
Synthesis for Dataset Augmentation of H&E Stained Images with
Semantic Segmentation Masks
Peter Sakalik
1
, Lukas Hudec
1 a
, Marek Jakab
1 b
, Vanda Bene
ˇ
sov
´
a
1 c
and Ondrej Fabian
2,3 d
1
Faculty of Informatics and Information Technologies, Slovak University of Technology, Ilkovicova 2, Bratislava, Slovakia
2
Clinical and Transplant Pathology Centre, Institute for Clinical and Experimental Medicine,
Videnska 9, Prague 4, Czechia
3
Department of Pathology and Molecular Medicine, 3rd Faculty of Medicine, Charles University and Thomayer Hospital,
Videnska 800, Prague 4, Czechia
Keywords:
Medical data, Annotated Data Synthesis, Generative Adversarial Networks.
Abstract:
The automatic analysis of medical images with the application of deep learning methods relies highly on
the amount and quality of annotated data. Most of the diagnostic processes start with the segmentation and
classification of cells. The manual annotation of a sufficient amount of high-variability data is extremely time-
consuming, and the semi-automatic methods may introduce an error bias. Another research option is to use
deep learning generative models to synthesize medical data with annotations as an extension to real datasets.
Enhancing the training with synthetic data proved that it can improve the robustness and generalization of the
models used in industrial problems. This paper presents a deep learning-based approach to generate synthetic
histological stained images with corresponding multi-class annotated masks evaluated on cell semantic seg-
mentation. We train conditional generative adversarial networks to synthesize a 6-channeled image. The six
channels consist of the histological image and the annotations concerning the cell and organ type specified in
the input. We evaluated the impact of the synthetic data on the training with the standard network UNet. We
observe quantitative and qualitative changes in segmentation results from models trained on different distribu-
tions of real and synthetic data in the training batch.
1 INTRODUCTION
A histology tissue sample can be very easily ab-
stracted as a non-stationary texture. The unique struc-
tures of a sample can be categorized as different tex-
ture classes. Therefore it is possible to apply sim-
ilar methods to texture synthesis in the research on
the synthesis of histology scans. Texture synthesis is
the process of algorithmically creating a texture ac-
cording to a small sample or a set of predefined char-
acteristics. Depending on the applications, the algo-
rithms must usually be both efficient and capable of
generating high-quality outputs with high variability.
The synthetic textures must be indistinguishable from
the original sample or at least deceive the human ob-
server. Over the years, plenty of texture synthesis
methods have been introduced.
a
https://orcid.org/0000-0002-1659-0362
b
https://orcid.org/0000-0002-4329-6417
c
https://orcid.org/0000-0001-6929-9694
d
https://orcid.org/0000-0002-0393-2415
One of the simplest methods is random sampling,
where the texture is sampled into tiles. They are then
pseudo-randomly selected and joined together. How-
ever, the result is rarely sufficient as the seams be-
tween the tiles may remain visible.
Traditional approaches can be classified as pixel-
based or patch-based depending on how large a sam-
ple is inserted into a new synthesized image. The
most successful approaches use Markov arrays, non-
parametric sampling, and tree-structured vector quan-
tization. A new texture is formed by finding and copy-
ing pixels with the most similar neighboring pixels to
the original texture. This technique limits the visible
seams on the borders. However, the usual drawback
is the repeatability of selected sampled patches.
Deep learning generative models have recently
overcome traditional methods’ generative quality. Ex-
tensive research has been done on generative adver-
sarial networks (GANs) applications, which achieved
significant results in various industrial and research
fields. Their power breaks into more quality-requiring
areas, such as the gaming industry and medicine.
Sakalik, P., Hudec, L., Jakab, M., Benešová, V. and Fabian, O.
Synthesis for Dataset Augmentation of HE Stained Images with Semantic Segmentation Masks.
DOI: 10.5220/0011679300003417
In Proceedings of the 18th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2023) - Volume 4: VISAPP, pages
873-880
ISBN: 978-989-758-634-7; ISSN: 2184-4321
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
873

One possible application is in pathology, which is
the main focus of this paper. Structures of patholog-
ical findings are small and in large amounts, which
makes it time-consuming for medical practitioners
to annotate them perfectly. Automatic or semi-
automatic annotation methods would be beneficial but
introduce errors that produce noisy labels. Also, the
semi-automatic methods usually require the initial-
ization of parameters that may also take a significant
amount of time.
The current cell segmentation datasets usually
suffer from insufficient data quantity and variability
because of the difficult annotation process. These
datasets play a significant role in researching deep-
learning models for cancer analysis, diagnosis, and
staging.
This paper presents an automatic approach using
GANs to generate many annotated synthetic data with
quality similar to real samples. The generated anno-
tated data can then be used for dataset augmentation
necessary for training deep learning models, which
makes models more robust, and better generalized.
GANs reduce manual preparation time.
The contribution of this paper is the following:
• The presented method is specified for synthesiz-
ing hematoxylin and eosin (H&E)-stained histo-
logical images.
• The generated images are accompanied by anno-
tation masks of cells of 4 classes.
• The model can generate visually organ-specific
tissue and cells.
• Evaluation of the influence of synthetic data aug-
mentations for semantic segmentation.
2 RELATED WORK
The quality of datasets depends on the accuracy of
the segmentation masks. In most domains, they
can be acquired by manual annotation or with semi-
supervised segmentation methods. For this reason,
different approaches have been developed to obtain
them. Some diagnostic tools often provide a semi-
supervised method for cell segmentation that can help
the user/annotator with guidance or a set of parame-
ters for automatic segmentation that can be later used
for diagnostic analysis and support. One such soft-
ware tool is QuPath (Bankhead et al., 2017), which
uses simple thresholding combined with normaliza-
tion and cell nucleus emphasizing. The user sets a
set of threshold values that define the color interval of
the hematoxylin purplish blue nucleus compared to
the pinkish eosin-stained extracellular matrix and cy-
toplasm. However, these simple segmentation meth-
ods may be insufficient for more complex images or
cell classes. More advanced approaches are based
on graph theory clustering methods or deep learn-
ing. Current deep-learning segmentation approaches
achieve state-of-the-art performance. Specifically,
these are the U-Net and U-Net++ architectures, re-
spectively. Unfortunately, both of them require anno-
tated images for training.
An alternative approach to acquiring annotated
data is synthesizing tissue images where the research
is open. As mentioned before, the histology tissue
can be abstracted as texture, we present several state-
of-the-art approaches for generating high-quality tex-
tures. The histology data are stored as the large res-
olution scans of the whole tissue, the Whole Slide
Images (WSI), and a smaller scale specific selection,
usually annotated in better detail, the Stain Tissue Mi-
croarray (TMA). There are known approaches to gen-
erate large-scale images, e.g., progressively growing
GANs (Beers et al., 2018;
ˇ
Stepec and Sko
ˇ
caj, 2020)
or specialized architectures for high-resolution image
generation like StyleGAN (Karras et al., 2019), and
BigGAN (Brock et al., 2018). The BigGAN is a net-
work containing 355.7 million parameters with a gen-
erator output of 256 × 256 pixels. The StyleGAN, on
the other hand, has only 26.2 million parameters and
the generator output resolution is 1024 ×1024 pixels.
Both architectures generate high-quality images.
Non-stationary texture synthesis with adversarial
expansion (Zhou et al., 2018) presents a generative
model that synthesizes texture by expanding the in-
put sample from k × k to 2k × 2k resolution. The ar-
chitecture consists of a generator and two discrimi-
nators. The first one takes care of discriminating be-
tween real and fake samples. The second one is a pre-
trained VGG-19 network and takes care of preserving
the stylistic similarity with the original texture. The
work proves a possibility to generate higher resolu-
tion, high-quality textures with preserved structures.
In addition to high-quality textures, generating re-
lated segmentation maps for dataset augmentation is
necessary. The MaterialGAN (Guo et al., 2020) in-
troduces a GANs modification for generating realis-
tic SVBRDF parameter maps. They used the dataset
from Deschaintre (Deschaintre et al., 2018) for train-
ing. It contains 155 SVBRDFs with a high resolu-
tion of 4096 × 4096 pixels. It was then augmented by
blending multiple SVBRDFs to generate 256 × 256
resolution patches at a random position, rotation, and
scaling. The generated result is 9-channeled with 3
channels for a fraction of incident light reflected from
the surface, 2 for the surface orientation of the geo-
VISAPP 2023 - 18th International Conference on Computer Vision Theory and Applications
874

metric object, 1 for the roughness, and 3 for a fraction
of incident light reflected from the surface. They used
StyleGAN 2 as the baseline architecture.
Specialized and modified GAN architectures can
have enough learning capacity to generate histolog-
ical data. A PathologyGAN (Quiros et al., 2019)
focuses on generating realistic histological images.
The variability of data is introduced from two dif-
ferent training datasets, H&E colorectal cancer tissue
from the National Cancer Center (NCT, Germany),
H&E breast cancer tissue from the Netherlands Can-
cer Institute (NKI, Netherlands), and Vancouver Gen-
eral Hospital (VGH, Canada). In total, it contains 86
whole slide images and 576 tissue microarrays. They
used BigGAN as the underlying architecture, which
they augmented with a mapping network from Style-
GAN, a style mixing regularization, and a relativistic
mean as a loss function for the discriminator.
StyleGAN is also used for prostate cancer data
synthesis (Daroach et al., 2022). However, the main
focus is on the trained latent space of the StyleGAN
to label the PCa regions according to the pathologist
annotations. The pathologist attached a label to each
of the model-generated realistically-looking patches.
These labels then defined the regions in the original
latent space from which sampled noise-generated his-
tology images were always of the latent-space class.
Therefore the StyleGAN-based solution is able to
synthesize sample patches of specified prostate can-
cer classes. However, they still required help from
a pathologist to annotate generated patches without
further medical information about the sample, which
may have introduced an error.
This paper presents a StyleGAN-based solution
for selectively synthesizing epithelial cells, lympho-
cytes, macrophages, and neutrophils in the lungs,
prostate, kidney, and breast. The result of our model
is an RGB image with a segmentation map of cells’
pixel positions and classes.
3 METHOD
The main goal of our method is to generate quality
histology images with associated cell multi-class seg-
mentation masks. GAN is the current, massively ap-
plied deep learning architecture framework suitable
for this problem. According to the related work,
GANs can generate non-stationary textures, medi-
cally valid histological data, related maps, and anno-
tated segmentation masks. We based our generator
architecture on StyleGAN.
Initial data are necessary to train the generator, so
we chose the MoNuSAC dataset (Verma et al., 2020).
It contains TMA images with their annotated segmen-
tation masks. The dataset consists of 4 cell types re-
sponsible for diagnosing stages and severity of lung,
prostate, breast, and kidney cancer. Each segmenta-
tion mask contains information about the classes of
cells and the organ. We use their initial color classes
of the cells: red, yellow, green, and blue for epithelial,
lymphocytes, macrophages, and neutrophils.
To validate the results and investigate the influ-
ence of the synthetic data used on training for seg-
mentation, we employed the standard segmentation
network UNet.
3.1 Generative Model
The tissue visuals depend on the organ, so the syn-
thesis method must preserve its tissue characteristics.
The standard input for the GAN network is sampled
Gaussian noise. Therefore, we extended the Style-
GAN architecture with an idea from Auxiliary Clas-
sifier GAN (ACGAN) (Odena et al., 2017). The or-
gan class is global information we represent by a one-
hot encoded vector, which sets the generator for the
intended organ visual. The cell classes are specific
to the location in the tissue. We do not pre-set the
segmentation mask defining the location of cells. We
use only the one-hot encoded vector to specify the ex-
pected classes the model should generate. To force the
generator to synthesize only specified classes is the
job of the discriminator. Also, to preserve the input
information about classes, we modify the ACGAN
approach and add the cell and organ information to
every 2n layer of the StyleGAN mapping network as
is shown In Figure 1. The result of the mapping net-
work is the style vector used in adaptive instance nor-
malization in generator layers. The generator archi-
tecture, random noise vector, constant vector of ones,
and blending alpha values for progressive growing are
the same as in the original StyleGAN paper.
The generator’s output and the discriminator’s in-
put is an image with 6 channels (2x RGB). The first
three channels present the generated histological im-
age, and the last three channels present the generated
segmentation mask of that image using the dataset’s
predefined colors per class. We need to modify the
discriminator to force the generator to train according
to the input class information. The standard regres-
sion is to distinguish real and fake images. Improved
Wasserstein loss is applied to reduce the chances of
Mode Collapse. The discriminator now requires also
two additional classifiers. One classifier classifies the
organ type - the class of the whole tissue segment,
which is activated by softmax on the output layer and
trained against multi-class categorical cross-entropy.
Synthesis for Dataset Augmentation of HE Stained Images with Semantic Segmentation Masks
875
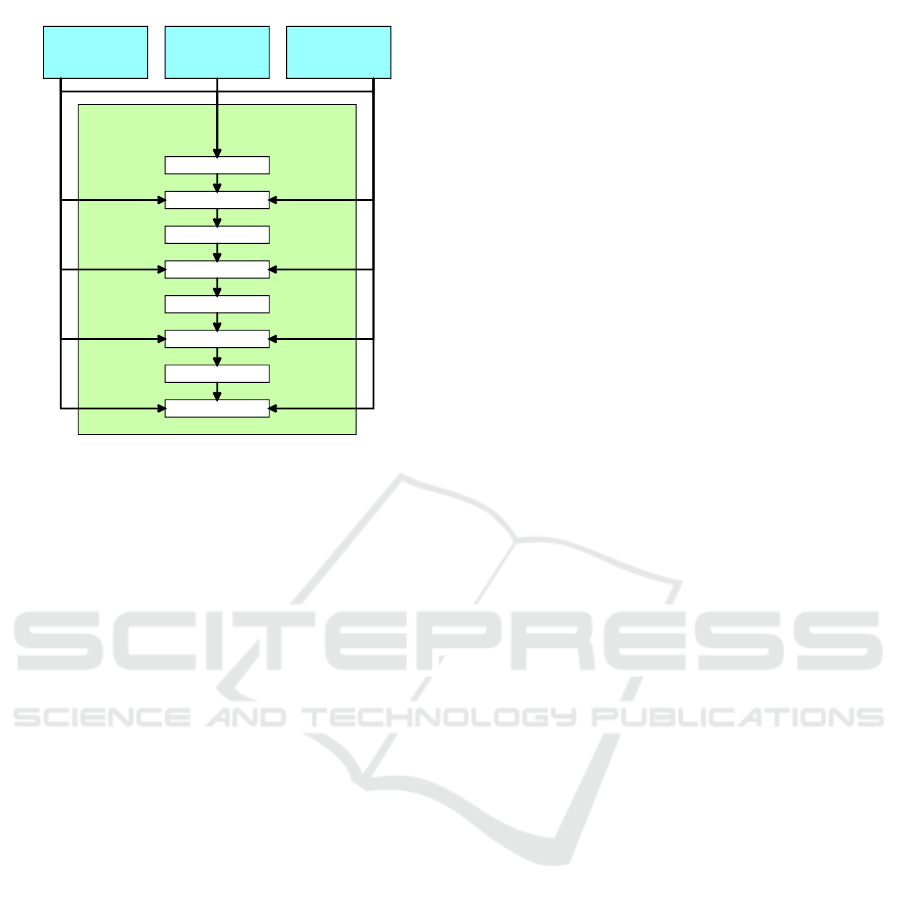
cell
information
latent vector
organ
information
2. layer
3. layer
4. layer
5. layer
6. layer
7. layer
8. layer
1. layer
mapping
network
Figure 1: A modified architecture of StyleGAN mapping
network with ACGAN class information input values.
The second classifier determines the classes of gener-
ated cells with segmentation masks. One image can
contain cells of multiple classes, so we use sigmoid
activation and multi-label binary cross entropy loss
function. These values are compared to the input one-
hot vectors. The architecture of a whole generative
adversarial framework is in Figure 2.
The generated annotation masks contain a certain
amount of noise, cell borders are sometimes unclear,
and there can be multiple labels on the area of the
same cell, even though the cell visual clearly repre-
sent only one class. We use simple post-processing
by color normalization and morphological transfor-
mation to increase the quality of segmentation masks.
The first step is color normalization to unify color seg-
ments into single-class clusters. Second, morpholog-
ical closing enlarges areas, fills holes in cells, and re-
moves unwanted details. Figure 3 visualizes the ef-
fect of post-processing of an example mask. Finally,
we transform the pixel color values into 4 classes of
regression for the computation of a loss function. A
black background is 0, red is 1, green is 2, blue is 3,
and yellow is encoded as 4.
4 EVALUATION
We evaluate both quantitative and qualitative results
of the generated histological images along with their
annotated segmentation masks. The impact of the
synthetic data generated by our GAN is discussed
over the segmentation results of the trained segmen-
tation model.
4.1 Synthesis of Histological Data and
Annotated Segmentation Masks
The presented images are the results of a model that
took 192 hours (8 days) to train. The time distribu-
tion over the training of individual resolutions is the
following: from 4 to 64 pixels took 24 hours, up to
128 pixels took 48 hours, and up to 256 pixels took
120 hours. Throughout the training, the values of all
loss functions were balanced, and we did not observe
any significant fluctuations. We expect that the longer
training with further progressive upsampling would
increase the quality of tissue and individual cells.
For the quantitative evaluation of our generative
model, we use standard metrics Frech
´
et Inception
Distance (FID), Inception Score (IS), and Kernel In-
ception Distance (KID). Accuracy and Recall mea-
sure the classification score of the discriminator. All
quantitative metrics are displayed in Table 1. The
PathologyGAN (Quiros et al., 2019) achieved an FID
of 32.05 on a different histology dataset to compare
the results to related work.
The qualitative evaluation took place in the pres-
ence of a pathologist. After several moments they
could determine that some images did not look realis-
tic, but others could not distinguish from real sam-
ples. In some cases, the generated cells’ structure
looked similar to the real samples. The sampled
cells contained the nucleus and preserved cytoplasm,
and the structural placement of cells also looked re-
alistic. However, to classify them, the pathologist
stated, they would require the original tissue sample
to see the whole tissue’s structure. Therefore, for the
dataset augmentation, we consider the results satis-
factory. Some examples are documented in Figure 4.
The post-processing of the segmentation maps helps
to improve the quality and cell-border accuracy and
to generate more exact cell types. The input infor-
mation about cell classes and organs provides strict
constraints for the generator.
4.2 Dataset Augmentation
We evaluated the impact of our generated synthetic
data on the performance of a segmentation network.
We use the MoNuSAC dataset and synthetic data gen-
erated by our trained generator. This section investi-
gates and compares the effect of the synthetic data on
the training and the segmentation predictions of UNet
model evaluated on the MoNuSAC test subset.
We use the focal loss function, which is often used
for training multi-label segmentation, to investigate
the effect of synthetic data on the training of segmen-
tation and prediction of multi-label masks.
VISAPP 2023 - 18th International Conference on Computer Vision Theory and Applications
876
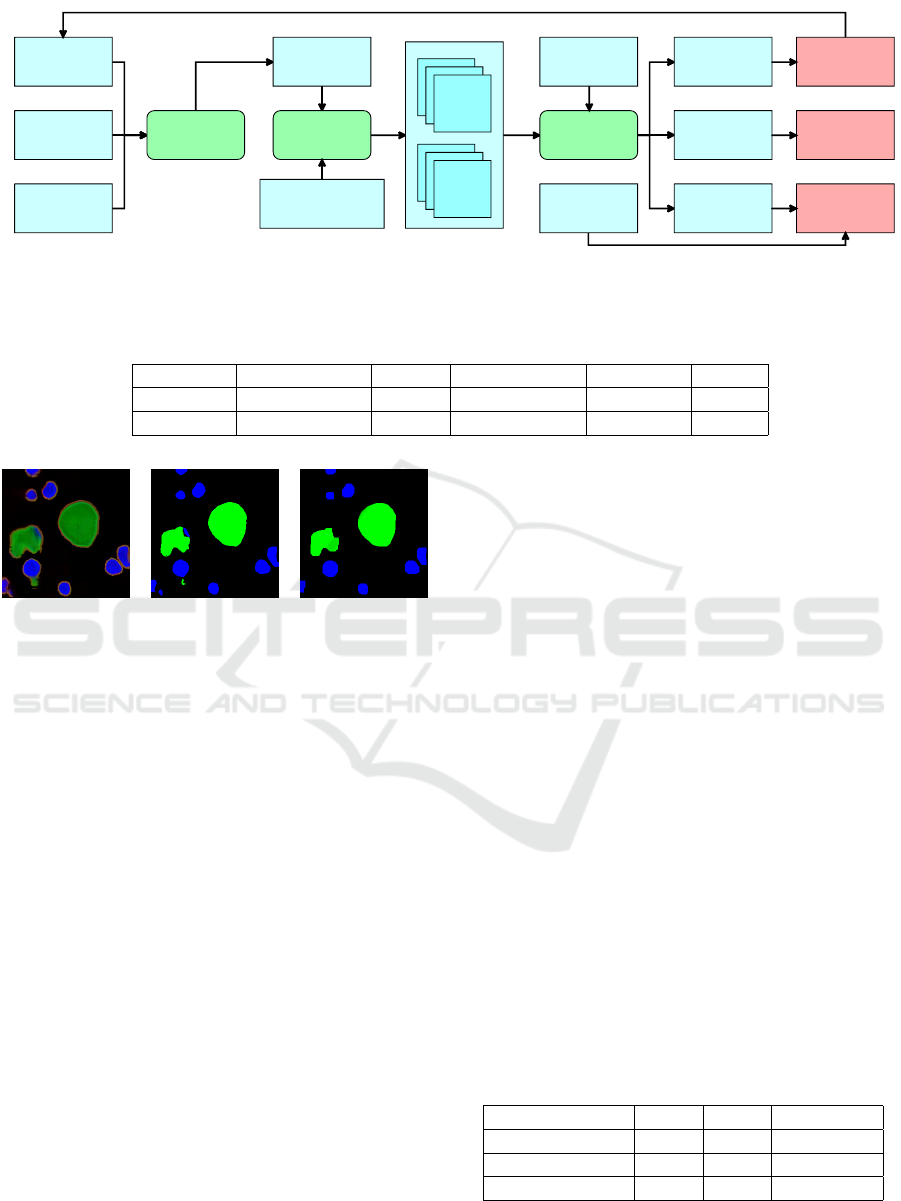
cell
information
latentvector
organ
information
mapping
network
stylevector
generator
RGB
image
RGB
mask
discriminator
real/false
classification
organ
classification
cell
classification
binarycross
entropy
Wasserstein
loss
categorical
crossentropy
realclass
alpha
noise,constant,
alpha
Figure 2: The architecture of used GAN framework. Generator inspired by StyleGAN. Discriminator modified to be auxiliary
classifier.
Table 1: Metrics for synthetic data and classification performance of discriminator. The low values of the discriminator do
not degrade the quality of the generated images.
Synthesis IS FID KID Accuracy Recall
Images 4.126 ± 0.185 84.973 0.049 ± 0.001 0.254 0.365
Masks 2.711 ± 0.067 51.211 0.034 ± 0.001 0.867 0.484
Figure 3: Post-processing of segmentation mask.
We train 3 models with the same architecture
on batches with different real and synthetic data
amounts. The datasets distributions of the 3 experi-
ments are following:
1. Vanilla baseline model with only real samples —
1 : 0 real:synthetic
2. A sample of synthetic data that preserves a major-
ity of real samples — 3 : 1 real:synthetic
3. Balanced dataset with the same distribution of real
synthetic data — 1 : 1 real:synthetic.
The training dataset contains 3356 samples. 1656
are from the MoNuSAC dataset, and 1700 images
are synthetic. To preserve the number of total train-
ing samples, the added amount of synthetic images is
compensated by removing the same amount of ran-
dom real samples. The test dataset contains 414 im-
ages, and all of them are from the MoNuSAC dataset.
To preserve the same training conditions, the train-
ing hyperparameters of every model were the same,
so the performance difference is affected only by the
synthetic data.
• number of epochs: 20,
• batch size: 32,
• number of steps: 51,
• optimizer: Adam,
• learning rate: 0.001.
The courses of tracked training are displayed in
Figure 5 and 6. Based on the results, we conclude that
the model with 1 : 1 equal distributions had the biggest
problems during training, leaving with the highest
loss and the lowest accuracy. The model with a 3 : 1
smaller sample of synthetic data performed similarly
to the vanilla model trained only on real data.
The qualitative results of the models are displayed
in Figure 7. The segmentation masks produced by the
model with a small amount of synthetic data achieved
the best performance and even overcame the preci-
sion of the model trained only on real samples. Unex-
pectedly the model with an equal distribution suffers
from over-segmentation, which makes it too inaccu-
rate compared to the other two.
The quantitative metrics, Intersection over Union
(IoU), Dice, and Hausdorff distance, are in Table 2.
The quantitative performance measurements confirm
the qualitative expectations that the model with an
equally distributed dataset performed the worst. The
model with an augmented dataset achieved similar
and slightly better results than the vanilla model.
Table 2: Classfication score depending on real to fake im-
ages rate in the training dataset.
UNET IoU Dice Hausdorff
model data 1 : 0 0.556 0.815 16.455
model data 3 : 1 0.577 0.817 14.736
model data 1 : 1 0.459 0.704 17.921
To evaluate the performance of the segmentation,
we analyzed the specific cell classes. Table 3 demon-
Synthesis for Dataset Augmentation of HE Stained Images with Semantic Segmentation Masks
877
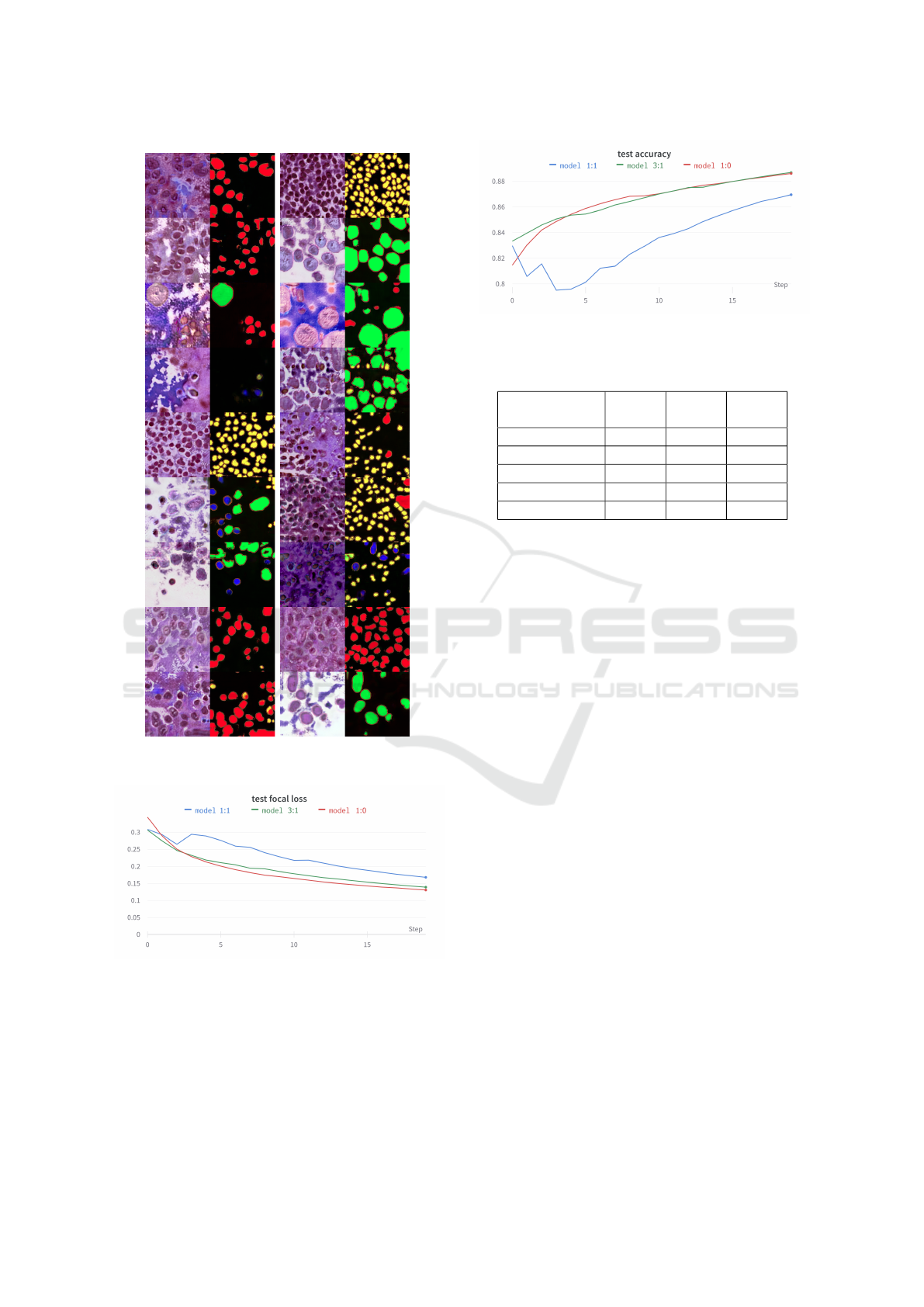
Figure 4: Direct results generated by our network without
the post-processing.
Figure 5: Progress of focal loss on a validation/test set dur-
ing the training.
strates that in individual cases, the model trained on
the dataset with a small amount of synthetic data even
achieved better segmentation than on only real data.
This could have happened by adding new data with
higher variability of shapes and structures than the
original dataset.
Figure 6: Progress of accuracy on a validation/test set dur-
ing the training.
Table 3: Dice scores for each cell class and background.
Classes
Model
1 : 1
Model
3 : 1
Model
1 : 0
Backround 0.820 0.861 0.877
Epithelial 0.401 0.577 0.570
Lymphocytes 0.203 0.362 0.360
Macrophages 0.418 0.514 0.406
Neutrophils 0.453 0.573 0.568
5 DISCUSSION
The proposed approach is tested and developed di-
rectly for histology data generation but can be used in
different segmentation application domains with hier-
archical data classes. Semantic cell segmentation al-
lows us to explore the approach’s benefits and weak-
nesses and can be forgiving for some irregularities.
Compared to the related approaches like TilGAN
(Saha et al., 2021) and PathologyGAN (Quiros et al.,
2019), our approach can generate precise segmenta-
tion masks of several cell classes in different organs.
A similar intermediate output can be found in the Un-
supervised training GANs for segmentation in (Ga-
dermayr et al., 2018), where the cycle GAN generates
segmentation masks of circular or ellipsoid regions.
Our approach uses supervised training, so the gener-
ated cells have various shapes depending on the an-
notated training data. The segmentation masks cell
shape quality is improved by post-processing, how-
ever, at times, there can be cells with multi-label an-
notations, especially when the cell regions are big-
ger. This is difficult to correct because more cells can
overlap at the same position since the tissue slice is a
3D volume. Also, the overall quality may be inferior
to the PathologyGAN, but their model has BigGAN
architecture which contains far more trainable param-
eters. Our model was trained using PyTorch (v1.11)
on a desktop computer with NVIDIA RTX 3090, and
the training took 24h for 4-64p, 33h for 128p, and 38h
for 256p, summing up to 95h total training time.
VISAPP 2023 - 18th International Conference on Computer Vision Theory and Applications
878

Figure 7: 2 left columns: Real images and ground truth an-
notations. 3 right columns: Segmentation predictions gen-
erated by our trained UNET models.
Compared to the state-of-the-art approaches, our
StyleGAN modification with ACGAN allows us to
control the generation of desired organ classes and
cell types. Unfortunately, it is impossible to define
the exact location of generated cells. However, we
do not see this as a disadvantage because the gener-
ated images have sufficient variability, and to augment
the dataset, it is not important to have total control
over cell positions, and it is easier to leave it unat-
tended. Additionally, without explicit control of cell
positions, the network can learn if there is a specific
spatial distribution of cells in the real data.
The current maximal size of generated tissue
patch is 256p, which is briefly large enough for most
of the segmentation networks. It is also possible to
increase the size of a generated patch by continu-
ing training with progressive growing or to merge the
generated patches by tiling.
The presented model was trained on the
MoNuSAC dataset with 36000 hand-annotated
cells, which is a large amount of manually made
annotations. However, we expect the generator to
learn the cell structures, tissue texture, and seg-
mentation masks even if they would be annotated
semi-automatically from, for example, QuPath
(Bankhead et al., 2017).
6 CONCLUSION
This study presented a novel approach to histological
datasets augmentation by generating images with cor-
responding annotations. The qualitative evaluation by
the pathologist concluded that the images look similar
to real tissue microarrays. Even though the synthetic
data may have unrealistic artefacts, the data can be
used to augment training datasets. We evaluated the
impact on training through several experiments and
three trainings where we observed the progress of the
Focal loss function and classification accuracy of cell
classes. The experiments proved that even a small
amount of synthetic data might improve the final per-
formance of a model. Also, the excessive amount of
synthetic data can add bias to the dataset and hurt the
generalization of the model.
In conclusion, we modified the StyleGAN archi-
tecture of auxiliary classification, so it is possible to
control the generated cell type and organ type. The
modification required adding two input layers to the
mapping network and two classifiers to the discrim-
inator. In order to better preserve the information
about the cell and organ type, we appended the input
information to every 2n layer of the mapping network.
We used MoNuSAC dataset and our synthetic
generated data to evaluate the impact of the data aug-
mentation on the training of the segmentation model.
We trained three models with different real and syn-
thetic data distributions. We set constant hyperparam-
eters for each training to maintain objectivity. The
model with a small amount of synthetic data achieved
better results than vanilla training on only real data.
Future studies should consider increasing synthe-
sized images’ quality and improving the model and
Synthesis for Dataset Augmentation of HE Stained Images with Semantic Segmentation Masks
879

output resolution. It could be beneficial for the pathol-
ogist to see the whole tissue structure, not only the
detail of some selected cells.
ACKNOWLEDGEMENTS
This work was partially supported by STU Grant
Scheme for support excellent teams of young re-
searchers and Cooperation (Financial support) with
Siemens Healthineers Slovakia.
REFERENCES
Bankhead, P., Loughrey, M. B., Fern
´
andez, J. A., Dom-
browski, Y., McArt, D. G., Dunne, P. D., McQuaid,
S., Gray, R. T., Murray, L. J., Coleman, H. G., et al.
(2017). Qupath: Open source software for digital
pathology image analysis. Scientific reports, 7(1):1–7.
Beers, A., Brown, J., Chang, K., Campbell, J. P., Ostmo, S.,
Chiang, M. F., and Kalpathy-Cramer, J. (2018). High-
resolution medical image synthesis using progres-
sively grown generative adversarial networks. arXiv
preprint arXiv:1805.03144.
Brock, A., Donahue, J., and Simonyan, K. (2018). Large
scale gan training for high fidelity natural image syn-
thesis. arXiv preprint arXiv:1809.11096.
Daroach, G. B., Duenweg, S. R., Brehler, M., Lowman,
A. K., Iczkowski, K. A., Jacobsohn, K. M., Yoder,
J. A., and LaViolette, P. S. (2022). Prostate cancer
histology synthesis using stylegan latent space anno-
tation. In Wang, L., Dou, Q., Fletcher, P. T., Spei-
del, S., and Li, S., editors, Medical Image Computing
and Computer Assisted Intervention – MICCAI 2022,
pages 398–408, Cham. Springer Nature Switzerland.
Deschaintre, V., Aittala, M., Durand, F., Drettakis, G., and
Bousseau, A. (2018). Single-image svbrdf capture
with a rendering-aware deep network. ACM Trans-
actions on Graphics (ToG), 37(4):1–15.
Gadermayr, M., Gupta, L., Klinkhammer, B. M., Boor, P.,
and Merhof, D. (2018). Unsupervisedly training gans
for segmenting digital pathology with automatically
generated annotations. ArXiv, abs/1805.10059.
Guo, Y., Smith, C., Ha
ˇ
san, M., Sunkavalli, K., and
Zhao, S. (2020). Materialgan: reflectance capture
using a generative svbrdf model. arXiv preprint
arXiv:2010.00114.
Karras, T., Laine, S., and Aila, T. (2019). A style-based
generator architecture for generative adversarial net-
works. In Proceedings of the IEEE/CVF conference
on computer vision and pattern recognition, pages
4401–4410.
Odena, A., Olah, C., and Shlens, J. (2017). Conditional
image synthesis with auxiliary classifier gans. In In-
ternational conference on machine learning, pages
2642–2651. PMLR.
Quiros, A. C., Murray-Smith, R., and Yuan, K. (2019).
Pathologygan: Learning deep representations of can-
cer tissue. arXiv preprint arXiv:1907.02644.
Saha, M., Guo, X., and Sharma, A. (2021). Tilgan: Gan
for facilitating tumor-infiltrating lymphocyte pathol-
ogy image synthesis with improved image classifica-
tion. IEEE access: practical innovations, open solu-
tions, 9:79829 – 79840.
ˇ
Stepec, D. and Sko
ˇ
caj, D. (2020). Image synthesis as a pre-
text for unsupervised histopathological diagnosis. In
International Workshop on Simulation and Synthesis
in Medical Imaging, pages 174–183. Springer.
Verma, R., Kumar, N., Patil, A., Kurian, N. C., Rane, S.,
and Sethi, A. (2020). Multi-organ nuclei segmentation
and classification challenge 2020. IEEE transactions
on medical imaging, 39(1380-1391):8.
Zhou, Y., Zhu, Z., Bai, X., Lischinski, D., Cohen-Or,
D., and Huang, H. (2018). Non-stationary texture
synthesis by adversarial expansion. arXiv preprint
arXiv:1805.04487.
VISAPP 2023 - 18th International Conference on Computer Vision Theory and Applications
880
