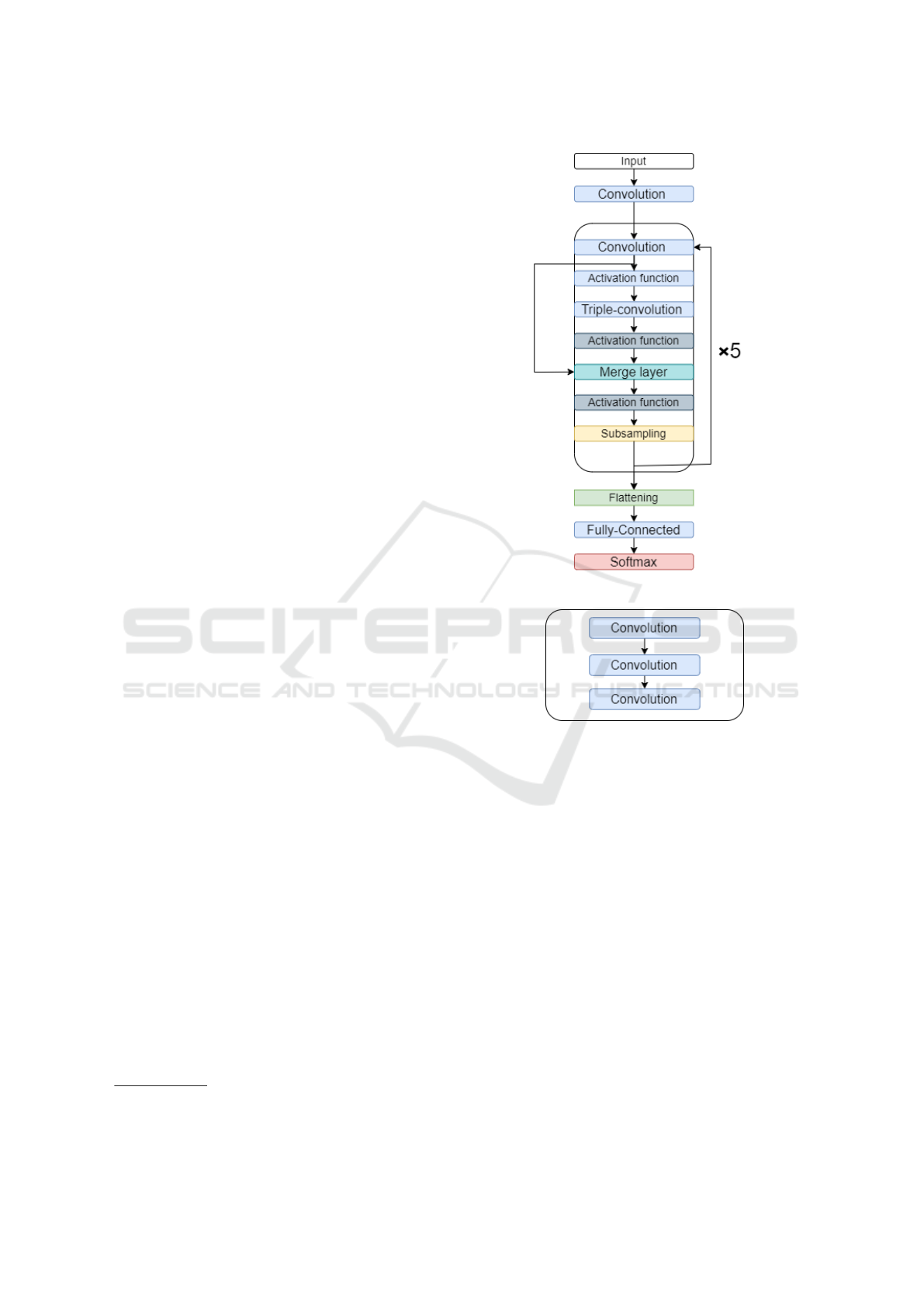
classification. The network architecture is shown in
Figure 1. The implementation of this methodology is
publicly available
1
and was coded in Python using
Tensorflow, Keras, and Numpy.
2.1 Dataset
In this study, we used the ECG Heartbeat Categoriza-
tion Dataset, freely available in the Internet
2
. We
used only the portion of the dataset derived from the
Physio Bank MIT-BIH Arrhythmia database (Mark
and Moody, 1988). This database consists of a 48
half-hour long ECG recordings from 47 subjects—
obtained with a Lead II ECG configuration—that was
band-pass filtered over the frequency range from 0.1
to 100Hz and digitized at 360 samples per second.
Furthermore, these recordings were interpreted and
validated by at least two cardiologists. The database
consists of annotations for both heartbeat class in-
formation and R-peak position information verified
by two or more expert cardiologists. The 17 beat
types can be grouped into five beat classes defined by
the Association of Advancement for Medical Instru-
mentation (AAMI) which follows the American Na-
tional Standard for Ambulatory ECGs (ANSI/AAMI
EC38:2007) recommendations.
1
https://github.com/Igor-Lopes-Souza/VISAPP-2023
2
https://www.kaggle.com/datasets/shayanfazeli/heartbeat
Figure 1: Neural network architecture.
Figure 2: Triple-convolution.
2.2 Preprocessing
The raw MIT-BIH signal is corrupted by myoelectric
interference, power line interference, and line drift.
To remove these noises, the raw ECG signal is fil-
tered using wavelet filters. The raw signal is decom-
posed by Daubechies wavelet 6 (db6) at six levels, and
wavelet coefficients from the third to the sixth level
were retained and used for signal reconstruction (Shi
et al., 2019). After noise removal, we segmented the
signal for heartbeats by taking advantage of informa-
tion from the positions of the R-peaks annotated in the
MIT-BIH arrhythmia database. Each heartbeat con-
sists of 300 samples: 149 before and 150 after the
R-peak position.
2.3 Classifier Architecture
Figure 1 shows the schematic of our CNN classi-
fier. The network is composed of convolutional lay-
ICPRAM 2023 - 12th International Conference on Pattern Recognition Applications and Methods
358
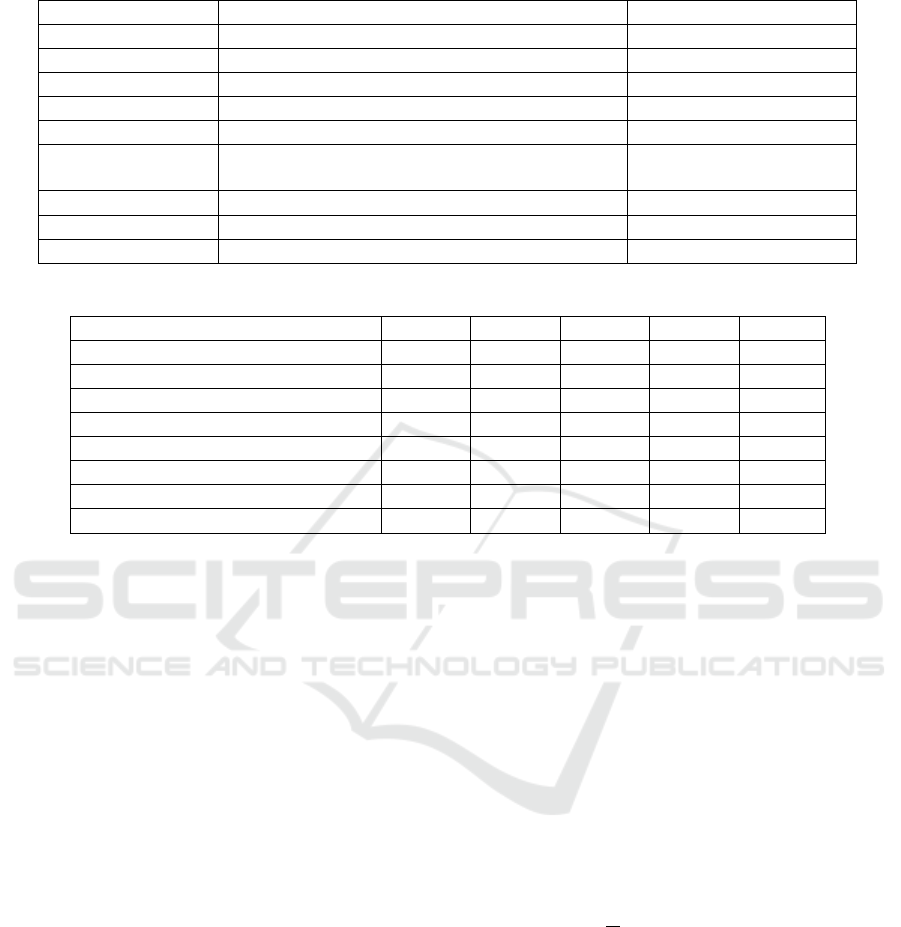
Table 1: Hyperparameter values chosen in classifier fine-tuning.
Parameters Values Chosen Value
Dropout 0.10, 0.25, 0.30, 0.50 0.50
Optimizer Adam, Adamax, SGD Adam
Activation function
Relu, Selu, Elu, Softmax, Softplus Relu
Batch size
100, 250, 500, 1000, 1500 500
Epochs
10, 25, 75, 125, 175, 300, 1000 75
Loss function
Binary cross-entropy, Categorical cross-entropy,
Poisson, Kullback-Leibler divergence, Huber
Categorical cross-entropy
Learning rate
0.01, 0.001, 0.0001, 0.00001 0.0001
β1
0.900, 0.990, 0.999 0.900
β2
0.900, 0.990, 0.999 0.990
Table 2: Comparison of the proposed algorithm classification using ventricular ectopic beats (VEB).
ACC SEN SPE PRE F1S
Martis (Martis et al., 2014)
99.45% 99.61% 99.99% 99.99% 99.8%
Proposed classifier
99.33% 99.59% 99.30% 99.12% 99.44%
Sellami (Sellami and Hwang, 2019)
99.48% 96.97% 99.87% 98.83% 97.80%
Acharya (Acharya et al., 2017)
94.03% 96.71% 91.54% 97.85% 97.27%
Zhai (Zhai and Tin, 2018)
99.10% 96.40% 99.50% 96.40% 96.40%
Yande (Xiang et al., 2018)
99.20% 93.70% 99.60% 94.80% 94.20%
Jiang (Jiang and Kong, 2007)
98.80% 94.30% 99.40% 95.30% 94.70%
Ince (Ince et al., 2009)
97.60% 83.60% 98.10% 87.40% 85.40%
ers, subsampling layers, fully connected layers, and
a softmax layer. The convolutional layers perform
the convolution operations on the output of a previ-
ous layer using the current convolution kernel (ω
ik
).
The merge layer adds two layers, in our case the sec-
ond convolution layer and the first activation function
of each execution. Usually, each convolution layer
is followed by a subsampling layer. However, to fa-
cilitate mapping between the heartbeat category and
its waveform, we use a triple-convolution structure
to achieve a more powerful fitting capability (Uchida
et al., 2018). Figure 2 shows the structure of a triple-
convolution layer sequence.
The subsampling layer was used to reduce by half
the input size of the next layer, compressing the size
of the ECG data, reducing the number of computa-
tions and extracting useful features, our max pooling
size is set to 5 with a stride of 2 in all pooling lay-
ers. The function max-pooling was used to obtain
the maximum value inside a region around each posi-
tion in the input matrix (Murray and Perronnin, 2014).
Fully connected layers were used to increase the num-
ber of nonlinear operations (Xu et al., 2019).
In this study, we use the ReLu function as an
activation function in both convolutional layers and
fully connected layers (Nair and Hinton, 2010; Girosi
et al., 1995). In the output layer, we use the acti-
vation function softmax to obtain the five heartbeat
categories (non-ectopic beat, supraventricular ectopic
beat, ventricular ectopic beat, fusion beat, and un-
known beat) (Nwankpa et al., 2018).
2.4 Training Method
The goal of training is to reduce the value of the loss
function L, i.e., to decrease the model loss and ad-
just the weights and biases so that Equation 1 fits the
model training set. The cross-entropy function is used
as the loss function (Xu and Liu, 2020):
We update the weights and offsets using the Adam
optimizer (Kingma and Ba, 2014). First, a batch
of samples was sent to calculate the gradient of the
Equation 1, and we set the batch size to 256:
g =
1
m
∇
θ
∑
i
L( f (x
(i)
;θ), y
(i))
!
. (1)
The g is the gradient value, m is the batch size, θ is
the parameter to be updated, f (x
(i)
;θ) is the heartbeat
type predicted by the i-th sample, y
(i)
is the actual type
of the i-th sample, and L is the loss function. The
m
t
and v
t
represent the first and second estimates of
the moment of the gradient. The ˆm
t
and ˆv
t
are the
corresponding bias corrections. The β
1
and β
2
are the
decay rates for the moment estimates, set to 0.900 and
0.990.
The regularization dropout (Hinton et al., 2012;
Srivastava et al., 2014) was used to avoid overfitting
Cardiac Arrhythmia Classification in Electrocardiogram Signals with Convolutional Neural Networks
359
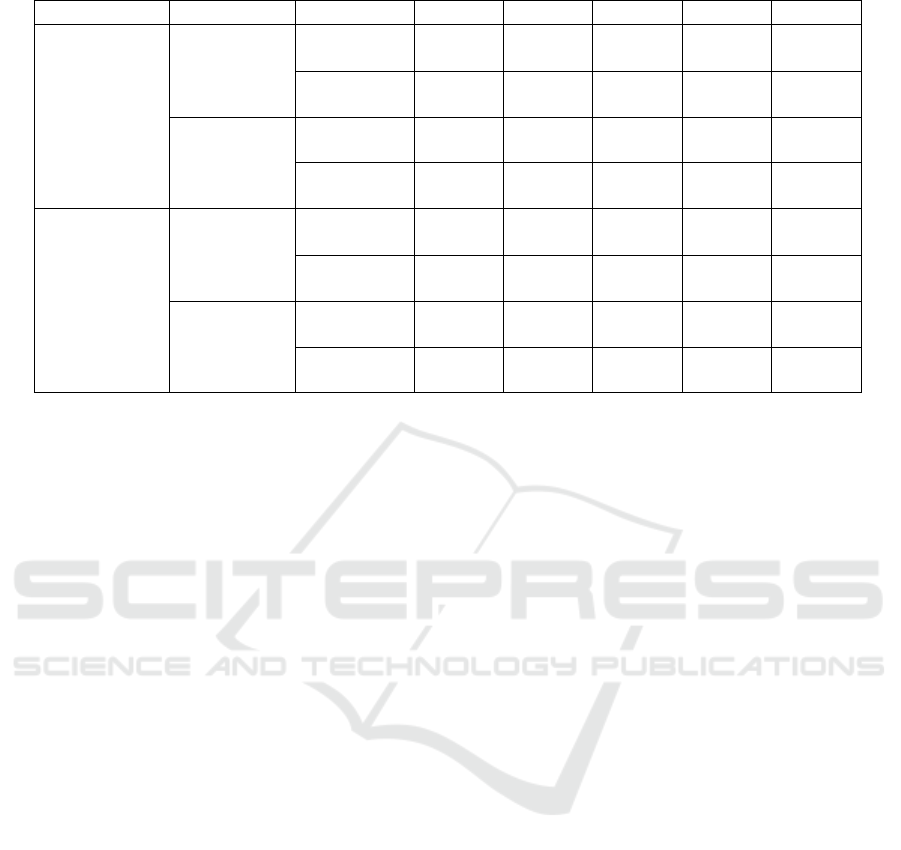
Table 3: Comparison of proposed implementations.
ACC SEN SPE PRE F1S
With
preprocessing
With
subsampling
Triple
convolution
99.33% 99.59% 99.30% 99.12% 99.44%
Simple
convolution
95.32% 95.73% 98.83% 96.39% 95.40%
Without
subsampling
Triple
convolution
95.40% 95.27% 94.70% 95.25% 95.35%
Simple
convolution
90.45% 90.14% 92.83% 90.89% 90.44%
Without
preprocessing
With
subsampling
Triple
convolution
89.65% 89.00% 97.41% 91.63% 89.50%
Simple
convolution
87.85% 83.25% 96.06% 90.63% 87.56%
Without
subsampling
Triple
convolution
86.68% 88.69% 96.80% 90.05% 86.67%
Simple
convolution
87.20% 90.40% 90.50% 89.20% 85.80%
and excessive specialization in the training dataset,
in the convolutional layer, and in the fully connected
layers. The dropout allows the weights of the hidden
layer neurons to be randomly set to zero during train-
ing, causing these nodes to be ignored. After defining
the architecture, fine-tuning was performed to obtain
the best number of epochs. The hyperparameters that
decreased the classifier training time and increased
accuracy and F1-score were obtained and displayed
in Table 1.
Since the best optimization method was obtained
with the Adam function, we also needed to optimize
the learning rate, β1 and β2 values. We tried the val-
ues 0.01, 0.001, 0.0001 and 0.00001 for the learning
rate. Furthermore, to obtain a variable learning rate,
we tested β1 and β2 with the values 0.900, 0.990 and
0.999. The best result was obtained with a 0.0001
learning rate, 0.900 β1 and 0.990 β2. We tested sev-
eral other loss functions to optimize the classifier, and
the categorical cross-entropy was the one that gen-
erated the best results. We needed to find the min-
imum number of epochs necessary to maximize the
accuracy. Excessive training could cause overfitting
and incapacity to generalize and evaluate new images.
We started the test with 100 epochs and increased this
value until 1500 epochs. This test showed that for 500
epochs or more, the accuracy remained stable.
3 RESULTS AND DISCUSSION
We performed classification experiments on 44
recordings from the MIT-BIH arrhythmia database,
among the 48 recordings obtained from 47 patients
studied by the BIH arrhythmia laboratory, and the
heartbeats were classified according to the recom-
mendation of the AAMI.
The training dataset contains a total of 375 rep-
resentative beats, including 75 from each class: type-
N, non-ectopic beats; type-S, supraventricular ectopic
beats; type-V, ventricular ectopic beats; type-F, fusion
beats and type-Q, unknown beats. The representative
beats are randomly sampled from each class of the
first 20 recordings (chosen in the range of 100 to 124)
from the MIT-BIH database. The neural networks are
trained with a total of 245 common training beats, and
a variable number of beats depending on the patient’s
heart rate, so less than 1% of the total beats are used
for training. The 24 unused recordings are used as test
patterns for performance evaluation.
Classification performance is measured using the
statistical error metrics found in the literature (Chen
et al., 2022): accuracy (ACC), sensitivity (SEN),
specificity (SPE), precision (PRE), and F1-score
(F1S). The F1-score measures the overall perfor-
mance of the beat classification, as shown in Table 2.
Our model was implemented using the Tensor-
Flow framework. The training time of each epoch
was approximately 5s, and the maximum epoch num-
ber was set to 75. Table 2 shows that the implemented
classification algorithm has an F1-score value compa-
rable to those of other studies obtaining better results,
presenting the second best in Table 2. We show only
the VEB in the comparison as it is the most commonly
used among other studies. Table 3 shows the results
of different convolutional architectures. In this study,
our best model achieved an accuracy of 99.33%, sen-
sitivity of 99.59%, specificity of 99.30%, precision of
99.12% and F1-score of 99.44%.
Figure 3 shows the confusion matrix of the clas-
ICPRAM 2023 - 12th International Conference on Pattern Recognition Applications and Methods
360

Figure 3: Confusion matrix for heartbeat classification on
the test set.
sification results of the test set. The model is able
to make accurate predictions and distinguish different
classes. The main reason behind this might be the fact
that we have used fine-tuning to optimize our model,
which allows us to better train our classifier.
4 CONCLUSIONS
In this study, we designed an ECG signals classifier
for cardiac arrhythmia detection using CNNs. The
proposed model achieved an accuracy of 99.33% and
an F1-score of 99.44% in the classification of ven-
tricular ectopic beats (VEB). In order to optimize our
model, we fine-tuned our variables and functions, the
selected values compose our final version of the clas-
sifier and are displayed in Table 1. Compared with the
methods in previous literature, our model performed
better in terms of VEB classification accuracy, and
F1-score.
The referenced authors in Table 2 achieved high
accuracy, sensitivity, specificity, precision and F1-
score with private datasets. On the other hand, our
study managed to obtain high results for these met-
rics with a public dataset. Our trained CNN heart-
beat classifier model can be used for real-life and real-
time applications. It can also be used to analyze other
biosignals by changing the training the dataset and
input size before use. Future work may refine this
approach with a better set of hyperparameter values
and different augmentation strategies. An F1-score of
99.00% is accurate enough for cardiovascular disease
detection in home devices. The method has the poten-
tial to be adapted to analyze other biosignals.
REFERENCES
Acharya, U. R., Oh, S. L., Hagiwara, Y., Tan, J. H., Adam,
M., Gertych, A., and San Tan, R. (2017). A deep
convolutional neural network model to classify heart-
beats. Computers in biology and medicine, 89:389–
396.
Al Rahhal, M. M., Bazi, Y., Almubarak, H., Alajlan, N.,
and Al Zuair, M. (2019). Dense convolutional net-
works with focal loss and image generation for elec-
trocardiogram classification. IEEE Access, 7:182225–
182237.
Chen, S. W., Wang, S. L., Qi, X. Z., Samuri, S. M., and
Yang, C. (2022). Review of ECG detection and clas-
sification based on deep learning: Coherent taxon-
omy, motivation, open challenges and recommenda-
tions. Biomedical Signal Processing and Control,
74:103493.
Girosi, F., Jones, M., and Poggio, T. (1995). Regulariza-
tion theory and neural networks architectures. Neural
computation, 7(2):219–269.
Hagiwara, Y., Fujita, H., Oh, S. L., Tan, J. H., San Tan, R.,
Ciaccio, E. J., and Acharya, U. R. (2018). Computer-
aided diagnosis of atrial fibrillation based on ecg sig-
nals: A review. Information Sciences, 467:99–114.
Han, C. and Shi, L. (2020). Ml–resnet: A novel network to
detect and locate myocardial infarction using 12 leads
ecg. Computer methods and programs in biomedicine,
185:105138.
Hinton, G. E., Srivastava, N., Krizhevsky, A., Sutskever, I.,
and Salakhutdinov, R. R. (2012). Improving neural
networks by preventing co-adaptation of feature de-
tectors. arXiv preprint arXiv:1207.0580.
Ince, T., Kiranyaz, S., and Gabbouj, M. (2009). A generic
and robust system for automated patient-specific clas-
sification of ECG signals. IEEE Transactions on
Biomedical Engineering, 56(5):1415–1426.
Jiang, W. and Kong, S. G. (2007). Block-based neural
networks for personalized ECG signal classification.
IEEE Transactions on Neural Networks, 18(6):1750–
1761.
Kingma, D. P. and Ba, J. (2014). Adam: A
method for stochastic optimization. arXiv preprint
arXiv:1412.6980.
Kiranyaz, S., Avci, O., Abdeljaber, O., Ince, T., Gabbouj,
M., and Inman, D. J. (2021). 1d convolutional neu-
ral networks and applications: A survey. Mechanical
systems and signal processing, 151:107398.
Kiranyaz, S., Ince, T., and Gabbouj, M. (2017). Personal-
ized monitoring and advance warning system for car-
diac arrhythmias. Scientific reports, 7(1):1–8.
Li, Z., Zhou, D., Wan, L., Li, J., and Mou, W. (2020). Heart-
beat classification using deep residual convolutional
neural network from 2-lead electrocardiogram. Jour-
nal of Electrocardiology, 58:105–112.
Lown, M., Brown, M., Brown, C., Yue, A. M., Shah, B. N.,
Corbett, S. J., Lewith, G., Stuart, B., Moore, M., and
Little, P. (2020). Machine learning detection of atrial
fibrillation using wearable technology. PLoS One,
15(1):e0227401.
Cardiac Arrhythmia Classification in Electrocardiogram Signals with Convolutional Neural Networks
361

Mant, J., Fitzmaurice, D. A., Hobbs, F. R., Jowett, S., Mur-
ray, E. T., Holder, R., Davies, M., and Lip, G. Y.
(2007). Accuracy of diagnosing atrial fibrillation on
electrocardiogram by primary care practitioners and
interpretative diagnostic software: analysis of data
from screening for atrial fibrillation in the elderly
(safe) trial. Bmj, 335(7616):380.
Mark, R. and Moody, G. (1988). Mit-bih arrhythmia
database directory. Cambridge: Massachusetts Insti-
tute of Technology.
Martis, R. J., Acharya, U. R., Adeli, H., Prasad, H., Tan,
J. H., Chua, K. C., Too, C. L., Yeo, S. W. J., and Tong,
L. (2014). Computer aided diagnosis of atrial arrhyth-
mia using dimensionality reduction methods on trans-
form domain representation. Biomedical signal pro-
cessing and control, 13:295–305.
McAloon, C. J., Boylan, L. M., Hamborg, T., Stallard, N.,
Osman, F., Lim, P. B., and Hayat, S. A. (2016). The
changing face of cardiovascular disease 2000–2012:
An analysis of the world health organisation global
health estimates data. International journal of car-
diology, 224:256–264.
Murray, N. and Perronnin, F. (2014). Generalized max
pooling. In Proceedings of the IEEE conference on
computer vision and pattern recognition, pages 2473–
2480.
Nair, V. and Hinton, G. E. (2010). Rectified linear units
improve restricted boltzmann machines. In Icml.
Nwankpa, C., Ijomah, W., Gachagan, A., and Marshall, S.
(2018). Activation functions: Comparison of trends in
practice and research for deep learning. arXiv preprint
arXiv:1811.03378.
Pollock, K. G., Sekelj, S., Johnston, E., Sandler, B., Hill,
N. R., Ng, F. S., Khan, S., Nassar, A., and Farooqui, U.
(2020). Application of a machine learning algorithm
for detection of atrial fibrillation in secondary care.
IJC Heart & Vasculature, 31:100674.
S
´
anchez, F. R. and Cervera, J. G. (2019). ECG classification
using artificial neural networks. In Journal of Physics:
Conference Series, volume 1221, page 012062. IOP
Publishing.
Schwab, P., Scebba, G. C., Zhang, J., Delai, M., and Karlen,
W. (2017). Beat by beat: Classifying cardiac arrhyth-
mias with recurrent neural networks. In 2017 Com-
puting in Cardiology (CinC), pages 1–4. IEEE.
Sellami, A. and Hwang, H. (2019). A robust deep convo-
lutional neural network with batch-weighted loss for
heartbeat classification. Expert Systems with Applica-
tions, 122:75–84.
Shi, H., Wang, H., Zhang, F., Huang, Y., Zhao, L., and Liu,
C. (2019). Inter-patient heartbeat classification based
on region feature extraction and ensemble classifier.
Biomedical Signal Processing and Control, 51:97–
105.
Shoemaker, M. B., Shah, R. L., Roden, D. M., and Perez,
M. V. (2020). How will genetics inform the clini-
cal care of atrial fibrillation? Circulation research,
127(1):111–127.
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I.,
and Salakhutdinov, R. (2014). Dropout: a simple way
to prevent neural networks from overfitting. The jour-
nal of machine learning research, 15(1):1929–1958.
Uchida, K., Tanaka, M., and Okutomi, M. (2018). Cou-
pled convolution layer for convolutional neural net-
work. Neural Networks, 105:197–205.
Xiang, Y., Luo, J., Zhu, T., Wang, S., Xiang, X., and Meng,
J. (2018). ECG-based heartbeat classification using
two-level convolutional neural network and rr interval
difference. IEICE TRANSACTIONS on Information
and Systems, 101(4):1189–1198.
Xie, Q., Wang, X., Sun, H., Zhang, Y., and Lu, X.
(2021). ECG signal detection and classification of
heart rhythm diseases based on ResNet and LSTM.
Advances in Mathematical Physics, 2021.
Xiong, Z., Stiles, M. K., and Zhao, J. (2017). Robust ecg
signal classification for detection of atrial fibrillation
using a novel neural network. In 2017 Computing in
Cardiology (CinC), pages 1–4. IEEE.
Xu, Q., Zhang, M., Gu, Z., and Pan, G. (2019). Over-
fitting remedy by sparsifying regularization on fully-
connected layers of cnns. Neurocomputing, 328:69–
74.
Xu, X. and Liu, H. (2020). ECG heartbeat classification
using convolutional neural networks. IEEE Access,
8:8614–8619.
Zhai, X. and Tin, C. (2018). Automated ECG classification
using dual heartbeat coupling based on convolutional
neural network. IEEE Access, 6:27465–27472.
Zhiqiang, W. and Jun, L. (2017). A review of object de-
tection based on convolutional neural network. In
2017 36th Chinese control conference (CCC), pages
11104–11109. IEEE.
Zhu, H., Cheng, C., Yin, H., Li, X., Zuo, P., Ding, J.,
Lin, F., Wang, J., Zhou, B., Li, Y., et al. (2020).
Automatic multilabel electrocardiogram diagnosis of
heart rhythm or conduction abnormalities with deep
learning: a cohort study. The Lancet Digital Health,
2(7):e348–e357.
ICPRAM 2023 - 12th International Conference on Pattern Recognition Applications and Methods
362
